Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2006; 2(3):110-116. doi:10.7150/ijbs.2.110 This issue Cite
Research Paper
An amphioxus LIM-homeobox gene, AmphiLim1/5, expressed early in the invaginating organizer region and later in differentiating cells of the kidney and central nervous system
1 Department of Biology, Kalamazoo College, Kalamazoo, Michigan 49006, U.S.A.
2 Scripps Institution of Oceanography, University of California at San Diego, La Jolla, California 92093-0202, U.S.A.
Received 2006-1-26; Accepted 2006-4-1; Published 2006-5-5
Abstract
A LIM-homeobox gene, AmphiLim1/5, from the Florida amphioxus (Branchiostoma floridae) encodes a protein that phylogenetic analysis positions at the base of a clade comprising vertebrate Lim1 and Lim5. Amphioxus AmphiLim1/5 is expressed in domains that are a composite of those of vertebrate Lim1 and Lim5, which evidently underwent subfunctionalization after duplication of an ancestral protochordate Lim1/5. During amphioxus development, transcription is first detected in the ectoderm of the blastula. Then, in the gastrula, a second expression domain appears in the mesendoderm just within the dorsal lip of the blastopore, a region known to have organizer properties in amphioxus. This mesendodermal expression corresponds to Lim1 expression in the Spemann organizer of vertebrates. At least one of the functions of vertebrate Lim1 in the organizer is to control the transcription of genes involved in cell and tissue movements during gastrulation, and a comparable early function seems likely for AmphiLim1/5 during gastrular invagination of amphioxus. Later embryos and larvae of amphioxus express AmphiLim1/5 in clusters of cells, probably motoneurons, in the anterior part of the central nervous system, in the hindgut, in Hatschek's right diverticulum (a rudiment of the rostral coelom), and in the wall of the first somite on the left side (a precursor of Hatschek's nephridium). In the early larva, expression continues in neural cells, in Hatschek's nephridium, in the wall of the rostral coelom, in the epidermis of the upper lip, and in mesoderm cells near the opening of the second gill slit. The developmental expression in Hatschek's nephridium is especially interesting because it helps support the homology between this amphioxus organ and the vertebrate pronephros.
Keywords: Spemann organizer, kidney, brain, cephalochordate, lancelet
1. Introduction
The cephalochordate amphioxus is widely regarded as the best available proxy for the protochordate ancestor of the vertebrates. Both the anatomy and the genetics of amphioxus are vertebrate-like, but simpler. For example, common anatomical features shared by amphioxus and basal vertebrates include a perforate pharynx, a notochord, and a dorsal nerve cord. Moreover, the amphioxus genome includes representatives of most vertebrate gene families, but usually with comparatively few genes per family. In sum, the morphology, genetics, and developmental mechanisms characterizing amphioxus can provide insights into the likely starting conditions as the vertebrates began to evolve from their protochordate ancestor.
In amphioxus, one important gene family that has not yet been widely studied encodes LIM-homeodomain proteins, which comprise two tandemly repeated LIM domains, a homeodomain, and a C-terminal transactivation domain. The LIM domains include characteristically spaced cysteine residues binding zinc ions to form zinc-finger like structures that function for protein-protein interactions, although not for DNA binding (in this way, differing from the more widely spaced, DNA-binding zinc fingers of such proteins as Snail and GATA). LIM domains can combine with a variety of proteins to form multiprotein complexes functioning as adaptors, competitors, autoinhibitors or localizers, and thereby play diverse roles in combinatorial developmental regulation.
For amphioxus, previous studies of LIM-homeobox genes have been limited to the islet subfamily [1] and the Lim3 subfamily [2]. The present paper concerns amphioxus AmphiLim1/5, which encodes a protein in the Lim1/5 subfamily (also known as the LIN-11 subfamily). It is likely that amphioxus has a single Lim1/5 gene representing the ancestral protochordate condition before gene duplication during vertebrate evolution produced two separate genes—Lim1 and Lim5 (sometimes alternatively named Lhx1 and Lhx5). We find that amphioxus AmphiLim1/5, is first expressed during gastrulation just within the dorsal lip of the blastopore, a region presumably homologous with the vertebrate Spemann organizer where Lim1 genes play important functional roles. Then, later in development, AmphiLim1/5 is expressed in subsets of cells in the central nervous system and in several restricted regions of coelomic epithelia, including Hatschek's nephridium. The expression of amphioxus AmphiLim1/5 and its vertebrate Lim1 orthologs during nephrogenesis helps strengthen the homology between the amphioxus Hatschek's nephridium and the vertebrate pronephros.
