Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2007; 3(2):85-90. doi:10.7150/ijbs.3.85 This issue Cite
Research Paper
A complete mitochondrial genome sequence of Asian black bear Sichuan subspecies (Ursus thibetanus mupinensis)
1. College of Life Science, China West Normal University, Nanchong 637002, China
2. Sichuan Agricultural University, Yaan 625014, China
3. Nanchong Agricultural Science Research Institute, Nanchong 637000, China
4. Sichuan Province Traditional Chinese Medicinal Materials Company Dujiangyan Raising Deer Field, Dujiangyan 611800, China
Received 2006-10-29; Accepted 2006-12-12; Published 2006-12-23
Abstract
We obtained the complete mitochondrial genome of U.thibetanus mupinensis by DNA sequencing based on the PCR fragments of 18 primers we designed. The results indicate that the mtDNA is 16 868 bp in size, encodes 13 protein genes, 22 tRNA genes, and 2 rRNA genes, with an overall H-strand base composition of 31.2% A, 25.4% C, 15.5% G and 27.9% T. The sequence of the control region (CR) located between tRNA-Pro and tRNA-Phe is 1422 bp in size, consists of 8.43% of the whole genome, GC content is 51.9% and has a 6bp tandem repeat and two 10bp tandem repeats identified by using the Tandem Repeats Finder. U. thibetanus mupinensis mitochondrial genome shares high similarity with those of three other Ursidae: U. americanus (91.46%), U. arctos (89.25%) and U. maritimus (87.66%).
Keywords: Ursus thibetanus mupinensis, mitochondrial genomes, sequencing, sequence analysis
1. Introduction
Mitochondria are vital subcellular organelles, responsible for the oxidizing reaction of the tricarboxylic acid cycle, the electron transfer and the energy metabolism in cells and have an independent genetic material called the mitochondria genome (mtDNA). Mammalian mtGenomes exist as closed circular strands and have a set of 13 protein-coding genes (NADH-ubiquinone oxidoreductase chain 1, 2, 3, 4L, 4, 5, 6, cytochrome c oxidase subunit 1, 2 and 3, ATP synthase F0 subunit 8 and ATP synthase F0 subunit 8), two rRNA genes (12s RNA and 16s RNA), and a full set of 22 tRNAgenes. The two strands that make up the genome are commonly known as the heavy strand (H-strand) and the light strand (L-strand) because of molecular weight differences as a result of different base compositions. Of the 13 protein-coding genes, 12 are on the H-strand and only one is on the L-strand. Noncoding regions are mainly limited to areas called the D-loop, thought to have functional roles in replication and transcription, and origin of replication of the L-strand (OL, thought to have a functional role in replication) [1]. The gene order is also highly conserved among most vertebrates [2]. During the last decades, mitochondrial genome sequence and gene arrangement comparisons were employed as powerful new tools for resolving ancient phylogenetic relationships [3, 4-8].
The black bear (Ursus thibetanus mupinensis) is listed in “The Convention on International Trade in Endangered Species of Wild Fauna and Flora “(CITES) the appendix I species, the national 2 levels of key protections wild animals and “China Red Data Book of Endangered Animals” V species [9]. There have been researches for mitochondria genome of Ursidae. Delisle and Strobeck obtained mitochondrial genome sequences of the Americas black bear (Ursus americanus), polar bear (Ursus maritimus) and brown bear (Ursus arctos) in 2002 [10]. Despite some studies of Asian black bear mitochondria individual genes such as the Cytb gene, there are no reports about the mitochondrial genome of the black bear, which live within the boundaries of Sichuan. In the current study, we report the complete mitochondrial genome sequence from a single Asian black bear Sichuan subspecies Ursus thibetanus mupinensis.
2. Materials and Methods
Skeletal muscle tissue samples of the Asian black bear Sichuan subspecies (Ursus thibetanus mupinensis) were obtained from black bears collected from Sichuan Province Traditional Chinese Medicinal Materials Company Dujiangyan Raising Deer Field.
DNA Extraction
Tissue sample was preserved in 70% ethanol until analysis. Total DNA was extracted from approximately 3×3×3 mm of the tissue using the conventional proteinase K/phenol/chloroform method [11] with some modifications as described by Masuda and Yoshida [12].
Primers Design, Polymerase Chain Reaction, Cloning of PCR Products and Sequencing
A series of primers were designed (Table 1), based on conserved regions identified from an alignment of published complete mitochondrial genomes from carnivores by Primer Premier 5.0: Ursus americanus (American black bear) (GenBank accession No. AF303109), Ursus maritimus (polar bear) (GenBank accession No. AF303111) and Ursus arctos (brown bear) (GenBank accession No. AF303110) [10]. PCR was using rTaq DNA polymerase (TAKARA) with 1–10μl DNA extracts in a total volume of 50μl.
According to PCR reaction condition exists difference because G, C and A, T basis Content and the base number of primers is different, we lead primers to be divided into 4 sets, the reaction condition of the same of set consistent. And established temperature steps a degree circulation in each set. It set circumstance with the reaction condition for corresponding describe respectively in table 2.
Primers designed with Primer Premier 5.0 for amplifying the complete mitochondrial genome of U.thibetanus mupinensis
| No. | Forward Primer | Reverse Primer | Product length (bp) | Forward Primer Sequence (5'-3') | Reverse Primer Sequence (5'-3') |
|---|---|---|---|---|---|
| 1 | 1-25 | 1072-1095 | 1095 | GATCACACATAACTGTGGTGTCATG | CGGAGACTTACATGTGTAATCTTG |
| 2 | 1036-1059 | 3622-3646 | 2611 | TAAAGGTTTGGTCCTAGCCTTCCC | AAGCCCTGTCTCTTGGGCAGTATTG |
| 3 | 2857-2882 | 3824-3849 | 993 | AGATTAAAAGAAGTAAAAGGAACTCG | GGCCCTACAATGTTTGGTCCTTTACG |
| 4 | 3725-3752 | 4657-4681 | 957 | ATGTTTATAATTAACACTATCTCACTAG | TTATATTTGGGGGGGAATGCTTGCT |
| 5 | 4181-4205 | 5207-5230 | 1050 | GTCCTACTAATGAATGGCTCATTCG | CAGAAGTGGAAGGGGGATAGGCCG |
| 6 | 4852-4878 | 5928-5953 | 1101 | ATACCCCGAAAATGTTGGTTTATCCCC | GGTCTTTTTAGCCTAAATCTCTAGTC |
| 7 | 5560-5583 | 6487-6514 | 955 | CCACAACAACACTATCACTGTCCC | GATTATCACGAATGCATGGGCAGTTACG |
| 8 | 6286-6310 | 7834-7854 | 1568 | CAGTCTAGTGCTTTTATCAGCCATT | AGTATAACGTAAGCGGGTTCT |
| 9 | 7202-7224 | 8188-8209 | 1008 | GGTATAGATGTCGACACACGAGC | GCCGGCAAGATGGTTCATACCG |
| 10 | 7958-7981 | 8728-8748 | 791 | CTTTGTCAGGGTTAAATTATAGGT | GGAGAAGTCTGCATTCTCAGT |
| 11 | 8671-8694 | 9588-9612 | 942 | CCTCAATACTATAAAATCATTGAG | GTTTGGTGAGTCATTAGGTGTTATC |
| 12 | 9588-9610 | 10434-10457 | 870 | GATAACACCTAATGACTCACCAA | GTTGATTGTTTCTTTCTGGATTAC |
| 13 | 10405-10427 | 11150-11178 | 774 | CAATTGACTTCCAATCAATTAGC | ATTTTTAGCATTGTAAGAG |
| 14 | 11147-11172 | 12522-12548 | 1402 | AATCTCTTACAATGCTAAAAATTATC | AAACTATATTTACAGTAAATGGGCCCC |
| 15 | 12196-12215 | 12947-12971 | 776 | GGTCTACAAACACTCCTTCC | AGCTTAGAGTTAGCTTTAGGGTTTG |
| 16 | 12703-12722 | 15134-15158 | 2456 | CAAAAAATTGGTGCAACTCC | GTTTTTCGGATGTTGGTCATTAAGG |
| 17 | 15139-15161 | 16261-16278 | 1140 | ATGACCAACATCCGAAAAACCCA | TCTTCATTTTGAGAGGTT |
| 18 | 16194-16215 | 158-183 | 858 | GCTAGCCTCCATCCTCTACTTC | CCCGTAACCATTGACTGAATAGCCCC |
Reaction condition of PCR for 18 pairs primerse (Group 1 primers to have: 2, 5, 7, 9, 18; Group 2 primers to have: 1, 3, 4, 6, 8, 10,12,15; Group 3 primers to have: 11,14,16; Group 4 primers to have: 13,17)
| Reaction condition | Group 1 | Group 2 | Group 3 | Group 4 |
|---|---|---|---|---|
| Denaturation temperature | 94℃ | 94℃ | 94℃ | 94℃ |
| Time | 5min | 5min | 5min | 5min |
| Cycleindex | 1 | 1 | 1 | 1 |
| Denaturation temperature | 94℃ | 94℃ | 94℃ | 94℃ |
| Time | 30s | 30s | 30s | 30s |
| Annealing temperature | 60-50℃ | 58-48℃ | 55-46℃ | 49-40℃ |
| Time | 30s | 30s | 30s | 30s |
| Primer extension temperature | 72℃ | 72℃ | 72℃ | 72℃ |
| Time | 90s | 90s | 90s | 90s |
| Cycleindex for each annealing temperature | 2 | 2 | 2 | 2 |
| Denaturation temperature | 94℃ | 94℃ | 94℃ | 94℃ |
| Time | 30s | 30s | 30s | 30s |
| Annealing temperature | 55℃ | 52℃ | 49℃ | 45℃ |
| Time | 30s | 30s | 30s | 30s |
| Primer extension temperature | 72℃ | 72℃ | 72℃ | 72℃ |
| Time | 90s | 90s | 90s | 90s |
| Cycleindex | 30 | 30 | 30 | 30 |
| Preservation temperature | 4℃ | 4℃ | 4℃ | 4℃ |
The PCR products were fractionated on 1.5% (W/V) agarose gel, and selected bands were purified using a gel extraction kit (Sangon, Shanghai, China). The purified PCR products were ligated into the pMD18-T vector (TakaRa) and transformed into JM109 competent cells. Bacteria were grown in LB-ampicillin agar. Cloned PCR products were sequenced by BGI Life Tech Co., Ltd.
Sequence Analysis
The gene sequences of U. thibetanus mupinensis mitochondrial genome were identified by sequence comparison with published Carnivora gene sequences and similarity analysis using DNASTAR. Start and stop codons were used to help defining the sequences of protein coding genes. Percentage of GC base component and Base skew overall were calculated to analyze the genome characters. Tandem repeat was found by Tandem Repeats Finder [13].
3. Results and Discussion
General features of U.thibetanus mupinensis mitochondrial genome
The complete mtDNA sequence has been determined for U. thibetanus mupinensis mtDNA (GenBank accession no. DQ402478). The mtDNA is 16 868 bp in size, shorter than those of U. maritimus and U. arctos, which are 17017bp and 17020bp in length, yet longer than that of U. americanus which is 16841bp in length. The size differences result from different lengths of the control region among these species. It encodes 13 protein genes, 22 tRNA genes, and 2 rRNA genes (Fig 1). Eight tRNA genes and one protein gene are located on the light strand. The overall base composition of U. thibetanus mupinensis mtDNA for the H-strand is: A, 31.2%; C, 25.4%; G, 15.5%; T, 27.9%. Guanine (G) is the rarest nucleotide and GC content is 40.9%. Nucleotide composition analysis reveals that the U. thibetanus mupinensis genome is biased towards AT rich; such an AT content is lower than that of U. americanus and higher than those of U. maritimus and U. arctos. The gene content and gene order of U. thibetanus mupinensis mtDNA is typical for vertebrates [14]. With respect to the length of intergenic spacers and overlaps, U. thibetanus mupinensis has a rather compact genome similar to the other three Ursidae (Table 3 and Table 4). The U.thibetanus mupinensis mitochondrial genome shares high similarity with those of the other three Ursidae: U. americanus (91.46%), U. arctos (89.25%) and U. maritimus (87.66%).
Vertebrate animal mitochondrial genomes deviate from a random usage of nucleotide. Saccone et al. (1999) [14] used the formula, base-skew= (A-T/A+T) or (G-C/G+C), to evaluate the degree of the base bias, and found all the values of GC-skew were negative while all the values of AT-skew were positive in amniotes [14]. The base bias overall (GC=-0.242; AT=0.056) of the U.thibetanus mupinensis is most similar to that of U. arctos (GC=-0.238; AT=0.053) (Table 4).
Components of U.thibetanus mupinensis Mitochondrial Genome
| Gene | Nucleotide number | Start codon | Stop codon | Size (bp) | aa | Strand1 |
|---|---|---|---|---|---|---|
| Control region | 1-967 | 967 | - | |||
| tRNA Phe | 968-1035 | 68 | H | |||
| 12S rRNA | 1036-2000 | 956 | H | |||
| tRNA Val | 2001-2066 | 66 | H | |||
| 16S rRNA | 2067-3646 | 1580 | H | |||
| tRNA Leu | 3648-3722 | 75 | H | |||
| NADH1 | 3725-4681 | ATG | TAA | 957 | 318 | H |
| tRNA Ile | 4680-4749 | 70 | H | |||
| tRNA Gln | 4747-4819 | 73 | L | |||
| tRNA Met | 4821-4889 | 69 | H | |||
| NADH2 | 4890-5933 | ATA | TAG | 1044 | 347 | H |
| tRNA Trp | 5932-5998 | 69 | H | |||
| tRNA Ala | 6007-6075 | 69 | L | |||
| tRNA Asn | 6076-6148 | 73 | L | |||
| Origin of L-strand replication | 6149-6181 | 35 | - | |||
| tRNA Cys | 6182-6248 | 67 | L | |||
| tRNA Tyr | 6248-6315 | 68 | L | |||
| COXI | 6317-7861 | ATG | TAA | 1545 | 514 | H |
| tRNA Ser | 7858-7929 | 72 | L | |||
| tRNA Asp | 7934-8000 | 67 | H | |||
| COXII | 8001-8684 | ATG | TAA | 684 | 227 | H |
| tRNA Lys | 8688-8755 | 68 | H | |||
| ATPase8 | 8758-8961 | ATG | TAA | 204 | 67 | H |
| ATPase6 | 8919-9599 | ATG | TAA | 681 | 226 | H |
| COXIII | 9599-10382 | ATG | Taa | 784 | 260 | H |
| tRNA Gly | 10383-10451 | 69 | H | |||
| NADH3 | 10452-10798 | ATC | TAa | 347 | 115 | H |
| tRNA Arg | 10799-10867 | 69 | H | |||
| NADH4L | 10868-11164 | ATG | TAA | 297 | 98 | H |
| NADH4 | 11158-12535 | ATG | Taa | 1378 | 458 | H |
| tRNA His | 12536-12604 | 69 | H | |||
| tRNA Ser | 12605-12663 | 59 | H | |||
| tRNA Leu | 12664-12733 | 70 | H | |||
| NADH5 | 12734-14562 | ATT | TAa | 1829 | 609 | H |
| NADH6 | 14538-15065 | ATG | TAA | 528 | 175 | L |
| tRNA Glu | 15066-15134 | 69 | L | |||
| Cytochrome b | 15139-16278 | ATG | AGA | 1140 | 379 | H |
| tRNA Thr | 16279-16348 | 70 | H | |||
| tRNA Pro | 16349-16413 | 65 | L | |||
| Control region | 16414-16868 | 455 | - |
1 H=heavy strand;L=light strand
The structure and annotation of the Ursus thibetanas mupinensis mitochondrial genome. (16S: 16S ribosomal RNA gene; 12S: 12S ribosomal RNA gene; ND1: NADH dehydrogenase subunit 1 gene; ND2: NADH dehydrogenase subunit 2 gene; ND3: NADH dehydrogenase subunit 3 gene; ND4L: NADH dehydrogenase subunit 4L gene; ND4: NADH dehydrogenase subunit 4 gene; ND5: NADH dehydrogenase subunit 5 gene; ND6: NADH dehydrogenase subunit 6 gene; COX1: Cytochrome c oxidase subunit 1 gene; COX2: Cytochrome c oxidase subunit 2 gene; COX3: Cytochrome c oxidase subunit 3 gene; ATP8: ATP synthase F0 subunit 8 gene; ATP6: ATP synthase F0 subunit 6 gene; Cyt B: Cytochrome b gene; Phe: Phenylalanine tRNA gene; Val: Valine tRNA gene; Leu: Leucine tRNA gene; Ile:Isoleucine tRNA gene; Gln: Glutamine tRNA gene; Met: Trp: Tryptophan tRNA gene; Ala: Alanine tRNA gene; Tyr: Tyrosine tRNA gene; Ser: Serine tRNA gene; Asn: Asparagine tRNA gene; Cys: Cysteine tRNA gene; Asp: Aspartic acid tRNA gene; Lys: Lysine tRNA gene; Gly: Glycin tRNA gene; Arg: Arginine tRNA gene; His: Histidine tRNA gene; Glu: Glutamic acid tRNA gene; Thr: Threonine tRNA gene; Pro: Proline tRNA gene)
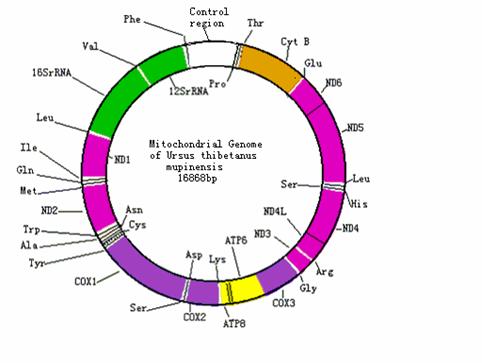
Protein coding genes
The mtDNA encodes 13 protein genes. Eight tRNA genes and one protein gene are located on the light strand [3]. We found the direction or the encoding-strand selection of the genes of U.thibetanus mupinensis is identical to the typical vertebrates (Table 3). Among 13 protein genes of U.thibetanus mupinensis, ten use ATG as start codon; NADH2, NADH3 and NADH5 use ATA, ATC and ATT as start codon, respectively. Some of these 13 protein genes are terminated with incomplete stop codons: NADH5 and NADH3 are terminated with TA; COXIII (cytochrome c oxidase subunit III) and NADH4 are terminated with T; the rest are terminated with TAA and AGA (Table 1). All these start codons and stop codons are almost the same with those of U. americanus, U. arctos and U. maritimus except that U. americanus, U. arctos and U. maritimus's ND5 use ATC as start codon, U. arctos and U. maritimus's ND4L use GTG as start codon and U. arctos and U. maritimus's ATP8 and COX2 are terminated with TAG. Presumably, these incomplete stop codons are accommodated post-transcriptionally in the mRNA maturation process, i.e. polyadenylation [15]. The 13 protein genes' nucleotide sequence and amino acid sequence of U.t. mupinensis mitochondrial genome shares high similarity with those of the other three Ursidae: U. americanus (88.7% to 93.7% and 80.6% to 100%), U. arctos (84.3% to 92.9% and 83.6% to 98.98%) and U. maritimus (84.8% to 93.4% and 82.1% to 98.98%) (Table 5).
