Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2007; 3(3):179-184. doi:10.7150/ijbs.3.179 This issue Cite
Review
Advances in Swine Biomedical Model Genomics
APDL, BARC, ARS, United States Department of Agriculture, Beltsville, MD, USA
Received 2007-2-2; Accepted 2007-2-10; Published 2007-2-10
Abstract
This review is a short update on the diversity of swine biomedical models and the importance of genomics in their continued development. The swine has been used as a major mammalian model for human studies because of the similarity in size and physiology, and in organ development and disease progression. The pig model allows for deliberately timed studies, imaging of internal vessels and organs using standard human technologies, and collection of repeated peripheral samples and, at kill, detailed mucosal tissues. The ability to use pigs from the same litter, or cloned or transgenic pigs, facilitates comparative analyses and genetic mapping. The availability of numerous well defined cell lines, representing a broad range of tissues, further facilitates testing of gene expression, drug susceptibility, etc. Thus the pig is an excellent biomedical model for humans. For genomic applications it is an asset that the pig genome has high sequence and chromosome structure homology with humans. With the swine genome sequence now well advanced there are improving genetic and proteomic tools for these comparative analyses. The review will discuss some of the genomic approaches used to probe these models. The review will highlight genomic studies of melanoma and of infectious disease resistance, discussing issues to consider in designing such studies. It will end with a short discussion of the potential for genomic approaches to develop new alternatives for control of the most economically important disease of pigs, porcine reproductive and respiratory syndrome (PRRS), and the potential for applying knowledge gained with this virus for human viral infectious disease studies.
Keywords: swine, humans, genomics, gene expression, biomedical model
1. Introduction
Swine have served as an important biomedical model for humans for decades; previous authors have summarized such models in more detail [1-5]. This manuscript is a short update of the diversity of swine biomedical models. Limitations on reference citations for this review has meant that only the most recent will be cited to cover the latest developments and the broadest range of current pig models. The review will discuss some of the genomic approaches used to probe these models. Advances using these genomic tools are described in the other reviews in this issue [6, 7].
Generally speaking, studying animal models of human disease helps scientists understand the mechanisms involved in the disease pathogenesis and thus provides tools for development of gene therapy to cure the disease/condition in humans. To date, the humanized mouse has been widely used to advance our understanding of human hematopoiesis, innate and adaptive immunity, autoimmunity, infectious diseases, cancer biology and regenerative medicine [8]. Unfortunately, the humanized mouse and many mouse disease models often do not faithfully mimic the relevant human conditions. Therefore, better animal models are needed. An example is the well developed swine atherosclerosis model which has facilitated analyses of disease progression and pathogenesis and testing of new therapeutics [9, 10].
This review will probe, as detailed examples, genomic studies of melanoma and of infectious disease resistance, highlighting issues to consider in designing such genomic studies. It will end with a short discussion of the potential for genomic approaches to develop new alternatives for control of viral infectious diseases, targeting porcine reproductive and respiratory syndrome virus (PRRSV), and the potential for applying knowledge gained with this virus for human infectious disease studies.
2. Advantage of the Swine as a Biomedical Model
As outlined in Table 1 the swine has been used as a major mammalian model for human biology [11]. The similarity in size, particularly for miniature pigs, and physiology, and in organ development and disease progression make the swine an ideal model. The ability to deliberately time studies, image internal vessels and organs using standard human technologies, and collect repeated peripheral samples and, at kill, detailed tissue samples, has meant that the pig is an excellent biomedical model for humans. The ability to use pigs from the same litter, or cloned or transgenic pigs, facilitates genetic mapping. Availability of numerous well defined cell lines, representing a broad range of tissues, will facilitate detailed testing of gene expression, drug susceptibility, etc. For genomics it is an asset that the pig genome has high sequence and chromosome structure homology with humans. With the swine genome sequence now well advanced there are increasingly improving genetic and proteomic tools for pigs.
Advantages of Swine as a Biomedical Model

3. Swine Biomedical Models
Swine have served as an important biomedical model for decades. Table 2 highlights some of the broad array of biomedical topics now addressed using swine models. Some of these studies already employ genomics approaches, such as the heart, transplantation and melanoma models; others are still in the early stages of affirming swine physiological parameters and utility as a human biomedical model. Each model will be impacted by the availability of the functional genomic tools and swine genome sequence and maps outlined in other reviews in this issue [6, 7].
Some of the best examples of the impact of swine as a biomedical model are found with atherosclerosis and diabetes, diseases that are increasingly important today as the US faces major problems with obesity [9, 10]. Research is underway in pigs to determine the role of genetic background and to identify nutritional, exercise, and drug approaches which will alleviate disease progression and prevent pathology. Advances in strategies to treat myocardial infarction are being pursued with cellular myoblast repair strategies [12] and tissue engineering of blood vessels [13]. Testing of emergency room treatment options, such as directed cardiopulmonary resuscitation (CPR) and ventricular fibrillation or cardiopulmonary bypass, which are difficult to assess in humans, are readily testable with pigs. Genomics will provide in depth analytic tools to probe these pig models in detail.