2. Methods
Ripe adults of the Florida amphioxus (Branchiostoma floridae) collected in Tampa Bay were stimulated to spawn electrically, and the embryos and larvae were raised in laboratory culture. An embryonic cDNA library was constructed [3] and screened at low stringency with a zebrafish Lim1a gene fragment encoding most of the LIM domains and the homeodomain. Eleven clones encoding the same LIM-homeodomain protein were obtained. The base sequence of the cDNA was blasted against genomic sequences in the amphioxus trace archives to determine the intron positions.
For phylogenetic analysis, the deduced amino acid sequence was aligned manually with other chordate LIM class orthologs (details available on request from the corresponding author). Neighbor-Joining phylogenetic trees were constructed from the resulting alignment with Clustal X [4] and corrected for multiple substitutions. The tree was rooted on amphioxus islet, a divergent LIM-homeodomain protein. Vertebrate protein sequences were Lim1, 2, and 5 orthologs from zebrafish (NP 571291, 042286, NP 571293), Xenopus laevis (p29674, p36200, P37137), mouse (CA125420, P50481, BAE24281), human (NP 005559, AAF17292, AAI09231); invertebrate chordate sequences were our amphioxus AmphiLim1/5 (DQ399521), amphioxus islet (AAF34717), amphioxus Lim3 (BAB91364), Ciona intestinalis Lim1/5 (BAE6535), and Ciona savignyi Lim1/5 (BAB68342).
Expression of AmphILim1/5 was determined by whole-mount in situ hybridization [5] of developmental stages fixed at frequent intervals after fertilization. The full-length clone was used as the template for synthesizing the riboprobe. Fertilization envelopes were removed with pins from pre-hatching stages to facilitate penetration of reagents. After being photographed as whole mounts, the specimens were counterstained pink in 1% Ponceau S in 1% aqueous acetic acid, dehydrated in ethanol, embedded in Spurr's resin, and prepared as 3.5 μm sections.
3. Results
Predicted protein structure and phylogenetic analysis
Our longest cDNA clone was 1888 bases long and encoded a predicted protein of 464 amino acids (Fig. 1) that included the following noteworthy motifs: two LIM domains, a homeodomain, an arginine-rich domain, and a tyrosine-rich domain. In LIM-homeodomain proteins that have been studied functionally in other animals, the LIM domains are sites of protein-protein interactions, and the homeodomain binds DNA. Moreover, at least for Xenopus Lim1, the protein is negatively regulated by its arginine-rich and tyrosine-rich domains; additionally, the tyrosine-rich domain plus its flanking regions is a strong transactivator [6]. It is possible that these domains in the amphioxus protein serve comparable functions, although functional studies would be required to confirm this.
Deduced amino acid sequence of AmphiLim1/5 from the Florida amphioxus, Branchiostoma floridae, with two LIM domains (each boxed), a homeodomain (single underlined), an arginine-rich region (double underlined), and a tyrosine-rich region (bracketed). The arrowheads correspond to intron positions in the base sequence.

Phylogenetic analysis (Fig.2) reveals that our amphioxus clone encodes a protein branching just basal to vertebrate Lim1 and Lim5 orthologs, but closer to these orthologs than to any if the other LIM-homeodomain proteins in the analysis. The amphioxus protein groups very robustly with vertebrate orthologs in both these vertebrate clades, indicating it represents the ancestral protochordate condition predating gene duplication events resulting in separate Lim1 and Lim5 clades in the vertebrates. Tree reconstruction with Maximum Likelihood [7] produced a nearly identical topology (data not shown). Phylogenetic analysis with a broader array of vertebrate LIM-homeodomain proteins (including Lim2, 4, 6, 7 and 9) also demonstrated that none of these cluster within the clade consisting of the amphioxus gene plus vertebrate Lim1 and Lim5 (data not shown). We thus name our gene AmphiLim1/5 (GenBank Accession Number DQ399521) and presume that it derives from a single ancestral cephalochordate gene that duplicated into Lim1 and Lim5 genes during early vertebrate evolution.
