Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2005; 1(1):19-23. doi:10.7150/ijbs.1.19 This issue Cite
Short Research Paper
No more than 14: the end of the amphioxus Hox cluster
1 Departament de Genètica, Facultat de Biologia, Universitat de Barcelona. Av. Diagonal, 645. E-08028, Barcelona, Spain.
2 Department of Zoology, University of Oxford. South Parks Road, Oxford, OX1 3PS, U.K.
3 Benaroya Research Institute at Virginia Mason, 1201 Ninth Avenue, Seattle, Washington 98101, U.S.A.
4 Division of Developmental Biology. National Institute for Medical Research. The Ridgeway, Mill Hill. London NW7 1AA, U.K.
Received 2004-9-1; Accepted 2004-10-18; Published 2005-1-5
Abstract
The Hox gene cluster has been a key paradigm for a generation of developmental and evolutionary biologists. Since its discovery in the mid-1980's, the identification, genomic organization, expression, colinearity, and regulation of Hox genes have been immediate targets for study in any new model organism, and metazoan genome projects always refer to the structure of the particular Hox cluster(s). Since the early 1990's, it has been dogma that vertebrate Hox clusters are composed of thirteen paralogous groups. Nonetheless, we showed that in the otherwise prototypical cephalochordate amphioxus (Branchiostoma floridae), the Hox cluster contains a fourteenth Hox gene, and very recently, a 14th Hox paralogous group has been found in the coelacanth and the horn shark, suggesting that the amphioxus cluster was anticipating the finding of Hox 14 in some vertebrate lineages. In view of the pivotal place that amphioxus occupies in vertebrate evolution, we thought it of considerable interest to establish the limits of its Hox gene cluster, namely resolution of whether more Hox genes are present in the amphioxus cluster (e.g., Hox 15). Using two strategies, here we report the completion and characterization of the Hox gene content of the single amphioxus Hox cluster, which encompasses 650 kb from Hox1 to Evx. Our data have important implications for the primordial Hox gene cluster of chordates: the prototypical nature of the single amphioxus Hox cluster makes it unlikely that additional paralogous groups will be found in any chordate lineage. We suggest that 14 is the end.
Keywords: gene clusters, Evx, gene duplication, vertebrate evolution
1. Introduction
Since Edward Lewis discovery of the Bithorax complex of Drosophila [1], the Hox gene cluster has captivated the imagination of developmental and evolutionary biologists. Hox genes are a subclass of homeobox transcription factors deeply involved in the regulation of body patterning in metazoans [2]. Nevertheless, what made Hox genes special among developmental regulators is not their function, but their organization in chromosomal clusters and, most specially, the phenomenon of spatial and temporal Colinearity. Genes at the 3' end of the cluster are expressed, and pattern, the anterior end of the embryos, whilst genes at the 5' end of the cluster pattern the posterior [3]. Mammalian genomes contain four Hox clusters, with a selection of 13 Hox paralogous groups (PG1 to PG13), originated by cluster duplication from a single Hox cluster early in vertebrate evolution [4]. Following duplication, gene loss in the particular duplicate clusters would account for the actual composition of the mammalian clusters. Since the early 1990's, it has been dogma that this ancestral cluster was composed of 13 genes, one of each paralogous group: two from the Anterior class (PG1-2), a single class 3 gene (PG3), five Central genes (PG4-8), and five from the Posterior class (PG9-13). Cephalochordates (amphioxus) are the closest extant relatives of vertebrates. Their genome is probably a direct descendant of the vertebrate ancestral genome that existed before the wide genome duplications events that concurred with the origin of vertebrates and early steps of vertebrate evolution [5]. The initial finding of the single amphioxus Hox gene cluster nicely fit with mammalian data: amphioxus had single members of all vertebrate PGs 1 to 10 [6]. Further chromosomal walking at the 5' end of the amphioxus cluster surprisingly showed the presence not only of 13 Hox genes, but a 14th Hox gene, AmphiHox14 [7]. The lack of a Hox14 gene in vertebrate Hox clusters and the faster rate of sequence divergence of the posterior class of Hox genes (so-called “posterior flexibility”) had made it difficult to distinguish between two evolutionary scenarios. Did the chordate ancestor possess a single Hox cluster with 14 genes? Or, as favored at the time, was AmphiHox14 a gene duplication novelty of the amphioxus genome? Recent findings by Powers and Amemiya [8] corroborate the amphioxus data and suggest that the first, and less favored, scenario proposed four years ago may hold true.
