ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2021; 17(1):119-133. doi:10.7150/ijbs.51628 This issue Cite
Review
Genome-to-phenome research in rats: progress and perspectives
1. Department of Animal Sciences, Washington State University, Pullman, WA 99164-7620.
2. Department of Psychology, Washington State University, Pullman, WA 99164-4820.
3. Department of Integrative Physiology and Neuroscience, Washington State University, Pullman, WA 99164-7620.
Abstract
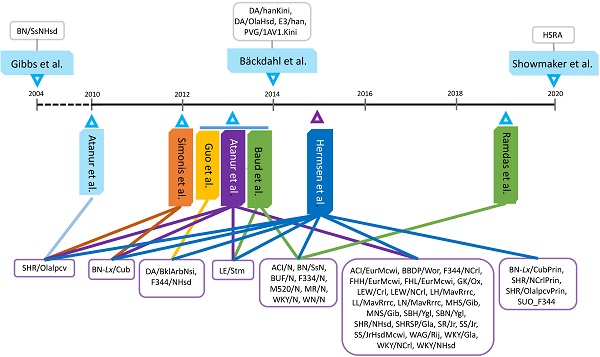
Because of their relatively short lifespan (<4 years), rats have become the second most used model organism to study health and diseases in humans who may live for up to 120 years. First-, second- and third-generation sequencing technologies and platforms have produced increasingly greater sequencing depth and accurate reads, leading to significant advancements in the rat genome assembly during the last 20 years. In fact, whole genome sequencing (WGS) of 47 strains have been completed. This has led to the discovery of genome variants in rats, which have been widely used to detect quantitative trait loci underlying complex phenotypes based on gene, haplotype, and sweep association analyses. DNA variants can also reveal strain, chromosome and gene functional evolutions. In parallel, phenome programs have advanced significantly in rats during the last 15 years and more than 10 databases host genome and/or phenome information. In order to discover the bridges between genome and phenome, systems genetics and integrative genomics approaches have been developed. On the other hand, multiple level information transfers from genome to phenome are executed by differential usage of alternative transcriptional start (ATS) and polyadenylation (APA) sites per gene. We used our own experiments to demonstrate how alternative transcriptome analysis can lead to enrichment of phenome-related causal pathways in rats. Development of advanced genome-to-phenome assays will certainly enhance rats as models for human biomedical research.
Keywords: Rat, Genome sequencing, Genetic variation, Phenome collection, Alternative transcriptomes, Genome-to-phenome