Neighbor-Joining phylogenetic analysis of amphioxus AmphiLim1/5 protein in the context of closely related LIM-homeodomain proteins from other chordates. Tree topology with bootstrap support based on 1000 replicates is constructed with Clustal_X.

Developmental expression of AmphiLim1/5
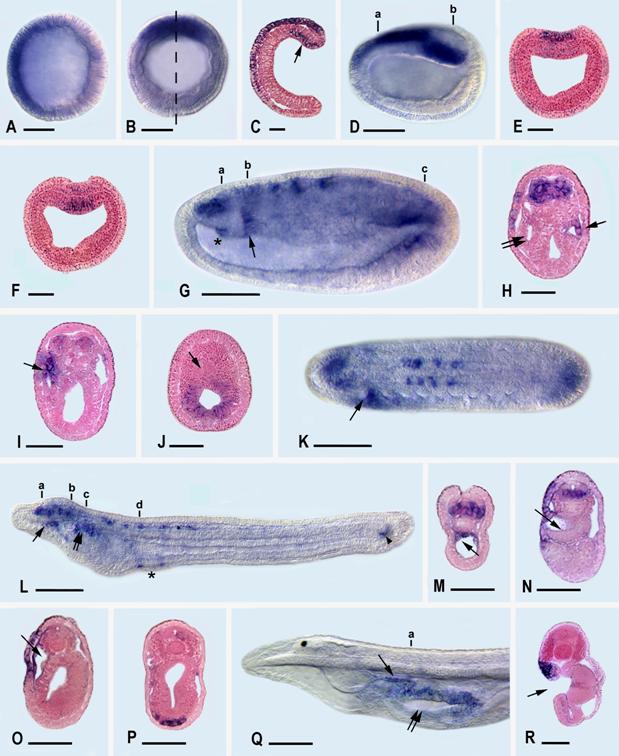
During the development of the Florida amphioxus, AmphiLim1/5 transcription is first detected at the late blastula stage (Fig. 3A) in the presumptive ectoderm cells of the animal hemisphere (recognizable because they are somewhat smaller than the presumptive mesendoderm cells of the vegetal hemisphere). By the mid gastrula stage (Fig. 3B,C), the ectodermal expression is detectable dorsally and anteriorly in the ectoderm, while a new expression domain appears the dorsal mesendoderm, just within the dorsal lip of the blastopore. At late gastrula, (Fig. 3D-F), ectodermal expression extends mid-dorsally where the anterior part of the neural plate is forming, and mesendodermal expression is still located posterodorsally.
By the mid-neurula stage (Fig. 3 G-K), AmphiLim1/5 is expressed far anteriorly in the central nervous system (the identities of these anterior nerve cells cannot readily be determined) and more posteriorly in segmentally arranged clusters of neural cells, most conspicuously at the levels of somites three through six. Most of these clustered neural cells are located ventrolaterally in the neural tube and are probably differentiating motoneurons. Expression is also detectable in a patch of epidermal cells in the left anterior region of the embryo, in the right (but not the left) diverticulum of Hatschek (Fig. 3H), and in the wall of the most anterior somite on the left side (Fig. 3 I). Posteriorly, the mesendodermal expression of the previous stage is now localized in the hindgut, but not the notochord (Fig. 3J). At this point we should add parenthetically that amphioxus embryos confusingly include a diversity of structures named after Hatschek. For instance, Hatschek's right diverticulum is destined to give rise to the rostral coelom, while the most anterior somite on the left side is destined, in part, to give rise to Hatschek's nephridium of the larva.
At the early larval stage (Fig. 3L-P), at 28 hr, just before the mouth opens, AmphiLim1/5 is transcribed segmentally in ventral and lateral cells along the anterior third of the dorsal nerve cord. Additional expression is detected in the wall of the rostral coelom (Fig. 3M), in epidermal cells along the anterior left side of the larva, in Hatschek's nephridium (Fig. 3O), in mesoderm cells in the region where the second gill slit will later penetrate the body wall (Fig. 3P), and in the chordaneural hinge of the neurenteric canal (tail bud). Later in the six-day larva (Fig. 3Q,R), when three gill slits have formed, expression is found only in Hatschek's nephridium and in the epidermal cells forming the upper lip of the mouth. At later larval stages no transcripts of AmphiLim1/5 were detectable by in situ hybridization of whole mounts.
AmphiLim1/5 expression in developing amphioxus. Whole mount side views with anterior toward left. Sections are viewed from posterior end of animal. Whole mount and section scale lines respectively 50 μm and 25 μm. A) Blastula with expression in animal hemisphere. B) Mid-gastrula in blastopore view with expression in dorsal quadrant. C) Section in plane of dashed line in B; expression in ectoderm and dorsal mesendoderm (arrow). D) Side view of late gastrula with expression in neural plate and in posterodorsal mesendoderm. E) Section through a in D showing expression in neural plate. F) Section through b in D showing expression in dorsal mesendoderm. G) Side view of mid-neurula with expression in central nervous system, hindgut, rudiment of Hatschek's right diverticulum (asterisk), and first somite on left side (arrow). H) Section through a in G showing expression in anterior neural plate, epidermis on left side, Hatschek's right diverticulum (single arrow), but not in Hatschek's left diverticulum (twin arrow). I) Section through b in G showing expression on epidermis on left side and in most anterior somite on left side (arrow). J) Section through c in G showing expression in hindgut, but not in notochord (arrow). K) Dorsal view of mid-neurula showing expression in central nervous system, hindgut, and first somite on left side (arrow). L) Side view of early larva with expression in cells of dorsal nerve cord, rostral coelom (single arrow), Hatschek's nephridium (twin arrows), mesoderm associated with rudiment of second gill slit (asterisk), and neurenteric canal (arrowhead) of tail bud. M) Section through a in L showing expression in ventral and lateral cells of central nervous system and in rostral coelom (arrow). N) Section through b in L showing expression in ventral and lateral cells of central nervous system, in epidermis on left side, but not in Hatschek's left diverticulum (arrow). O) Section through c in L showing expression in epidermis on left side and in wall of Hatschek's nephridium (arrow). P) Section through d in L with expression in mesoderm cells near rudiment of second gill slit. Q) Side view of anterior end of later larva with expression in Hatschek's nephridium (single arrow) and in epidermis bordering upper lip of mouth (twin arrows). R) Section through a in Q, at level of mouth (arrow), showing expression in upper lip epidermis and in underlying Hatschek's nephridium.
4. Discussion
LIM-homeobox genes in amphioxus
Several hundred genes have by now been described in the LIM superfamily, with representatives in slime moulds, flowering plants, invertebrates, and vertebrates. The commonest LIM genes are LIM-only genes (i.e. lacking a homeobox) that encode proteins predominantly localized either in the nucleus to influence gene expression or in the cytoplasm to influence the cytoskeleton. By contrast, LIM-homeobox genes, although first to be discovered, are in the minority within the superfamily. For example, the human genome includes about a hundred LIM-only genes, but only twelve LIM-homeobox genes, two in each of six subfamilies (which are apterous, LHX6/7, LMX, islet, Lim-3, and Lim1/5 = LIN-11). Amphioxus lags far behind vertebrates in the number of known LIM genes—to date, only a single LIM-only gene and three LIM-homeobox genes [1,2, present study] have been described. However, in the near future, when the annotated genome sequence for the Florida amphioxus becomes available, dozens of additional amphioxus LIM genes will probably come to light. It will be especially interesting to see whether amphioxus LIM-homeobox genes will be distributed among all of the six subfamilies now known for vertebrates and for Caenorhabditis. If one assumes that the genomic organization of extant amphioxus reflects that of the protochordate ancestor of the vertebrates, each LIM-homeobox subfamily in amphioxus will likely include only a single gene. This would be consistent with an increase to two genes per subfamily in vertebrates as a result of extensive gene duplication (or perhaps whole genome duplication) during chordate evolution.
LIM-homeobox genes in tunicates, hemichordates and sea urchins
In the ascidian tunicate, Ciona intestinalis, five LIM-homeobox genes have been found in the genome, one in each subfamily except for Lhx6/7 [8]. To date, the expression of tunicate LIM-homeobox genes is adequately known only for the Lim3 subfamily [9], which cannot usefully be compared with our results for a Lim1/5 subfamily gene. In an enteropneust hemichordate, the expression of a somewhat divergent Lim1/5 gene is limited to a region of developing epidermis, presumably in developing nerve cells [10]. It is likely that the hemichordate Lim1/5 is involved in nerve cell differentiation and maintenance in parallel with its homologs in chordates. Finally, sea urchins have a Lim1 gene (again, somewhat divergent) that is expressed in the vegetal plate of the early gastrula, where invagination is about to take place [11]. Thus, sea urchin Lim1 may have an early function comparable to its homologs in amphioxus and vertebrates, where Lim1 in the organizer evidently directs expression of downstream genes involved in cell and tissue movements of gastrulation (see below). No sea urchin Lim1 expression was detected in other embryonic tissues, although this may have been a false negative due to the weakness of the in situ staining.
Vertebrate Lim1 and presumably AmphiLim1/5 associated with the organizer
In vertebrates, the earliest detectable expression of Lim1 begins at the gastrula stage in the amphibian Spemann organizer or its homologs in other vertebrate classes—that is, in the dorsal blastoporal lip and dorsal mesoderm of Xenopus [12-16], in the shield of zebrafish [17,18], and in the node/anterior primitive streak/anterior visceral endoderm of mice [19-21]. For amphioxus, the region of where AmphiLim1/5 is first expressed in the mesendoderm is just within the dorsal lip of the blastopore, a tissue that has been shown experimentally to have organizer properties [22]. For vertebrate Lim1, and presumably for amphioxus Lim1/5, expression in the organizer plays a key role in regulating genes that direct gastrulation movements and/or help establish the body axes. In the organizer gene network, vertebrate Lim1 is positioned downstream from Siamois [14], nodal [17], activin [23], VegT [24], and retinoic acid signaling [25]. In turn, vertebrate Lim1 regulates goosecoid [26], cerberus [27], iroquois [28] and other genes [29], including some (like protocadherin and angiomotin) involved in cell and tissue movements during gastrulation [16,30]. The expression domains of homologs for many of these vertebrate genes have not yet been described for amphioxus, except for amphioxus goosecoid [31] and AmphiNodal [32]. Early in amphioxus development, the expression domains of the amphioxus homologs of Lim1/5, goosecoid, and nodal are congruent in the mesendoderm just within the dorsal lip of the blastopore of the gastrula, although their expression domains later begin to diverge as the neurula stage progresses.
During later embryonic development, expression of amphioxus AmphiLim1/5 and vertebrate Lim1 can be followed continuously from the region of the dorsal blastoporal lip—via notochordal expression in some vertebrates [18], but via hindgut expression in amphioxus—to the chordaneural hinge of the tail bud [present results, 33]. This pattern is consistent with the concept that the chordate tail bud, and at least part of the gene network operating there, derives from the organizer during development.
Vertebrate Lim1 and AmphiLim1/5 expressed in the central nervous system
Amphioxus AmphiLim1/5 and vertebrate Lim1 are expressed toward the anterior end of the developing central nervous system, as are LIM-homeobox genes in general. In amphioxus the neural expression domain can be traced back in development to expression in the animal hemisphere of the blastula, whereas the vertebrate neural expression domains arise de novo during the neurula stage. This difference probably reflects the combination of vertebrate Lim1 and Lim 5 functions in Amphioxus Lim1/5. In the developing vertebrate central nervous system, Lim1 transcription begins in relatively coarse patterns that apparently correspond to functional subdivisions (e.g. forebrain prosomeres) along the neuraxis [34]. During subsequent neural development in vertebrates, Lim1 transcription becomes progressively restricted to small cell clusters or even to single isolated neurons that transcribe various combinations of different LIM-homeobox genes to help establish and maintain their differentiated cell identities [13,18,35]. Presumably the neural expression of amphioxus AmphiLim1/5 in individual clusters of cells is also involved in their differentiation and maintenance as particular nerve cell types.
Vertebrate Lim1 and AmphiLim1/5 expressed in mesodermal derivatives
Mesodermal expression of Lim1 in vertebrates begins in lateral mesoderm and then becomes restricted to intermediate mesoderm. Subsequently, the mesodermal expression in the intermediate mesoderm becomes associated with the forming pronephros in zebrafish [18] and Xenopus [25] and with the forming mesonephros in mice [13]. In amphioxus, AmphiLim1/5 transcription begins in the mesoderm comprising the wall of the first somite on the left side of the embryo. During the next few hours of development, the AmphiLim1/5-expressing wall of this somite evaginates as a posteriorly directed tube, known as Hatschek's nephridium. The correspondence in LIM gene expression between the pronephros of basal vertebrates and Hatschek's nephridium of amphioxus helps support the homology between these organs that was originally suggested on morphological grounds. There are two additional mesodermal structures expressing AmphiLim1/5 in amphioxus that appear to have no counterparts in vertebrate embryos: one is the lining of the rostral coelom and the other is a cluster of mesoderm cells near the opening of the second gill slit.
Vertebrate Lim5 and amphioxus AmphiLim1/5 are expressed early in the ectoderm and later in the central nervous system
Just as for amphioxus AmphiLim1/5, Xenopus and zebrafish Lim5 [36]—although not mouse Lim5 [37]—are initially expressed widely, but transiently, in the ectoderm at the blastula stage. At the blastula stage, at least one function of Xenopus Lim5 is evidently the regulation of cell-cell adhesion in the ectoderm [38]. As vertebrate development proceeds, expression of vertebrate Lim5 genes rapidly becomes restricted to the anterior neural plate and subsequently to scattered regions of the brain and anterior spinal cord, again just as for AmphiLim1/5. During the differentiation of the central nervous system, mouse Lim5 is essential for regulating proliferation of nerve cell precursors and their subsequent differentiation and migration in the hippocampus [39].
Likely origin of vertebrate Lim1 and Lim-5 by subfunctionalization following duplication of a single Lim1/5 gene in a protochordate ancestor
When the developmental expression domains of vertebrate Lim1 and Lim5—well studied for Xenopus, zebrafish, and mice [12-23,36-39]—are considered in the aggregate, the overall pattern is closely comparable to the developmental expression of amphioxus AmphiLim1/5. This is to be expected from the general model for gene duplication-degeneration-complementation [40]: amphioxus AmphiLim1/5 is probably similar to an ancestral protochordate gene that split into vertebrate Lim1 and Lim5 genes, each of which underwent subsequent subfunctionalization. Thus, the ectodermal function of the ancestral gene was retained in vertebrate Lim5, while the functions related to the organizer, tail bud, and nephrogenic mesoderm were retained in vertebrate Lim1 genes. Moreover, vertebrate Lim1 and Lim5 are known to specify at least partially different subsets of nerve cells in the central nervous system [13,18,25,34,36,37,39], and it is possible that future, detailed study of the AmphiLim1/5-expressing nerve cells in amphioxus will show that they encompass representatives of both subsets of vertebrate neurons.
Acknowledgements
We are indebted to Dr. John M. Lawrence for providing us with laboratory space at the University of South Florida. This research was supported by National Science Foundation research grants NSF 0110540 to JAL and IBN-04-16292 to NDH and LZH.
Confict of interest
The authors have declared that no conflict of interest exists.
References
1. Jackman WR, Langeland JA, Kimmel CB. islet reveals segmentation in the amphioxus hindbrain homolog. Dev Biol. 2000 ;220:16-26
2. Wang Y, Zhang PJ, Yasui K, Saiga H. Expression of Bblhx3, a LIM-homeobox gene, in the development of amphioxus Branchiostoma belcheri tsingtauense. Mech Dev. 2002 ;117:315-319
3. Langeland JA, Tomsa JA, Jackman WR, Kimmel CB. An amphioxus snail gene: expression in paraxial mesoderm and neural plate suggests a conserved role in patterning the embryo. Dev Genes Evol. 1998 ;208:569-577
4. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997 ;25:4876-4882
5. Holland LZ, Holland PWH, Holland ND. Revealing homologies between body parts of distantly related animals by in situ hybridization between body parts of distantly related animals by in situ hybridization to developmental genes: amphioxus versus vertebrates. In: (ed.) Ferraris JD, Palumbi SR. Molecular Zoology, Advances, Strategies and Protocols. New York, Wiley. 1996:267-282
6. Hiratani I, Mochizuki T, Tochimoto N, Taaira M. Functional domains of the LIM homeodomain protein Xlim-1 involved in negative regulation, transactivation, and axis formation in Xenopus embryos. Dev Biol. 2001 ;229:456-467
7. Schmidt HA Strimmer K, Vigron M, von Haeseler A. TREE-PUZZLE maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002 ;18:502-504
8. Dehal P. et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002 ;298:2157-2167
9. Satou Y, Imai KS, Satoh N. Early embryonic expression of a LIM-homeobox gene CS-lhx3 is downstream of β-catenin and responsible for endoderm differentiation in Ciona savignyi embryos. Development. 2001 ;128:3559-3570
10. Lowe CJ, Wu M, Salic A, Evans L, Lander E, Stange-Thomann N, Gruber CE, Gerhart J, Kirschner M. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003 ;113:853-865
11. Kawasaki T, Mitsunaga-Nakatsubo K, Takeda K, Akasaka K, Shimada H. Lim1 related homeobox gene (HPLim1) expressed in sea urchin embryos. Dev Growth Differen. 1999 ;41:273-282
12. Taira M, Jamrich M, Good PJ, Dawid IB. The LIM domain-containing homeobox gene Xlim1 is expressed specifically in the organizer region of Xenopus gastrula embryos. Genes Dev. 1992 ;6:356-366
13. Barnes JD, Crosby JL, Jones CM, Wright CVE, Hogan BLM. Embryonic expression of Lim-1, the mouse homolog of Xenopus XLim-1, suggests a role in lateral mesoderm differentiation and neurogenesis. Dev Biol. 1994 ;161:168-178
14. Kodjabachian L, Karavanov AA, Hikasa H, Hukriede NA, Aoki T, Taira M, Dawid IB. A study of Xlim1 function in the Spemann-Mangold organizer. Int J Dev Biol. 2001 ;45:209-218
15. Xanthos JB, Kofron M, Tao QH, Schaible K, Wylie C, Heasman J. The roles of three signaling pathways in the formation and function of the Spemann Organizer. Development. 2002 ;129:4027-4043
16. Hukriede NA, Tsang TE, Habas R, Weeks DL, Tam PPL, Dawid IB. Conserved requirement of Lim1 function for cell movements during gastrulation. Dev Cell. 2003 ;4:83-94
17. Toyama R, O'Connell ML, Wright CVE, Kuehn MR, Dawid IB. nodal induces ectopic goosecoid and lim1 expression and axis duplication in zebrafish. Development. 1995 ;121:383-391
18. Toyama R, Dawid IB. lim6, a novel LIM homeobox gene in the zebrafish: comparison of its expression pattern with lim1. Dev Dynam. 1997 ;209:406-417
19. Shawlot W, Behringer RR. Requirement for Lim1 in head-organizer function. Nature. 1995 ;374:425-430
20. Perea-Gómez A, Shawlot W, Sasaki H, Behringer RR, Ang SL. HNF3β and Lim1 interact in the visceral endoderm to regulate primitive streak formation and anterior-posterior polarity in the mouse embryo. Development. 1999 ;126:4499-4511
21. Nishioka N, Nagano S, Nakayama R, Kiyonari H, Ijiri T, Taniguchi K, Shawlot W, Hayashizaki Y, Westphal H, Behringer RR, Matsuda Y, Sakoda S, Kondoh H, Sasaki H. Ssdp1 regulates head morphogenesis of mouse embryos by activating the Lim1-Ldb1 complex. Development. 2005 ;132:2535-2546