Detailed analyses of the HoxA cluster of the coelacanth (Latimeria menadoensis) and the HoxD cluster of the horn shark (Heterodontus francisci) have revealed the existence of a Hox14 gene between the group 13 gene and the Evx1 and Evx2 genes, respectively [8]. Phylogenetic analyses clearly show these genes to be closely related, implying that the last common ancestor of gnathostomes had 14 Hox paralogous groups [9]. Posterior flexibility creates uncertainty concerning orthology of AmphiHox14 with coelacanth and horn shark Hox14 genes, although the genomic organization, the transcription orientation and, distinctively, the sharing of an intron position within the homeobox [8], are suggestive of orthology.
Fifteen years after vertebrate Hox13, and four years after amphioxus Hox14, are there hopes for finding a chordate Hox15 gene? The answer is probably not, as here we report the end of the amphioxus Hox cluster.
2. Methods
Fluorescence in situ hybridization to Branchiostoma floridae metaphase chromosomes and interphase nuclei was performed using cosmid probes from AmphiHox12/AmphiHox13 and AmphiEvxB genomic regions as described in [10]. Chromosomal walking at the 5' end of AmphiHox14 was performed from a single animal lambda genomic library [7]. An amphioxus PAC library was probed with AmphiEvx clones and with a probe from the most 5' extreme phage obtained in the lambda library walking. The PAC ends from positive clones were cloned in plasmids and probed against the lambda Hox and the previously isolated Evx cosmid clones [11]. Linkage between PACs from either screening was determined by end-cloning and crosshybridizations. Clones isolated were subsequently probed under low stringency TMAC hybridization conditions [12] with the degenerate oligonucleotide S02 (128x degeneracy, 17mer [6]). Faintly hybridizing bands were subcloned and sequenced.
3. Results
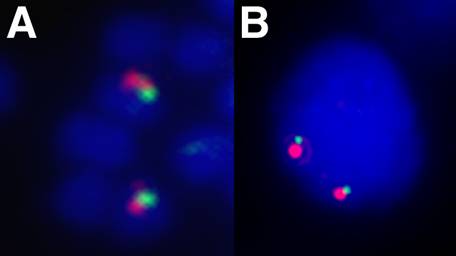
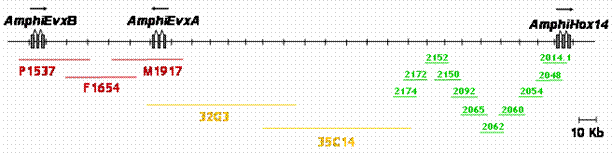
The 5' end of a Hox gene cluster is usually demarcated by the presence of linked Evx genes [13]. Evx genes are tightly linked to the Hox gene cluster in a range of organisms, namely mammals, coelacanth, teleost and chondrichthyan fishes, and cnidarians, and this linkage seems to represent the ancestral condition of the primordial metazoan Hox cluster. We sought to establish whether more Hox genes were present in the amphioxus Hox cluster and whether we had reached its end, as indicated by linkage to Evx. Two independent strategies were used to test linkage between AmphiHox genes and the two tandem amphioxus Evx genes [11]. First, two-color in situ hybridization on amphioxus metaphase chromosomes and interphase nuclei [10], using cosmid probes from AmphiHox12/AmphiHox13 and AmphiEvxB genomic regions, showed co-localization on the same chromosome (Fig. 1A). Moreover, the close association of both signals (Fig. 1B) suggests that they are separated at most by a few hundred kilobases (kb). And second, we performed chromosome walking from AmphiHox14 and Evx regions using B. floridae lambda, cosmid, and PAC libraries. This revealed that AmphiEvxA (which shows canonical Evx expression [11]) is more proximal to AmphiHox14, and separated from the Hox cluster by about 200 kb (Fig. 2). Furthermore, the inverted transcriptional orientation of AmphiEvxA with respect to AmphiHox genes is in agreement with that of vertebrate clusters [14]. Notably, we extensively searched for additional Hox genes between AmphiHox14 and AmphiEvxA by TMAC hybridization [12] using as a probe the degenerate oligonucleotide S02 (128x degeneracy, 17mer), which recognizes the third helix of the homeobox [6]. Low-stringency hybridization conditions were used to detect sequences with up to three mismatches with the degenerate probe [12] in order to assure detection of even the most divergent Hox-like homeoboxes. Although several faintly-hybridizing bands were cloned and sequenced, in all cases these regions contained non-coding sequences with only serendipitous similarity to the SO2 probe. From these data, we conclude that no additional homeoboxes are present beyond AmphiHox14 in the amphioxus genome. In summary, our data indicate that the complete Hox cluster of amphioxus extends for 450 kb from Hox1 to Hox14, and 650 kb from Hox1 to Evx, and contains only 14 Hox genes.
4. Discussion
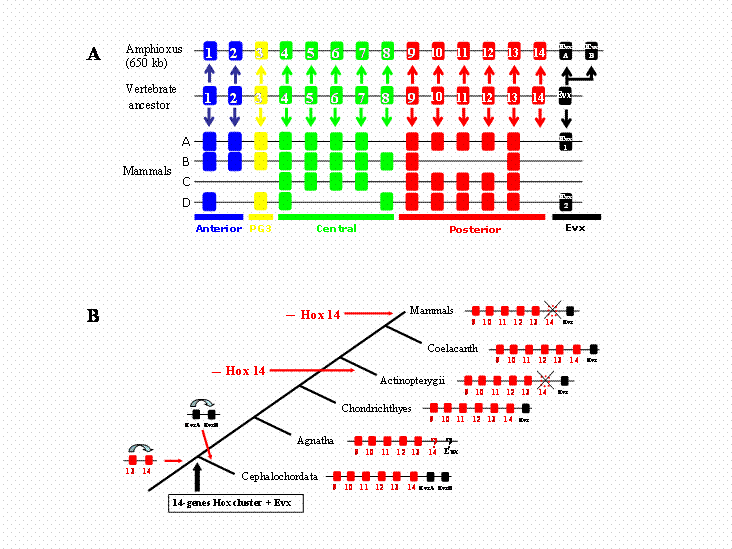
In view of the pivotal place that amphioxus occupies in vertebrate evolution, we thought it of considerable interest to establish the limits of its Hox gene cluster, namely resolution of whether more Hox genes are present in the single amphioxus Hox cluster (e.g., Hox 15). Using two strategies, we show here that the amphioxus Hox cluster (up to AmphiHox14) is linked to AmphiEvx, and that there are no additional Hox genes beyond AmphiHox14. The amphioxus Hox cluster encompasses 650 kb; this is about four times the size of a mammalian cluster [15]. Posterior flexibility does not clearly resolve whether AmphiHox Posterior genes are pro-orthologous of vertebrate genes, altough we favour a one-to-one ortology relationship (Fig. 3A). The recent finding of Hox 14 in some vertebrate lineages [8] however challenges the amphioxus data. We suggest as the most parsimonious hypothesis that the ancestor of vertebrates, prior to cluster duplication, possessed a single Hox cluster with 14 Hox genes linked to an Evx gene (Fig. 3B). PG14 was subsequently lost in some vertebrate lineages, namely actinopterigian fishes and mammals, whilst was maintained in chondrichthyes and coelacanth. Such ancestral vertebrate cluster would have a Hox content completely matching the amphioxus cluster. Hence, the prototypical nature of the single amphioxus Hox cluster makes it unlikely that additional paralogous groups will be found in any chordate lineage. We suggest that 14 is the end.
Acknowledgements
This research was funded by grant BMC2002-03316 (Ministerio de Ciencia y Tecnología, Spain), and by the Departament d'Universitats, Recerca i Societat de la Informació de la Generalitat de Catalunya (Distinció per la Promoció de la Recerca Universitaria) to JGF, and by grants NSF IBN-0321461 and NIH U01 HG02526 to CTA. CM and JG holded and hold respectively a CIRIT (Generalitat de Catalunya) PhD fellowship. L.F.C.C. was a GABBA PhD student funded by Fundação para a Ciência e a Tecnologia, Portugal (BD/21818/99).
Conflict of interest
The authors have declared that no conflict of interest exists.