Comparisons of mitochondrial genome features in four Ursidae
| Genome character | Black bear | American black bear | Polar bear | Brown bear |
|---|---|---|---|---|
| GC base component | 40.9% | 40.6% | 41.3% | 41.3% |
| Base skew overall | GC=-0.242 AT=0.056 | GC=-0.235 AT=0.048 | GC=-0.235 AT=0.053 | GC=-0.238 AT=0.053 |
The 37 gene's similarity comparison at the nt and aa level from between U.thibetanus mupinensis mtDNA with Ursus Americanus, Ursus maritimus and Ursus arctos
| Gene | Ursus Americanus (American black bear) | Ursus maritimus (polar bear) | Ursus arctos (brown bear) | |||
|---|---|---|---|---|---|---|
| nt | aa | nt | aa | nt | aa | |
| tRNA Phe | 89.71% | 88.24% | 88.24% | |||
| 12S rRNA | 95.85% | 95.45% | 95.13% | |||
| tRNA Val | 96.97% | 95.45% | 92.42% | |||
| 16S rRNA | 96.01% | 94.75% | 95.25% | |||
| tRNA Leu | 97.33% | 100% | 100% | |||
| NADH1 | 92% | 97% | 90% | 97% | 90% | 97% |
| tRNA Ile | 98.57% | 98.57% | 98.57% | |||
| tRNA Gln | 97.26% | 97.26% | 97.26% | |||
| tRNA Met | 98.55% | 97.10% | 100% | |||
| NADH2 | 93% | 96% | 91% | 95% | 91% | 96% |
| tRNA Trp | 92.54% | 95.52% | 95.59% | |||
| tRNA Ala | 100% | 97.10% | 97.10% | |||
| tRNA Asn | 97.26% | 97.26% | 98.63% | |||
| tRNA Cys | 100% | 100% | 100% | |||
| tRNA Tyr | 100% | 92.65% | 95.59% | |||
| COXI | 92.4% | 97.5% | 90.7% | 97.7% | 90.3% | 96.9% |
| tRNA Ser | 94.44% | 97.22% | 97.22% | |||
| tRNA Asp | 100% | 100% | 100% | |||
| COXII | 93.7% | 98.2% | 89.2% | 96.5% | 89.0% | 96.0% |
| tRNA Lys | 95.59% | 100% | 98.53%% | |||
| ATPase8 | 88.7% | 80.6% | 84.3% | 83.6% | 84.8% | 82.1% |
| ATPase6 | 90.0% | 100% | 88.8% | 94.3% | 89.1% | 94.3% |
| COXIII | 91.2% | 98.1% | 92.9% | 98.9% | 93.4% | 98.5% |
| tRNA Gly | 91.30% | 89.86% | 91.30 | |||
| NADH3 | 91% | 96% | 93% | 97% | 93% | 97% |
| tRNA Arg | 92.75% | 92.75% | 94.20% | |||
| NADH4L | 89% | 100% | 89% | 98.98% | 88% | 98.98% |
| NADH4 | 91% | 94% | 89% | 94% | 89% | 95% |
| tRNA His | 94.20% | 95.65% | 95.65% | |||
| tRNA Ser | 96.61% | 96.61% | 94.92% | |||
| tRNA Leu | 98.57% | 97.14% | 97.14% | |||
| NADH5 | 90% | 95% | 90% | 93% | 90% | 93% |
| NADH6 | 90% | 94% | 90% | 94% | 90% | 95% |
| tRNA Glu | 95.65% | 97.10% | 95.65% | |||
| Cytochrome b | 90.7% | 96.3% | 90.9% | 94.7% | 89.8% | 94.7% |
| tRNA Thr | 81.43% | 75.71% | 75.71% | |||
| tRNA Pro | 78.46% | 75.38% | 75.38% | |||
RNA genes
There are 22 tRNA genes identified in the U. thibetanus mupinensis genome, with length ranges from 59 to 75 bp. The 12S rRNA and 16S rRNA genes are 965 bp and 1 580bp in length, respectively. These are typical for mammalian mitochondrial genomes [16-19]. The 24 RNA genes of U.t. mupinensis mitochondrial genome shares high similarity with those of the other three Ursidae: U. americanus (78.46% to 100%), U. arctos (75.38% to 100%) and U. maritimus (75.38% to 100%), too (Table 5).
Control region
The stem-and-1oop structure of origin for the putative light-strand replication of U.thibetanus mupinensis located between tRNA-Asn and tRNA-Cys, is 35bp in size, which is the same size and location detected in U. americanus, U. arctos and U. maritimus. The sequence of the control region (CR) of U. thibetanus mupinensis located between tRNA-Pro and tRNA-Phe is 1422 bp in size and consists of 8.43% of the whole genome. This percentage is slightly lower than that of U. maritimus (9.25%) and U. arctos (9.27%), and higher than that of U. americanus (8.28%). Its GC content is 51.9%, which is lower than U. americanus's (52.6%) and higher than U. maritimus's (48.6%) and U. arctos's (48.2%). By using the Tandem Repeats Finder [14], we have found a 6bp tandem repeat (ACGTGT) and two 10bp tandem repeats (ACGTGTACGT and TACGTGTACG). But U. maritimus and U. arctos contain two 10bp tandem repeats (U. maritimus and U. arctos have the same two 10bp tandem repeats which are CGTACGCATA and CGCACGTGTA) and no 6bp tandem repeat, while U. americanus contains a 6bp tandem repeat (ACGTGT) and no two 10bp tandem repeats. The sequence of control region of U.thibetanus mupinensis shares 90% identity with that of U. americanus, 71% with that of U. arctos, and 68% with that of U. maritimus.
Acknowledgements
This work is supported by the Key Chinese National Natural Science Foundation (30570275), Application Technology Project in Sichuan Province (2006J13-057), Key Scientific Research Foundation of Educational Committee of Sichuan Province (2004A101) and Key Discipline Construction Project in Sichuan Province (SZD0420).
Conflict of interest
The author has declared that no conflict of interest exists.
References
1. Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 1997 ;66:409-435
2. Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999 ;27(8):1767-1780
3. Boore JL. Complete mitochondrial genome sequence of Urechis caupo, a representative of the phylum Echiura. BMC Genomics. 2004 ;5(1):67
4. Zardoya R, Meyer A. Complete mitochondrial genome suggests diapsid affinities of turtles. Proc Natl Acad Sci USA. 1998 ;95(24):14226-14231
5. Janke A, Erpenbeck D, Nilsson M. et al. The mitochondrial genomes of the iguana (Iguana iguana) and the caiman (Caiman crocodylus): Implications for amniote phylogeny. Proc. R. Soc. Lond. B Biol. Sci. 2001 ;268(1467):623-631
6. Zhou KY. Molecular phylogenetics of amphibians and reptiles. Zoological Research. 2001 ;22(5):397-405
7. Joshua SR, Jennifer CA, Christopher CA. et al. Molecular systematics of primary reptilian lineages and the tuatara mitochondrial genome. Mol. Phylogenet. Evol. 2003 ;29(2):289-297
8. Wu XB, Wang YQ, Zhou KY. et al. The complete mitochondrial genome sequence of Chinese alligator and the phylogeny of crocodiles [J]. Chinese Science Bulletin. 2003 ;48(18):1954-1958
9. Wang S. China Red Data Book of Endangered Animals: Mammalia. Beijing: Science Press. 1998
10. Delisle I, Strobeck C. Conserved primers for rapid sequencing of the complete mitochondrial genome from carnivores, applied to three species of bears. Mol. Biol. Evol. 2002 ;19(3):357-361
11. Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual 2nd edition. New York: Cold Spring Harbor Laboratory Press. 1989
12. Masuda R, Yoshida MC. A molecular phylogeny of the family Mustelidae (Mammalia, Carnivora), based on comparison of mitochondrial cytochrome b nucleotide sequences. Zool. Sci. 1994 ;11:605-612
13. Benson G. Tandem Repeats Finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999 ;27:573-580
14. Saccone S, Giorgi CD, Gissi C. Evolutionary genomics in Metazoa: The mitochondrial DNA as a model system. Gene. 1999 ;238(1):195-209
15. Ojala D. et al. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981 ;290:470-474
16. Hiendleder S. et al. The complete mitochondrial DNA sequence of the domestic sheep (Ovis aries) and comparison with the other major ovine haplotype. J. Mol. Evol. 1998 ;47:441-448
17. Anderson S. et al. Sequence and organization of the human mitochondrial genome. Nature. 1981 ;290:457-465
18. Lin CS. et al. Complete nucleotide sequence of pig (Sus scrofa) mitochondrial genome and dating evolutionary divergence within Artiodactyla. Gene. 1999 ;236:107-114
19. Parma P. et al. The complete nucleotide sequence of goat (Capra hircus) mitochondrial genome. Goat mitochondrial genome. DNA Seq. 2003 ;14:199-20
Author contact
![]() Correspondence to: Wan-ru Hou, College of Life Science, China West Normal University, 44# Yuying Road, 637002, Nanchong, China. Phone/Fax: +86-0817-2314311. E-mail: hwr168com.cn.
Correspondence to: Wan-ru Hou, College of Life Science, China West Normal University, 44# Yuying Road, 637002, Nanchong, China. Phone/Fax: +86-0817-2314311. E-mail: hwr168com.cn.

 Global reach, higher impact
Global reach, higher impact