All aspects of reproductive function have been studied in the pig, from the basics of maternal-fetal interactions [14], embryo development [15-17] and the impact of transgenesis [18, 19] to sperm function and quality [20, 21]. Basic sperm biology, such as chromosome positioning during spermatogenesis [22], as well as semen transmission of infectious disease [23, 24] are under active investigation (Table 2). Major transcriptional genomic and mapping efforts are underway in the pig model [14, 18] and should reveal important pathways regulating reproductive function.
Swine Biomedical Models
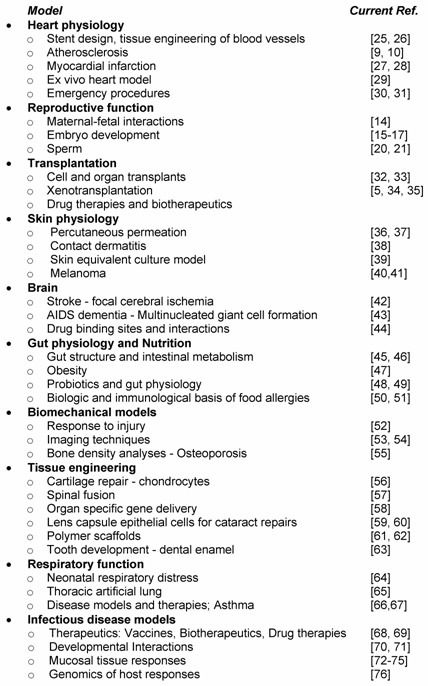
Because of the size and physiologic similarity of pigs to humans, the pig has become a model of choice for tissue engineering and imaging studies. A range of imaging techniques has been developed with pigs as an early pre-human validation model. For example, sentinel node detection is increasingly important for cancer therapeutics; the pig model has informed techniques for laparoscopic colon visualization and resection procedures [53, 54]. Tissue engineering using polymer scaffolds [61] have targeted areas as diverse as alternatives for knee meniscus cartilage and artificial bladder construction [62].
Swine skin studies have been very important, the swine melanoma model has been particularly informative. Targeted studies analyzing percutaneous permeation with different chemicals will influence international biodefense efforts as well as responses to biological toxins [77]. Gut physiology and intestinal development following probiotics have been pursued in pigs both as a means of decreasing antibiotic usage in pig feed as well as an important human model. Assessments have focused on probiotic strain selection, timing and dosing and the effects of these particularly on neonatal gut physiology.
4. Genomics and Melanoma: A case study
The swine melanoma model is a well established spontaneous melanoma model and one of the best developed for genomic approaches. The Sinclair melanoma and Melanoma-bearing Libechov Minipig (MeLiM) have been studied in detail using immune analyses, focusing on the role of tumor infiltrating lymphocytes, the potential effects of natural killer (NK) and γδ T cells in tumor regression. Imaging and sentinel lymph node (SLN) mapping will enhance these studies [54]. Comparative studies of normal melanocytes with localized tumor cells should reveal tumor specific regulatory pathways [78]; indeed laser capture microdissection studies revealed loss of the 13q36-49 chromosomal region in nodular melanoma cells [79].
Early mapping studies in the Sinclair swine melanoma model determined that a single dose of a specific “B” swine leukocyte antigen (SLA) haplotype on SSC7 was required for tumor initiator [80]; complex segregation analysis identified a second locus [81]. These were followed by more detailed QTL studies using the MeLiM model which identified numerous melanoma candidate loci [40]; interestingly human candidate genes CDK4 and BRAF were not susceptibility genes in this model. Zhi-Qiang [41] continued these studies identifying QTL for the synthetic trait, development of melanoma on SSC1, 2, 13, 15 and 17. Their detailed phenotyping of 331 pigs revealed highly significant QTLs (p < 0.001/p < 0.05 respectively, chromosome-/genome-wide levels) for precise disease traits. These included SSC10 42.0 cM for ulceration; SSC12 95.6 cM for presence of melanoma at birth; SSC13 81.0 cM for lesion type; SSC16 45.3 cM and SSC17 44.8 cM for number of aggressive melanomas; and the SSC1 MeLiM MC1R*2 allele for black coat color predisposing to melanoma. As outlined in Table 3 these studies affirmed that more exact mapping of complex traits such as tumor growth and regression are improved when very detailed phenotypic information is collected on a large group of animals.
