10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2005; 1(3):110-113. doi:10.7150/ijbs.1.110 This issue Cite
Short Research Communication
Translocation of a 190-kb mitochondrial fragment into rice chromosome 12 followed by the integration of four retrotransposons
1 Genetic Diversity Department, National Institute of Agrobiological Sciences, Tsukuba, Ibaraki, Japan
2 Laboratory of Plant Molecular Genetics, Graduate School of Agricultural and Life Sciences, University of Tokyo, Tokyo, Japan
Received 2005-5-6; Accepted 2005-6-8; Published 2005-7-1
Abstract
A 190-kb mitochondrial DNA sequence interrupted by seven foreign DNA segments was identified in rice chromosome 12. This fragment is the largest mitochondrial fragment translocated into the rice nuclear genome. The sequence is composed of a 190-kb segment of mitochondrial origin corresponding to 38.79% of the mitochondrial genome, 45 kb comprising four segments of retrotransposon origin, and 13 kb comprising three segments of unknown origin. The 190-kb sequence shows more than 99.68% similarity to the current mitochondrial sequence, suggesting that its integration into the nucleus was quite recent. Several sequences in the 190-kb segment have been rearranged relative to the current mitochondrial sequence, suggesting that the past and present arrangements of the mitochondrial genome differ. The four retrotransposons show no mutual sequence similarity and are integrated into different locations, suggesting that their integration events were independent, frequent, and quite recent. A fragment of the mitochondrial genome present in the nuclear genome, such as the 248-kb sequence characterized in this study, is a good relic with which to investigate the past mitochondrial genome structure and the behavior of independent retrotransposons during evolution.
Keywords: mitochondria, nuclear, genome, integration, retrotransposon
1. Introduction
It is well accepted that present-day mitochondria and chloroplasts resulted from the integration of living organisms into an ancestral mother cell [1]. During evolution, most genes originally encoded by the endosymbiont have been translocated to the nuclear genome. However, most processes of gene translocation are still unclear. In recent studies, many mitochondrial genes that have been translocated into the nuclear genome have been successfully identified using molecular techniques. For instance, the respiratory mitochondrial gene cox2, which is ubiquitously found in the mitochondrial genome, is absent in the mitochondrial genome of the cowpea but has been translocated to the nuclear genome [2]. In rice, rps11 was the such organellar gene identified as having translocated to the nucleus. An rps11-related sequence is present in the rice mitochondrial genome but it is non-functional because of an internal stop codon in the original reading frame. A functional rps11 gene has been identified in the rice nuclear genome and is well characterized [3]. However, such successful gene translocations are rare events. More than 500 translocation events have been identified in which a mitochondrial sequence transferred into the rice nuclear genome [4]. The fate of the longest fragment, comprising 248 kb, identified in these events is further characterized in this study.
2. Materials and Methods
2.1 Program
The available rice genome database (http://rgp.dna.affrc.go.jp/) and the BLAST programs in the RiceBLAST BLAST Search Service (http://riceblast.dna.affrc.go.jp/) were used in this study. The parameters used in the BLASTN programs were the default values.
3. Results and Discussion
3.1 Identification of a 248-kb mitochondrial sequence in rice chromosome 12
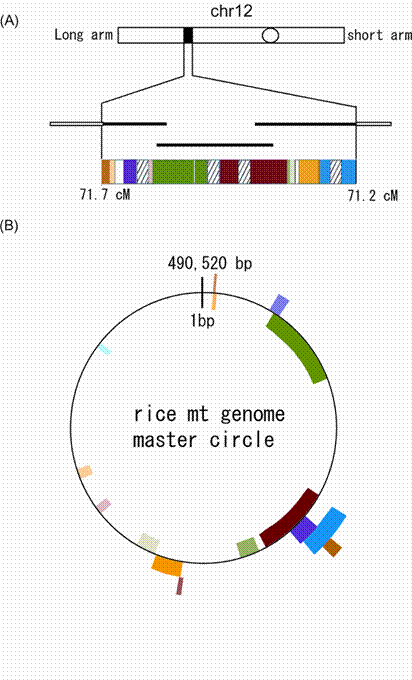
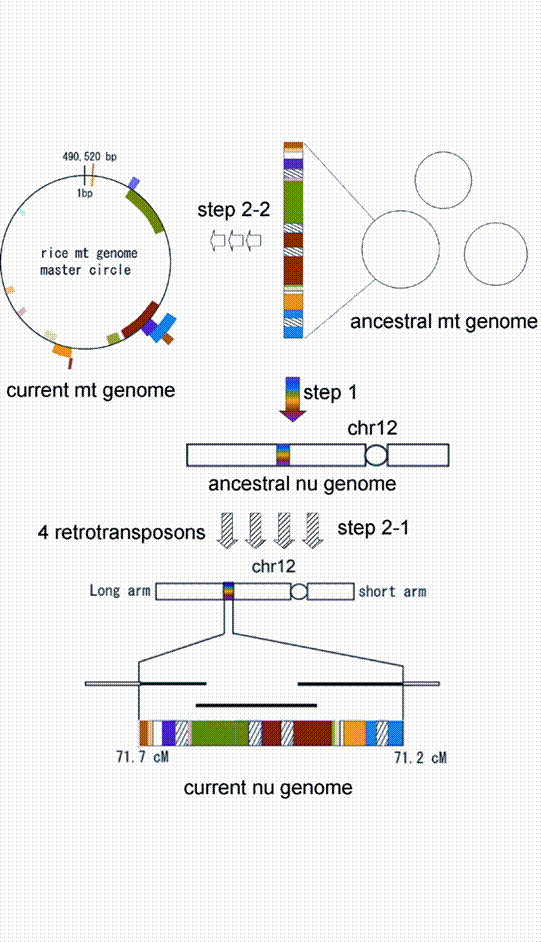
The complete mitochondrial genome of rice has been sequenced [5]. Most of the nuclear genome sequence is also available for rice (http://rgp.dna.affrc.go.jp/). DNA sequences that have translocated from the mitochondrial genome to the nuclear genome have been identified at over 500 loci [4]. The longest of these sequences, a 248-kb mitochondrial fragment, was identified on chromosome 12 (Fig. 1). The 248-kb mitochondrial DNA fragment in the nuclear genome is designated '248-kb numtDNA' in this study. The sequence starts at position 20493130 and ends at 20741523, corresponding to 71.2-71.7 centimorgans, on chromosome 12. The 248-kb numtDNA was contiguous across three overlapping bacterial artificial chromosome (BAC) clones: AL731738, AL713949, and AL732377. In the 248-kb numtDNA, seven segments of DNA comprising 58 kb are of non-mitochondrial origin. The remaining 190-kb numtDNA is of mitochondrial origin, corresponding to 38.79% of the mitochondrial genome. It has 99.68% identity with the original mitochondrial sequence [5], strongly suggesting that the sequence transferred from the mitochondrial to the nuclear genome very recently. However, the arrangement of DNA segments is very different in the 248-kb numtDNA and the present mitochondrial genome. This suggests that the past mitochondrial DNA conformation was quite different from the present conformation, rather than that DNA fragments from different mitochondrial positions integrated into the same chromosomal locus or that DNA rearrangements have occurred within the nuclear genome. Mutation is much faster in nuclear genome than in the mitochondrial genome [6]. If the mutation rate is 5-30 × 10-9 per site per year in the plant nucleus [6], the translocation of the 190-kb numtDNA is estimated to have happened about 1.0-6.4 x 105 years ago. The similarities between each part of the mitochondrial fragments are shown in Table 1.
A 620-kb mitochondrial sequence has been identified in the Arabidopsis nuclear genome [7]. Our study and the previous Arabidopsis study suggest that the plant nuclear genome is flexible enough to accept into chromosomes more than 190 kb of foreign DNA sequence at a time.
3.2 Recent integration of four retrotransposons into the 190-kb numtDNA
Seven segments comprising 58 kb within the 248-kb numtDNA are inferred to be of non-mitochondrial origin because no homology was identified between them and the present rice mitochondrial genomic sequence. Four of these segments were found to be retrotransposon-related sequences in that they contain 3982-bp, 4228-bp, 3155-bp, and 3237-bp long terminal repeats (LTR). The total lengths of the four retrotransposons are 10.7 kb, 11.3 kb, 12.1 kb, and 10.7 kb, respectively (Table 1). One of them is a Ty3-gypsy like retrotransposon on the basis of its structure [8], but the other three are class I retrotransposons that had not yet been categorized. To determine the possible ancestral retrotransposons, we conducted NCBI BLASTX searches [9] using internal sequences from the three retrotransposons as queries. Among the retrotransposons extracted from the analysis, we found several retrotransposons encoding full sets of proteins, including capsid-like protein, endonuclease, integrase, protease, reverse transcriptase, and Rnase-H [8]. A comparison of the orders of these protein-encoding sequences strongly suggests that all the retrotransposons are Ty3-gypsy like retrotransposon. Therefore, we conclude that all four retrotransposons in the 248-kb numtDNA were derived from a Ty3-gypsy like retrotransposon. Disruption of the three retrotransposons may have occurred during translocation, in the process of reverse transcription or integration. Because the migration of the 190-kb numtDNA into the nuclear genome was quite recent and no corresponding retrotransposon sequence was identified in the original mitochondrial sequence, it is highly likely that the transposition of the four retrotransposons into the numtDNA occurred very recently. It also suggests that the four retrotransposons were active at the time of integration. In short, these retrotransposons integrated into the 190-kb numtDNA since 1.0-6.4 x 105 years ago. tos17 is an endogenous copia-like retrotransposon of rice that is activated in tissue culture. The four retrotransposon segments are different from tos17 and are actually integrated into the numtDNA, so they are possibly another kind of active retrotransposon, but their activation triggers are not known.
An NCBI BLAST search [9] was conducted to determine the origins of the three remaining segments that were not mitochondrially derived (Table 1). Sequence unknown 1 (8719 bp) contained 2003 bp and 606 bp of the rice mitochondrial and chloroplast sequences, respectively, whereas no information about the remaining 6110-bp sequence is available. Sequences unknown 2 (1118 bp) and unknown 3 (3436 bp) are similar to maize mitochondrial sequences of 358 bp and 478 bp, respectively. These data suggest that the two sequences were present in the mitochondrial genome in the past but have been lost from the present form of the rice mitochondrial genome.
In conclusion, the 248-kb numtDNA is a good relic sequence with which to determine the past mitochondrial genome structure and the behavior of independent retrotransposons during evolution. DNA segments of foreign origin, like numtDNA, should make a considerable contribution to the study of genome evolution.
Evolutionary information on the DNA segments in the 248-kb numtDNA
| Position on chr12 | Position on mt genome | Similarity | Remarks | |
|---|---|---|---|---|
| 20493130-20505933 | 174769-182286 | 1) 99.89% | ||
| 336511-341789 | 1) 99.47% | |||
| 20505934-20514652 | Unknown 1 | |||
| 20514653-20527904 | 177831-190726 | 1) 99.88% | ||
| 317744-317382 | 1) 100% | |||
| 20527905-20538625 | 2) 100% | *1 Retrotransposon | ||
| 20538626-20542684 | 317387-313326 | 1) 99.73% | ||
| 92237-51835 | 1) 99.98 | |||
| 20542685-20583082 | ||||
| 20583083-20584200 | Unknown 2 | |||
| 20584201-20596624 | 51835-40090 | 1) 99.42 | ||
| 7395-6728 | 1) 99.70% | |||
| 20596625-20607940 | 2) 99.91% | *2 Retrotransposon | ||
| 20607941-20626615 | 6733-6114 | 1) 99.84% | ||
| 165164-183215 | 1) 99.83% | |||
| 20626616-20638693 | 2) 99.97% | *3 Retrotransposon | ||
| 20638694-20673702 | 183211-208532 | 1) 99.12% | ||
| 212838-222652 | 1) 99.75% | |||
| 20673703-20677265 | 281779-285341 | 1) 99.69% | ||
| 20677266-20682489 | 416262-421493 | 1) 99.90% | ||
| 20682490-20685923 | Unknown 3 | |||
| 20685924-20716720 | 273647-254621 | 1) 99.83% | ||
| 7395-6111 | 1) 100% | |||
| 165164-175799 | 1) 98.41% | |||
| 20716721-20727451 | 2) 99.57% | *4 Retrotransposon | ||
| 20727452-20741523 | 175795-189870 | 1) 99.87% |
Positions on chromosome 12 (chr12) and the mitochondrial (mt) genome and their sequence similarities are shown. 1) comparing the mt genome against the nuclear genome. 2) comparing one part of the LTR against the other part of the LTR in the same retrotransposon. *3) Ty3-gypsy like retrotransposon; *1, *2, *4 thought to be Ty3-gypsy like retrotransposons
Acknowledgements
This work was partly supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (grant 15208001) to N.T. and K.K. We thank Drs. F. Quetier, T. Nishikawa and A. Miyao for helpful discussions.
Conflict of interest
The authors have declared that no conflict of interest exists.
References
1. Gray MW. The endosymbiont hypothesis revisited. Int Rev Cytol. 1992 ;141:233-357
2. Nugent JM, Palmer JD. RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell. 1991 ;66(3):473-81
3. Kadowaki K, Kubo N, Ozawa K. et al. Targeting presequence acquisition after mitochondrial gene transfer to the nucleus occurs by duplication of existing targeting signals. EMBO J. 1996 ;15(23):6652-61
4. Richly E, Leister D. NUMTs in sequenced eukaryotic genomes. Mol Biol Evol. 2004 ;21(6):1081-4
5. Notsu Y, Masood S, Nishikawa T. et al. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics. 2002 ;268(4):434-45
6. Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 1987 ;84(24):9054-8
7. Stupar RM, Lilly JW, Town CD. et al. Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: implication of potential sequencing errors caused by large-unit repeats. Proc Natl Acad Sci U S A. 2001 ;98(9):5099-103
8. Kumar A, Bennetzen JL. Plant retrotransposons. Annu Rev Genet. 1999 ;33:479-532
9. Altschul SF, Madden TL, Schaffer AA. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997 ;25(17):3389-402
Figures
Schematic representation of the 248-kb mitochondrial sequence on chromosome 12 and the corresponding sequences on a master circle of rice mitochondrial genome. (A) Three contiguous BAC clones carrying mitochondrial sequences are indicated by bold lines. cM, centimorgans. A black box and a circle represent a mitochondrial sequence and a centromere on chromosome 12, respectively. Four striped boxes show retrotransposon sequences. Three white boxes indicate sequences of unknown origin. (B) A presumptive mitochondrial master circle of the rice mitochondrial genome. Sequences with strong similarity are shown in the same colors
A model for the evolution of the 248-kb numtDNA on rice chromosome 12. Step 1, integration of a 190-kb mitochondrial fragment into chromosome 12. Step 2-1, integration of four retrotransposons into the 190-kb mitochondrial fragment. Step 2-2, recombination events within the mitochondrial genome
Author contact
![]() Corresponding address:
Corresponding address:
Koh-ichi Kadowaki, Ph.D, National Institute of Agrobiological Sciences, 2-1-2 Kannondai, Tsukuba, Ibaraki 305-8602, Japan. Tel: +81-29-838-7449 Fax: +81-29-838-7408 e-mail: kadowakigo.jp

 Global reach, higher impact
Global reach, higher impact