22. Tung TC, WU SC, Tung YYF. Experimental studies on the neural induction in amphioxus. Sci Sinica. 1962 ;11:805-820
23. Rebbert ML, Dawid IB. Transcriptional regulation of the Xlim-1 gene by activin is mediated by an element in intron I. Proc Natl Acad Sci USA. 1997 ;94:9717-9722
24. Xanthos JB, Kofron M, Tao Q, Schaible K, Wylie C, Heasman J. The roles of three signaling pathways in the formation and function of the Spemann organizer. Development. 2002 ;129:4027-4043
25. Taira M, Otani H, Jamrich M, Dawid IB. Expression of the LIM class homeobox gene Xlim-1 in pronephros and CNS cell lineages of Xenopus embryos is affected by retinoic acid and exogastrulation. Development. 1994 ;120:1525-1536
26. Mochizuki T, Karavanov AA, Curtiss PE, Ault KT, Sugimono N, Watabe T, Shiokawa K, Jamrich M, Cho KW, Dawid IB, Taira M. Xlim-1 and LIM binding protein 1 cooperate with various transcription factors in the regulation of the goosecoid promoter. Dev Biol. 2000 ;224:470-485
27. Yamamoto S, Hisaka H, Ono H, Taira M. Molecular link in the sequential induction of the Spemann organizer: direct activation of the cerberus gene by Xlim-1, Xnot2, Mix. 1, and Siamois, immediately downstream from Nodal and Wnt signaling, Dev Biol. 2003 ;257:190-204
28. Kudoh T, Dawid IB. Role of the iroquois3 homeobox gene in organizer formation. Proc Natl Acad Sci USA. 2001 ;98:7852-7857
29. Shimono A, Behringer RR. Isolation of novel cDNAs by subtractions between the anterior mesoderm of single mouse gastrula stage embryos. Dev Biol. 1999 ;209:369-380
30. Tam PP, Khoo PL, Wong N, Tsang TE, Behringer RR. Regionalization of cell fates and cell movements in the endoderm of the mouse gastrula and the impact of loss of Lhx1(Lim1) function. Dev Biol. 2004 ;274:171-187
31. Neidert AH, Panopoulou G, Langeland JA. Amphioxus goosecoid and the evolution of the head organizer and predhordal plate. Evol Dev. 2000 ;2:303-310
32. Yu JK, Holland LZ, Holland ND. An amphioxus nodal gene (AmphiNodal) with early summetrical expression in the organizer and mesoderm and later asymmetrical expression associated with left-right axis formation. Evol Dev. 2002 ;4:418-425
33. Beck CW, Slack JM. Analysis of the developing Xenopus tail bud reveals separate phases of gene expression during determination and outgrowth. Mech Dev. 1998 ;72:41-52
34. Moreno N, Bachy I, Rétaux S, González A. LIM-homeodomain genes as developmental and adult genetic markers of Xenopus forebrain functional subdivisions. J Comp Neurol. 2004 ;472:52-72
35. Appel B, Korzh V, Glasgow E, Thor S, Edlund T, Dawid IB, Eisen JS. Motoneuron fate specification revealed by patterned LIM homeobox expression in embryonic zebrafish. Development. 1995 ;121:4117-4125
36. Toyama R, Curtiss PE, Otani H, Kimura M, Dawid IB, Taira M. The LIM class homeobox gene lim5: implied role in CNS patterning in Xenopus and zebrafish. Dev Biol. 1995 ;170:583-593
37. Sheng HZ, Bertuzzi S, Chiang C, Shawlot W, Taira M, Dawid I, Westphal H. Expression of murine Lhx5 suggests a role in specifying the forebrain. Dev Dynam. 1997 ;208:266-277
38. Houston DW, Wylie C. The Xenopus LIM-homeodomain protein Xlim5 regulates the differential adhesion properties of early ectoderm cells. Development. 2003 ;130:2695-2704
39. Zhao YG, Sheng HZ, Amini R, Grindberg A, Lee E, Huang SP, Taira M, Westphal H. Control of hippocampal morphognesis and neuronal differentiation by the LIM homeobox gene Lhx5. Science. 1999 ;284:1155-1158
40. Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999 ;151:1531-1545
Author contact
![]() Corresponding address:
Corresponding address:

 Global reach, higher impact
Global reach, higher impact