Lessons learned from Swine Melanoma Studies

5. Swine as infectious disease models
As scientists evaluate methods to prevent infectious diseases and test new therapies and vaccines the pig is an ideal choice. Approaches include probing mucosal tissue responses in respiratory [72], reproductive [23, 24], neurological [73], and intestinal infections [74, 75], testing biotherapeutics and drug therapies, probing the effects of disease on development [70, 71] and testing therapies for specific ailments, e.g., asthma [66, 67]. Major efforts to determine the genomics of host responses using transcription and proteomic analyses are in early stages [82] Transgenic and mapping approaches [76, 83] will help to affirm specific gene and allele involvement. Table 4 outlines some of the major issues to consider as one attempts to use swine models for biomedical studies.
Utilizing genomics for Swine Models - Issues to Consider

6. Pig anti-Viral Responses: the response to Porcine Reproductive and Respiratory Syndrome virus
Regulation of immune responses and genetic resistance to infectious viral diseases is an area of concern for human and swine. Porcine Reproductive and Respiratory Syndrome (PRRS) is caused by the PRRS virus, an enveloped, single-stranded positive-sense RNA virus. When present in a herd, PRRSV causes increased abortions, stillbirths, mummies and chronic respiratory problems in pigs resulting in >$560 million losses in the US each year [84]. As an RNA virus with an evolving genome, PRRSV is particularly problematic due to slow development of protective immunity to homologous challenge and lack of protection against heterologous virus challenge. Thus it is a major target for swine research; information gained from swine studies will inform human infectious disease studies, particularly for analyses of viral persistence and of factors relevant to prevention of congenital and seminal transmission pathways.
Major efforts are underway to identify factors regulating PRRSV immunity, persistence and transmission. Tests involve probing local mucosal anti-viral responses (Petry et al., submitted). Detailed cellular analyses have assessed gene and protein expression. As with human disease studies analyses of cultured cells, e.g., infected MARC cells [85], swine macrophages [86, 87], or samples from infected pigs [88], have expanded our knowledge of the impact of timing and level of viral infection on gene expression and pathway involvement. Future work will determine whether RNAi approaches will be effective. More detailed gene expression analyses are underway using long oligo and Affymetrix arrays, testing both pooled and individual animal samples, as well as mucosal samples at death. An important issue is the timing of mucosal sample collection. Results can be disappointing if mucosal samples are collected only after viral levels begin to resolve. The peak of anti-viral immunity (and relevant gene expression) may be much earlier; however, that may be at a time when the actual viral levels may be difficult to evaluate and thus complicate comparative analyses. Therefore, it is important to affirm preliminary results with hypothesis driven repeated analyses and with translation of gene expression and array results into protein expression data.
Genetic variation does exist in resistance/susceptibility to viral infections and has been proven for swine resistance/susceptibility to PRRS [87, 89]. Although to date there is limited knowledge of host genes determining PRRSV resistance; some candidate genes have been identified (Petry et al., submitted). More detailed studies are required to determine whether naturally disease resistant pigs can be identified and why do [some] pigs stay healthy even with PRRS? What set of factors (detailed phenotype) truly correlate with lower PRRSV burden? What is the potential for sampling peripheral blood cells, serum or saliva for preinfection predictive studies of genetically determined virus resistance phenotype?
An international PRRS Genomics Consortium, of university, government, and company based scientists, has been established to assess host genetics of PRRS resistance/susceptibility. The goal is to develop a large, publicly available disease sample and dataset from thousands of pigs from relevant commercial lines infected with PRRSV and from which a detailed phenotype have been collected. Access to samples will be dependent on data sharing. The end user performs his/her analysis on the appropriate sample and returns the data to the consortium. It is hoped that the data generated by the Consortium will verify the genetic variation in pigs responding to PRRSV infection, will reveal factors influencing health, survivability and growth, and will identify the relative importance of different phenotypes, and their heritability, in predicting responses to PRRSV infection. Overall this data should enable breeders to produce healthier pigs with improved resistance to PRRSV and help animal health companies to develop improved vaccines and alternative anti-PRRSV therapeutics. This data should help identify new critical control factors in human responses to viral infections.
Conflict of interest
The author declares that no conflict of interest exists.
References
1. Information Resources on Swine in Biomedical Research 1990-2000. 2000 Swindle MM and Smith AC. www.nal.usda.gov/awic/pubs/swine/swine.htm
2. Schook L, Beattie C, Beever J, Donovan S, Jamison R, Zuckermann F, Niemi S, Rothschild M, Rutherford M, Smith D. Swine in biomedical research: creating the building blocks of animal models. Anim Biotechnol. 2005;16:183-90
3. Schook LB, Tumbleson ME. Advances in Swine in Biomedical Research. Springer Publishing Corp. 2004
4. Vodicka P, Smetana K Jr, Dvorankova B, Emerick T, Xu YZ, Ourednik J, Ourednik V, Motlik J. The miniature pig as an animal model in biomedical research. Ann N Y Acad Sci. 2005;1049:161-71
5. Ibrahim Z, Busch J, Awwad M, Wagner R, Wells K, Cooper DK. Selected physiologic compatibilities and incompatibilities between human and porcine organ systems. Xenotransplantation. 2006;13:488-99
6. Tuggle CK, Wang Y-F, Couture O. Advances in Swine Transcriptomics. Int J. Biol Sci. 2007;3:132-152
7. Rothschild MF, Hu Z-L, Jiang Z. Advances in QTL Mapping in Pigs. Int J. Biol Sci. 2007;3:192-197
8. Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nat Rev Immunol. 2007;7:118-130
9. Turk JR, Henderson KK, Vanvickle GD, Watkins J, Laughlin MH. Arterial endothelial function in a porcine model of early stage atherosclerotic vascular disease. Int J Exp Pathol. 2005;86:335-45
10. Turk JR, Laughlin MH. Physical activity and atherosclerosis: which animal model? Can J Appl Physiol. 2004;29:657-83
11. Porcine Genomic Sequencing Initiative. Rohrer et al. www.genome.gov/Pages/Research/Sequencing/SeqProposals/PorcineSEQ021203.pdf
12. Opie SR, Dib N. Surgical and catheter delivery of autologous myoblasts in patients with congestive heart failure. Nat Clin Pract Cardiovasc Med. 2006;3(Suppl 1):S42-5
13. Solan A, Niklason L. Age effects on vascular smooth muscle: an engineered tissue approach. Cell Transplant. 2005;14:481-8
14. Green JA, Kim JG, Whitworth KM, Agca C, Prather RS. The use of microarrays to define functionally-related genes that are differentially expressed in the cycling pig uterus. Reprod Suppl. 2006;62:163-76
15. Sun QY, Nagai T. Molecular mechanisms underlying pig oocyte maturation and fertilization. J Reprod Dev. 2003;49:347-59
16. Rohrer GA, Wise TH, Ford JJ. Deciphering the pig genome to understand gamete production. Reprod Suppl. 2006;62:293-301
17. Tayade C, Black GP, Fang Y, Croy BA. Differential gene expression in endometrium, endometrial lymphocytes, and trophoblasts during successful and abortive embryo implantation. J Immunol. 2006;176:148-56
18. Gerrits RJ, Lunney JK, Johnson LA, Pursel VG, Kraeling RR, Rohrer GA, Dobrinsky JR. A vision for artificial insemination and genomics to improve the global swine population. Theriogenology. 2005;63:283-299
19. Prather RS. Nuclear remodeling and nuclear reprogramming for making transgenic pigs by nuclear transfer. Adv Exp Med Biol. 2007;591:1-13
20. Strzezek J, Wysocki P, Kordan W, Kuklinska M. Proteomics of boar seminal plasma - current studies and possibility of their application in biotechnology of animal reproduction. Reprod Biol. 2005;5:279-90
21. Lavitrano M, Busnelli M, Cerrito MG, Giovannoni R, Manzini S, Vargiolu A. Sperm-mediated gene transfer. Reprod Fertil Dev. 2006;18:19-23
22. Foster HA, Abeydeera LR, Griffin DK, Bridger JM. Non-random chromosome positioning in mammalian sperm nuclei, with migration of the sex chromosomes during late spermatogenesis. J Cell Sci. 2005;118(Pt 9):1811-20
23. Wasilk A, Callahan JD, Christopher-Hennings J, Gay TA, Fang Y, Dammen M, Reos ME, Torremorell M, Polson D, Mellencamp M, Nelson E, Nelson WM. Detection of US, Lelystad, and European-like porcine reproductive and respiratory syndrome viruses and relative quantitation in boar semen and serum samples by real-time PCR. J Clin Microbiol. 2004;42:4453-61
24. Prieto C, Castro JM. Porcine reproductive and respiratory syndrome virus infection in the boar: a review. Theriogenology. 2005;63:1-16
25. Bedoya J, Meyer CA, Timmins LH, Moreno MR, Moore JE. Effects of stent design parameters on normal artery wall mechanics. J Biomech Eng. 2006;128:757-65
26. Gyongyosi M, Strehblow C, Sperker W, Hevesi A, Garamvolgyi R, Petrasi Z, Pavo N, Ferdinandy P, Csonka C, Csont T, Sylven C, Declerck PJ, Pavo I Jr, Wojta J, Glogar D, Huber K. Platelet activation and high tissue factor level predict acute stent thrombosis in pig coronary arteries: prothrombogenic response of drug-eluting or bare stent implantation within the first 24 hours. Thromb Haemost. 2006;96:202-9
27. Ambrose JA. Myocardial ischemia and infarction. J Am Coll Cardiol. 2006;47(11 Suppl):D13-7
28. Boluyt MO, Cirrincione GM, Loyd AM, