10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2006; 2(2):79-86. doi:10.7150/ijbs.2.79 This issue Cite
Research Paper
Expression of AmphiNaC, a new member of the amiloride-sensitive sodium channel related to degenerins and epithelial sodium channels in amphioxus
Department of Biology, University of Genoa, Genoa, Italy
Received 2006-1-30; Accepted 2006-3-22; Published 2006-4-10
Abstract
Degenerins and amiloride-sensitive Na+ channels form a new family of cationic ion channels (DEG/NaC). DEG/NaC family emerged as common denominator within a metazoan mechanosensory apparatus. In this study, we characterized a new member of such family in amphioxus, Branchiostoma floridae. The AmphiNaC cDNA sequence encodes a protein showing amino acid residues characteristic of DEG/NaC family, such as two hydrophobic domains surrounding a large extracellular loop that includes cystein-rich domains; nevertheless its predicted sequence is quite divergent from other family members. AmphiNaC is expressed at early larval stage in some putative sensory epidermal cells in the middle of the body and in neurons of the posterior cerebral vesicle, as well as in some ventrolateral and mediolateral neurons of the neural tube. In late larvae, AmphiNaC expression is maintained in some neurons of the neural tube, and it is expressed in putative sensory epidermal cells of rostrum and mouth. The analysis of AmphiNaC gene expression pattern suggests that it might be involved in neurotransmission and sensory modulation.
Keywords: amiloride-sensitive sodium Na+, degenerins, Branchiostoma floridae, developmental expression, sensory neurons
1. Introduction
The degenerins and amiloride-sensitive Na+ channels (DEG/NaC) is a large family of cationic ion channels associated with highly diversified functions in many organisms [1]. Members of the DEG/NaC superfamily have been found in nematodes, flies, snails, and vertebrates [1, 2]. The DEG/NaC gene family shares common properties including permeability to Na+, inhibition by diuretic amiloride and voltage-independent gating. The family includes channels that are constitutively active, such as the epithelial Na+ channel (ENaC) associated with transepithelial Na+ transport and Na+ homeostasis, as well as taste perception [3], the Drosophila amiloride-sensitive Na+ channels, dGNaC1/RPK [4] involved in gametogenesis and early development, and the dmdNaC1/PPK probably linked to proprioceptive signals and salt taste [4, 5]. Moreover, the DEG/NaC family comprises ligand-gated channels such as the Phe-Met-Arg-Phe-NH (FMRFamide) peptide gated Na+ channel (FaNaC), cloned in the snail Helix aspersa [6], and presumably involved in neuronal modulation, and the Caenorhabditis elegans degenerins thought to form a mechanosensory complex [7]. The DEG/NaC family also includes the neuronal acid sensing ion channels (ASICs) identified in mammals and zebrafish [2, 8]. ASICs are proton-gated channels, which are expressed in brain and sensory neurons, and in mammals they seem to be involved in pain perception and mechanosensation [9, 10]. More recently, a novel related gene called BLINaC has been identified in liver, intestine and brain of mammals [11]. BLINaC is only 30% identical to ASICs and it is not activated by an extracellular acidification, although it is able to form an amiloride-sensitive Na+-selective channel.
The DEG/NaC channels family shares a common topology: such channels span the membrane twice and have intracellular N and C termini with a large cystein-rich extracellular loop. The position of many cystein residues is well conserved, suggesting that intramolecular disulfide bonds are necessary to make three dimensional structure of the extracellular domain acting as sensor or receptor for extracellular stimuli. Furthermore, these channels are composed of multiple subunits, as homo- or hetero-multimers.
Despite the widespread importance of mechanotransduction in biology, remarkably little is known about the molecules that mediate mechanical signalling in amphioxus, the closest living invertebrate relative to the vertebrates. Therefore, we cloned a member of DEG/NaC channels superfamily and we studied its expression during amphioxus development.
2. Methods
In order to isolate the AmphiNaC sequence in the amphioxus Branchiostoma floridae we performed a BLAST search to the Trace Archives of Whole Genome Sequence (NCBI) using the FaNaC sequence from Helix aspersa. We isolated a partial sequence from the embryos cDNA library (kindly given by Jim Langeland) using specific primers (For1: 5'- atgagctgttcctacaacgg -3'; Rev1: 5'- aaggtaagacacctttacc -3'). After that, we extend the sequence to the 3'- and 5'-end using specific primers (For2: 5'- agtcctacgtcaccaccatgacg -3'; Rev2: 5'- agtccttggcctctggaggacgc -3') in conjunction with universal vector primers. Several isolated clones were analysed and sequenced, and the corresponding deduced cDNA sequence of AmphiNaC was submitted to GenBank under accession number (DQ374391). We also performed analysis of amphioxus genomic sequences in the Trace Archives but we have not found any more long open reading frame.
In order to infer evolutionary relationship between AmphiNaC and FaNaC/DEG/ENaC/ASIC genes family of vertebrates and invertebrates, a phylogenetic analysis was done using the neighbouring-joining method implemented in ClustalX. The phylogenetic tree was visualized with TREEVIEW.
The spatial expression pattern of the AmphiNaC was examined by whole mount in situ hybridization on amphioxus developmental stages collected in Tampa Bay, FL. The probes used in the experiments corresponding to +1 bp to +1985 bp of the AmphiNaC cDNA sequence and were produced by PCR using primers For3 and Rev3 (5'- actgatccaacatggcaggaag -3'; 5'- tgaggtgaggtgagatacaac -3'). The amplified fragment was cloned into pCRII plasmid (Invitrogen, CA) and used to synthesize antisense and sense RNA probes using T7 and Sp6 polymerase following the instructions supplied with the DIG RNA labelling kit (Roche Diagnostics SpA, Italy).
3. Results
The AmphiNaC receptor cDNA sequence from B. floridae is 2587-bp long including a coding region of 1326-bp, and a 3' untranslated region 1261-bp long with a polyadenylation signal 20 bases upstream from the poly(A) tail. An in frame stop codon upstream from the putative start codon was also found (Fig.1). The putative longest open reading frame codes for 442 amino acids. AmphiNaC deduced protein presents two putative transmembrane regions near N-terminus and C-terminus, as predicted with the TMpred program (available at
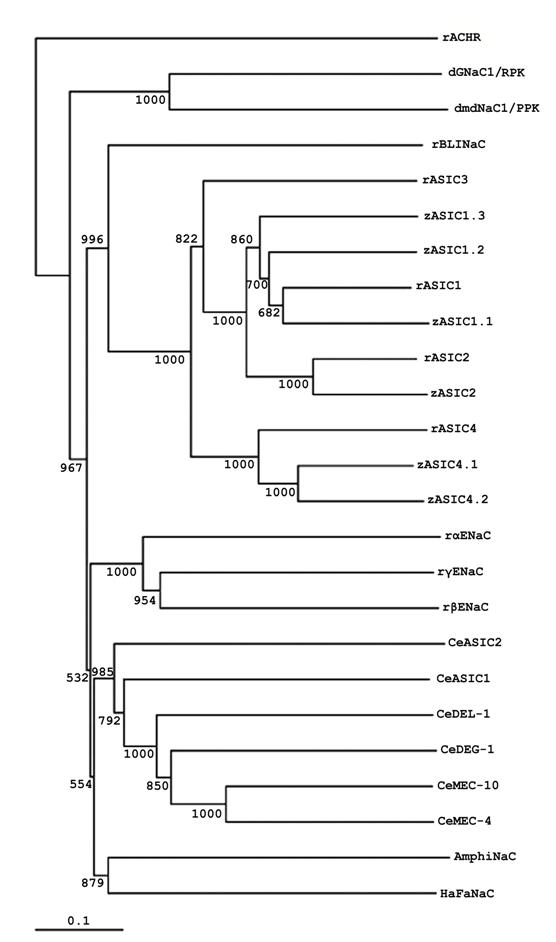
The sequence of AmphiNaC indicates that it is a member of DEG/NaC superfamily. Nevertheless, it is not a member of the degenerins, ASIC, ENaC, or Drosophila amiloride-sensitive Na+ channel branches: AmphiNaC is missing certain conserved sequences, and a phylogenetic analysis place it in a new branch of the family together with the high divergent sequence of HaFaNaC (Fig.2). At present it is not possible to argue if AmphiNaC is a FMRF-peptide gated Na+ channel or it has some specific functional activity or it forms a functional membrane channel. Furthermore, it is interesting that at present no mammalian orthologs of C. elegans degenerins, Drosophila RPK/dGNaC1 and PPK/dmdNaC1 genes or FaNaC have not yet been identified, therefore it seems reliable that a divergence of the different DEG/NaC subfamilies occurred early in evolution.
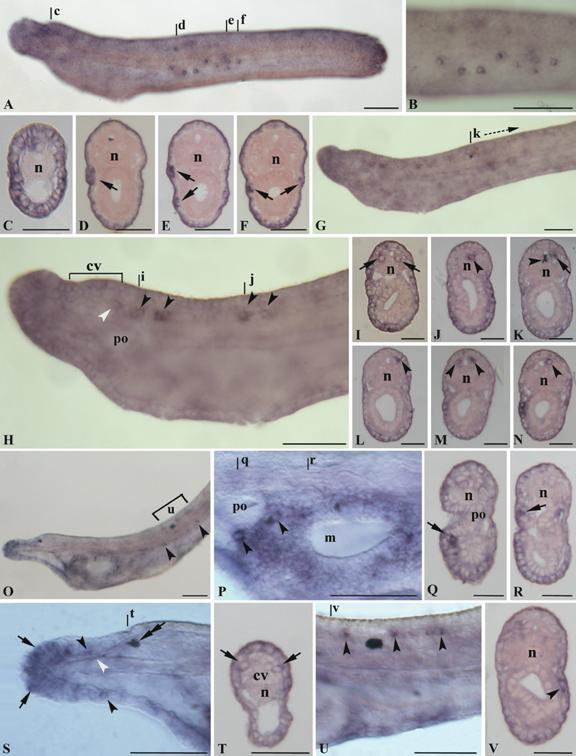
No expression signal of AmphiNaC was observed in oocytes and early stages embryos. The earliest detectable expression begins at early larval stage in the epidermis. Such expression was found in the rostrum and in some other epidermal cells scattered in the larval body, and a strong expression was detected in some large ventrolateral epidermal cells located in the middle of the body (Fig. 3A-F). Such cells are found on the right and left walls and in some cases appear arranged in groups of more than one cell (Fig. 3D-E). In early larvae, AmphiNaC expression was also detected in few cells of the posterior region of the cerebral vesicle, probably corresponding to elements of the infundibular organ (Fig. 3H), and in some ventrolateral and floor plate neurons of the nerve cord (Fig. 3H-J). In particular, the labelled ventrolateral neurons, located just behind the cerebral vesicle, are large paired nerve cell bodies (as visible in cross section, Fig. 3I) and could correspond to the large paired descending interneurons (LPN) of the primary motor centre (PMC). Behind the first pigment spot, the AmphiNaC labelled neurons were mainly located in mediolateral position (Fig. 3G, K-N).
In three-gill slit larva an expression was present in various epidermal cells of rostrum; in particular some of the most anterior tip cells expressing AmphiNaC could correspond to the primary sensory cells (Fig. 3O, S). Furthermore, transcripts were also detectable in the rostral nerves (Fig. 3S). Several labelled epidermal cells were also located around the mouth region and near the preoral pit (Fig. 3O-R), and some scattered epidermal cells were visible below the myotomes in the half of the larval body (Fig. 3O, V). Two large labelled cells, below and behind the preoral pit, have distinct morphology from that of the surrounding epidermis cells and could be assigned to the large extrinsic neurons described by Lacalli and coworkers [12]. Further AmphiNaC expressing cells around the oral region could represent intrinsic neurons or the precursors of the oral spine cells. The expression in the nervous system persists exclusively in some cells of the dorsal neural tube just near the first pigment spot (Fig. 3O, U), whereas a new expression appears in some cells of the frontal eye (Fig. 3S-T). Ultrastructural studies have shown similarities among frontal eye and paired eyes of vertebrates [13]. The amphioxus frontal eye consists of a pigment cup, and at least four rows of cells. The cells of the first two rows are simple sensory neurons and they are probably photoreceptors, whereas those of the last two rows are more typically nerve-like cells. AmphiNaC is expressed in two symmetrical cells probably corresponding to the first two rows.
AmphiNaC nucleotide and amino acid sequence. The nucleotide and the predicted protein sequences of AmphiNaC are shown. Stop codon upstream of the first methionine is indicated by an asterisk, and the 3' end stop codon and the polyadenylation signal are underlined. Putative transmembrane domains (MI and MII) are outlined in dark. Cystein residues are grey shaded.

Phylogenetic tree of AmphiNaC and DEG/NaC proteins. Phylogenetic analysis of AmphiNaC with several DEG/NaC family members isolated from Drosophila (dGNaC1/RPK, AAC38824; dmdNaC1/PPK, AAC38823), C. elegans (CeMEC-4, T29859; CeMEC-10, AAB07694; CeDEG-1, AAB07574; CeDEL-1, NP_510380; CeASIC1, AAB93309; CeASIC2, NP_492099), Helix aspersa (HaFaNaC, CAA63084), Danio rerio (zASIC1.1, CAE81918; zASIC1.2, CAE81919; zASIC1.3, CAE81920; zASIC2, CAE81921; zASIC4.1, CAE81922; zASIC4.2, CAE81923), Rattus norvegicus (rASIC1, AAB53002; rASIC2, Q62962; rASIC3, O35240; rASIC4, NP_071570; rBLINaC, CAB54072; rαENaC, NP_113736; rβENaC, NP_036780; rγENaC, NP_058742). We used the rattus sequence of nicotinic acetylcholine receptor (rACHR, AAA40664) as outgroup. The numbers near the branches are bootstrap values based on 1,000 replicates. For the alignment and construction of phylogenetic tree highly divergent sequences at the N and C termini as well as in the proximal part of the extracellular loop has been deleted.
Gene expression of AmphiNaC in amphioxus larvae. Anterior of whole mounts toward left, cross sections, counterstained pink, viewed from the posterior end of animal; whole mount scale lines = 50 µm and sections scale lines = 25 µm. A, 26-h larva with the epidermis in focus. AmphiNaC expression was found in some epidermal sensory cells in the flanks of the body. B, Enlargement of the preceding specimen where at least eight labelled epidermal sensory cells was visible. C, Cross section through level c in A, with AmphiNaC diffuse expression in cells of the anterior ectoderm. D-F, Cross sections trough levels d-f in A showing expression in some large cells located in the epidermal layer (arrows). G, In 28-h larva AmphiNaC transcripts were visible in the anterior ectoderm, in few neurons of the posterior cerebral vesicle, and in some neurons along the nerve cord. H, Enlargement of the anterior two-thirds of the preceding specimen where are better visible some neurons in the posterior cerebral vesicle (white arrowhead), as well as in some scattered neurons located in the nerve cord (arrowheads). I and J, Cross sections through levels i and j in H, where are visible a pair of labelled symmetrical ventrolateral cells (arrows) and a single labelled neuron in the floor plate (arrowhead). K-N, Cross sections starting from k, at level shown in G. AmphiNaC expression was found in a single neuron located in mediolateral position (arrow) (K) near to the first pigment spot (arrowhead), and in some mediolateral neurons (arrowheads) (L-N) of the neural tube. O, Lateral view of the three-gill slit