References
1. Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978 ;276:565-570
2. Gellon G, McGinnis W. Shaping animal body plans in development and evolution by modulation of Hox expression patterns. Bioessays. 1998 ;20:116-125
3. Duboule D, Dollé P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989 ;8:1497-1505
4. Holland PWH, Garcia-Fernàndez J. Hox genes and chordate evolution. Dev Biol. 1996 ;173:382-395
5. Furlong RF, Holland PWH. Were vertebrates octoploid? Philos Trans R Soc Lond B Biol Sci. 2002 ;357:531-544
6. Garcia-Fernàndez J, Holland PWH. Archetypal organization of the amphioxus Hox gene cluster. Nature. 1994 ;370:563-566
7. Ferrier DEK, Minguillón C, Holland PWH, Garcia-Fernàndez J. The amphioxus Hox cluster: deuterostome posterior flexibility and Hox14. Evol Dev. 2000 ;2:284-293
8. Powers TP, Amemiya CT. Evidence for a Hox14 paralog group in vertebrates. Curr Biol. 2004 ;14:R183-184
9. Ferrier DEK. Hox genes: Did the vertebrate ancestor have a Hox14? Curr Biol. 2004 ;14:R210-211
10. Castro LFC, Holland PWH. Fluorescent in situ hybridisation to amphioxus chromosomes. Zoolog Sci. 2002 ;19:1349-1353
11. Ferrier DEK, Minguillón C, Cebrián C, Garcia-Fernàndez J. Amphioxus Evx genes: implications for the evolution of the Midbrain-Hindbrain Boundary and the chordate tailbud. Dev Biol. 2001 ;237:270-281
12. Wood WI, Gitschier J, Lasky LA, Lawn RM. Base composition-independent hybridization in tetramethylammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc Natl Acad Sci U S A. 1985 ;82:1585-1588
13. Minguillón C, Garcia-Fernàndez J. Genesis and evolution of the Evx and Mox genes and the extended Hox and ParaHox gene clusters. Genome Biol. 2003 ;4:R12-
14. Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998 ;282:1711-1714
15. Boncinelli E, Somma R, Acampora D, Pannese M, D'Esposito M, Faiella A, Simeone A. Organization of human homeobox genes. Hum Reprod. 1988 ;3:880-886
Figures
Chromosomal linkage between Hox and Evx in amphioxus. Fluorescence in situ hybridization to B. floridae metaphase chromosomes (A) and interphase nuclei (B) showing co-localization of Hox (green) and Evx (red) genes. Cosmid probes containing the AmphiHox12/13 genomic region and AmphiEvxB region were used as described in [10].
Genome map of the posterior end of the amphioxus Hox cluster, and linkage to AmphiEvx genes. Overlapping lambda (green), PAC (yellow) and cosmid (red) clones are shown below the map. Transcriptional orientation is indicated above the genes.
Evolutionary history of Hox clusters in the vertebrate lineage. A) Structure of the amphioxus and mammalian Hox clusters, and deduced cluster structure inferred for the last common ancestor of Cephalochordates and Vertebrates. Hox paralogous group 14 genes were lost in the lineage leading to mammals, and the two amphioxus Evx genes arose from a tandem duplication event in the amphioxus lineage. B) Duplications and losses of posterior Hox and Evx genes in the vertebrate lineage. The consensus cluster is shown for selected vertebrate lineages. A single duplication event (Hox13/Hox14) needs to be assumed in the vertebrate stem lineage. Further duplications in the cephalochordate lineage and gene losses in actinopterygian fishes and mammalians results in the actual gene content of the clusters.
Author biography
Carolina Minguillón obtained her Ph. D. from the University in Barcelona in 2002. Her main achievements involved the partial characterisation of the Hox and Evx genomic regions and the study of somitogenesis (by virtue of the analyses of Mox and hairy genes) in the cephalochordate amphioxus. Currently, she is an EMBO postdoctoral Fellow at the laboratory of Dr. Malcolm Logan at the National Institute for Medical Research, Mill Hill, London. Her research focus on the function of certain T-box transcription factors in vertebrate limb development.
Chris T. Amemiya, Ph.D. (from Texas A&M University), B.S. (Purdue University), Postdoc 1 (Tampa Bay Research Institute), Postdoc 2 (Lawrence Livermore National Laboratory). Current: Member, Benaroya Research Institute at Virginia and Professor of Biology, University of Washington. Past: Associate Professor of Human Genetics, Boston University School of Medicine. Interests: Evolution and Development, vertebrate comparative genomics, Hoxgenes, evolution of the vertebrate immune system
Peter Holland, MA (Oxford), PhD (London), DSc (Reading), FRS, is the Linacre Professor of Zoology at the University of Oxford, UK. He was awarded the Scientific Medal of the Zoological Society of London in 1996, the De Snoo Medal in 1999 and the Genetics Society Medal in 2004. He has an unhealthy obsession with homeobox genes and their evolution.
Jordi Garcia-Fernàndez, PhD (University of Barcelona), EMBO, Human Frontiers and EU postdoctoral fellow (Oxford). He is currently Associate professor of Genetics at the University of Barcelona, and holds the Distinction of the Generalitat de Catalunya for the Promotion of University Research (young investigator). He has been linked to homeobox genes from an evolutionary point of view since the beginning (Hox, ParaHox...). His team is concentrated now in the invertebrate/vertebrate transition, using amphioxus as a model, and homeobox and other developmental genes as tools to understand what happened there.
![]() Corresponding address:
Corresponding address:
Dr. Jordi Garcia-Fernàndez. e-mail: jgarciaub.es. Tel. +34-934034437 Fax. +34-934034420

 Global reach, higher impact
Global reach, higher impact