larva shows expression in the rostrum, in cells lining the mouth, and in some epidermal cells (arrowheads). P, Enlargement of the mouth region of the preceding specimen where AmphiNaC expression was found in some cells around the preoral pit (arrowheads) and in several epidermal cells lining the opening mouth. Q-R, Cross sections trough levels q and r in P show a signal in some cells located in the epidermis just near the preoral pit (arrow) (Q), and in epidermal cell lining the mouth (arrow) (R). S, Anterior rostral region of the specimen in O where AmphiNaC transcripts were conspicuous in several epidermal cells located around the anterior tip of the rostrum (arrows) and in dorsal and ventral sides of the rostral epithelium (arrowheads). An expression was also found in rostral nerves (white arrowhead). A double arrow indicates the pigment spot of the frontal eye complex. T, Cross section through level shown in S with expression in a pair of symmetrical cells of the most anterior region of frontal eye complex (arrows). U, Enlargement of the body portion (u bracket in O) at level of the first pigment spot where expression was found in some neurons of the dorsal neural tube (arrowheads). V, Scattered epidermal cells are still found in the middle of the body as visible in cross section (level v in U) where a single labelled large cell body occurs in the epidermis (arrowhead). cv, cerebral vesicle; po, preoral pit; m, mouth; n, notochord.
4. Discussion
Amphioxus belongs to the subphylum Cephalochordata, which is classified in the Chordata together with the Urochordata and the Vertebrates. The body plan of amphioxus is similar to, but simpler than, that of vertebrates. Several studies on amphioxus CNS have shown that its organization is similar to that of vertebrates excepted for neural crest and telencephalon that are likely vertebrate innovations. Little is known about the differentiation of the amphioxus peripheral nervous system (PNS) and in particular about the sensory modalities of the peripheral receptors. Several studies have outlined that amphioxus has at least two types of sensory receptors: type I primary sensory neurons (having axons projecting to CNS) and type II secondary sensory neurons (axonless with a peculiar microvillar collar around the cilium). More recently, data have shown that some sensory neural markers such as AmphiCOE and AmphiHu are expressed at neurula stages in scattered epidermal cells on the flanks of the body probably representing precursors of primary sensory neurons [14, 15]. Differentiated primary sensory neurons appear down the flanks of early larva, whereas secondary sensory neurons, widely distributed on the body, start at early metamorphic larvae [16, 17, 18, 19, 20, 21]. The expression of AmphiNaC in some epidermal cells on the flanks larval body remembers the expression of AmphiCoE and AmphiHu in the primary epidermal sensory neurons.
Additional types of primary and secondary sensory cells have been also described in association with mouth and preoral ectoderm [12]. The epithelium around the mouth is characterized by two types of peripheral nerve cells in 8-12 day larva: intrinsic, embedded in the oral nerve plexus and extrinsic lying in the preoral ectoderm [12]. The first type borders the oral region, whereas the second one is found near the preoral pit. Further types of sensory cells consisting of clusters of flask-shaped uniciliate cells, called oral spines, are distributed around the edges of the mouth. The oral spines are secondary sensory cells and display the morphological features of mechanoreceptors.
AmphiNac is expressed in the extrinsic cells and in some cell types around the mouth. However, for the latter it is difficult to establish what type of cells they are, because our results become from younger larvae. Nevertheless, more extensive descriptions of the expression pattern of AmphiNaC in amphioxus late larvae and adults will be necessary to provide additional information about the labelled cells located at the margin of the mouth.
Despite of the large amount of microscopic anatomy data on the PNS, it remains quite controversial the possible function of such sensory receptors, although mechanoreception and chemoreception have been postulated [22, 23, 24]. More recently, amphioxus gene expression data on a G protein coupled receptor (AmphiGPCR1) related to the olfactory receptor family genes, support the existence of a chemosensory subpopulation in sensory cells located in the rostrum of adults [25].
Our results show that AmphiNaC is expressed in several epidermal cells located in the most anterior tip of the rostral region that are considered to be only primary sensory neurons, and in axons of the rostral nerves. Interestingly, the sensory pathways reported in vitally stained amphioxus by Holland and Yu [18] show that the rostrum sensory cells send axons via the first rostral nerves into the ventral part of the cerebral vesicle. Therefore, such data support the involvement of AmphiNaC in some integrative sensory pathways. The rostrum of young larvae seems to be very sensitive to touch and that is in accord with the presence of primary sensory neurons at the rostral tip [26]. On the other hand, several genetic, molecular, and electrophysiological studies have suggested that several DEG/NaC members are implicated in mechanotransduction and neurotransmission in nematodes, flies and mammals [27, 28]. For instance, mutations of degenerins in C. elegans lead to a disruption of touch sensation and/or coordinated activities [29, 30]. The central component of the mechanotransduction apparatus in C. elegans is the putative mechanosensitive ion channel that includes degenerins MEC-4 and MEC-10 subunits in touch neurons, and UNC-8 and DEL-1 subunits in motoneurons [27, 30, 31]. Furthermore, studies have suggested that some ENaC subunits in the skin may be components of a mechanosensory receptor for touch [32, 33], as well as some ASIC genes may be involved in mechanosensation [34]. The ASICs gene subfamily is expressed in CNS and PNS, and has a conserved role in the processing of sensory information in vertebrates [8, 35]. In particular in zebrafish, ASICs genes are expressed in the trigeminal ganglion, the posterior and anterior lateral line ganglia innervating trunk and head neuromasts, the otic sensory neurons, and some nuclei of midbrain and hindbrain involved in integration of sensory inputs [8]. Interestingly, AmphiNaC is expressed in several types of PNS neurons: namely in some flanks epidermal cells of early larvae and in several sensory neurons located mainly in the rostrum and in the mouth of late larvae.
About the expression of AmphiNaC in the CNS, we have found transcripts in elements of the infundibular organ and in different types of neurons in the nerve cord. In the latter, two large paired neurons expressing AmphiNaC, could correspond to the interneurons of the PMC [36]. The PMC, a locomotory control system, is located just behind the cerebral vesicle and contains a variety of neurons such as somatic motoneurons and three pairs of large symmetrical interneurons (LPNs). At the same time, AmphiNaC is also found in mediolateral neurons of the nerve cord just behind the first pigment spot. A similar expression has been described for amphioxus islet gene and it has been suggested that such islet-expressing cells could correspond to sensory neurons by analogy with zebrafish islet genes that label sensory Rohon-Beard neurons in the dorsal neural tube [37, 38, 39].
A recent finding has shown that ASIC2 in mice is expressed in retina where it acts as a negative modulator of rod phototransduction, and that functional ASIC2 channels are important for the maintenance of retinal integrity [40]. Similarly, AmphiNaC is expressed in some cells of the anterior cerebral vesicle probably corresponding to some photoreceptor cells of the frontal eye.
In conclusion, the present paper does not provide any direct evidence that AmphiNaC is related to a particular function, first of all because the literature reported by several authors shows that channels of this large superfamily serve diverse roles, even if the expression pattern shown by AmphiNaC in different sensory cell types suggests that it might be involved in the mechanosensation and/or sensory modulatory pathways.
Acknowledgements
We thank Skip Pierce and John M. Lawrence (Department of Biology, USF, Tampa, FL) for the use of laboratory space and equipment; Ray Martinez and Marilyn Wetzel (Department of Biology, USF, Tampa, FL) for logistic support. This research was supported by MIUR (COFIN 2004 grant 2004057732)
Conflict of interest
The authors have declared that no conflict of interest exists.
References
1. Kellenberg S, Schild L. Epithelial sodium channel/Degenerin family of ion channels: A variety of functions for a shared structure. Physiol Rev. 2002 ;82:735-767
2. Waldmann R, Lazdunski M. H+-gated cation channels: neuronal acid sensors in the NaC/DEG family of ion channels. Curr Opin Neurobiol. 1998 ;8:418-424
3. Garty H, Palmer L. Epithelial sodium channels: function, structure and regulation. Physiol Rev. 1997 ;77:359-396
4. Adams CM, Anderson MG, Motto DG. et al. Ripped pocket and pickpocket, novel Drosophila DEG/ENaC subunits expressed in early development and in mechanosensory neurons. J Cell Biol. 1998 ;140:143-152
5. Liu L, Leonard AS, Motto DG. et al. Contribution of Drosophila DEG/ENaC genes to salt taste. Neuron. 2003 ;39:133-146
6. Lingueglia E, Champigny G, Lazdunski M. et al. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature. 1995 ;378:730-733
7. Tavernarakis N, Driscoll M. Molecular modelling of mechanotransduction in the nematode Caenorhabditis elegans. Annu Rev Physiol. 1997 ;59:659-689
8. Paukert M, Sidi S, Russell C. A Family of Acid-sensing Ion Channels from the Zebrafish. J Biol Chem. 2004 ;279(18):18783-18791
9. Waldmann R, Bassilana F, de Weille J. et al. Molecular cloning of a non-inactivating proton-gated Na+ channel specific for sensory neurons. J Biol Chem. 1997 ;272(34):20975-8
10. Waldmann R, Lazdunski M. H(+)-gated cation channels: neuronal acid sensors in the NaC/DEG family of ion channels. Curr Opin Neurobiol. 1998 ;8:418-24
11. Sakai H, Lingueglia E, Champigny G. et al. Cloning and functional expression of a novel degenerin-like Na+ channel gene in mammals. J Physiol. 1999 ;519(2):323-33
12. Lacalli TC, Gilmour THJ, Kelly SJ. The oral nerve plexus in amphioxus larvae: function, cell types and phylogenetic significance. Proc Roy Soc Lond B. 1999 ;266:1461-1470
13. Lacalli TC. Frontal eye circuitry, rostral sensory pathways and brain organization in amphioxus larvae: evidence from 3D reconstructions. Philosophical Transactions of the Royal Society of London B. 1996 ;351:243-263
14. Mazet F, Masood S, Luke GN. et al. Expression of AmphiCoe, an amphioxus COE/EBF gene, in the developing central nervous system and epidermal sensory neurons. Genesis. 2004 ;38:58-65
15. Satoh G, Wang Y, Zhang P. et al. Early development of amphioxus nervous system with special reference to segmental cell organization and putative sensory cell precursors: a study based on the expression of pan-neuronal marker gene Hu/elav. J Exp Zool. 2001 ;291:354-364
16. Stokes MD, Holland ND. Embryos and larvae of a lancelet, Branchiostoma floridae, from hatching through metamorphosis: growth in the laboratory and external morphology. Acta Zool. 1995 ;76:105-120
17. Lacalli TC, Hou SF. A reexamination of the epithelial sensory cells of amphioxus (Branchiostoma). Acta Zool. 1999 ;80:125-134
18. Holland ND, Yu JK. Epidermal receptor development and sensory pathways in vitally stained amphioxus (Branchiostoma floridae). Acta Zool. 2002 ;83:309-319
19. Lacalli TC. Sensory systems in amphioxus: a window on the ancestral chordate condition. Brain Behav Evol. 2004 ;64:148-62
20. Bone Q. A note on the innervation of the integument in amphioxus, and its bearing on the mechanism of cutaneous sensibility. Q J Microscopi Sci. 1960 ;101:371-379
21. Lacalli TC. Sensory pathways in amphioxus larvae I. Constituent fibres of the rostral and anterodorsal nerves, their targets and evolutionary significance. Acta Zool. 2002 ;83:149-166
22. Schulte E, Riehl R. Elektronenmicroskopische Untersuchungen an den Oralcirren und der Haut von Branchiostoma lanceolatum. Helgoländer Wissenschaftliche Meeresuntersuchungen. 1977 ;29:337-357
23. Bone Q, Best ACG. Ciliated sensory cells in amphioxus (Branchiostoma). J Mar Biol Assoc UK. 1978 ;58:479-486
24. Baatrup E. Primary sensory cells in the skin of amphioxus Branchiostoma lanceolatum. Acta Zool. 1981 ;62:147-157
25. Satoh G. Characterization of novel GPCR gene coding locus in amphioxus genome: gene structure, expression, and phylogenetic analysis with implications for its involvement in chemoreception. Genesis. 2005 ;41:47-57
26. Stokes MD. Larval locomotion of the lancelet, Branchiostoma floridae. J Exp Biol. 1997 ;200:1661-1680
27. Tavernarakis N, Driscoll M. Molecular modelling of mechanotransduction in the nematode Caenorhabditis elegans. Annu Rev Physiol. 1997 ;59:659-689
28. Waldmann R, Lazdunski M. H(+)-gated cation channels: neuronal acid sensors in the NaC/DEG family of ion channels. Curr Opin Neurobiol. 1998 ;8:418-24
29. Huang M, Chalfie M. Gene interactions affecting mechanosensory transduction in Caenorhabditis elegans. Nature. 1994 ;367:467- 470
30. Tavernarakis N, Shreffler W, Wang S, Driscoll M. unc-8, a DEG/ENaC family member, encodes a subunit of a candidate mechanically gated channel that modulates C. elegans locomotion. Neuron. 1997 ;18:107-119
31. Tavernarakis N, Driscoll M. Degenerins at the core of the metazoan mechanotransducer? Ann NY Acad Sci. 2001 ;940:28-41
32. Awayda MS, Ismailov II, Berdiev BK. et al. A cloned renal epithelial Na+ channel protein displays stretch activation in planar lipid bilayers. Am J Physiol. 1995 ;268:C1450-9
33. Drummond HA, Abboud FM, Welsh MJ. Localization of beta and gamma subunits of ENaC in sensory nerve endings in the rat foot pad. Brain Res. 2000 ;884:1-12
34. Price MP, Lewin GR, McIlwrath SL. et al. The mammalian sodium channel BNC1 is required for normal touch sensation. Nature. 2000 ;407:1007-1011
35. Chen CC, England S, Akopian AN. et al. A sensory neuron-specific, proton-gated ion channel. Proc Natl Acad Sci USA. 1998 ;95:10240-10245
36. Lacalli TC, Kelly SJ. Ventral neurons in the anterior nerve cord of amphioxus larvae. I. An inventory of cell types and synaptic patterns. J Morphol. 2003 ;257:190-211
37. Jackman WR, Langeland JA, Kimmel CB. Islet reveals segmentation in the amphioxus hindbrain homolog. Dev Biol. 2000 ;220:16-26
38. Jackman WR, Kimmel CB. Coincident iterated gene expression in the amphioxus neural tube. Evol Dev. 2002 ;4:366-74
39. Bardet PL, Schubert M, Horard B. et al. Expression of estrogen-receptor related receptors in amphioxus and zebrafish: implications for the evolution of posterior brain segmentation at the invertebrate-to-vertebrate transition. Evol Dev. 2005 ;7:223-33
40. Ettaiche M, Guy N, Hofman P. et al. Acid-Sensing Ion Channel 2 Is Important for Retinal Function and Protects against Light-Induced Retinal Degeneration. J Neurosci. 2004 ;24(5):1005-1012
Author contact
![]() Corresponding address:
Corresponding address:

 Global reach, higher impact
Global reach, higher impact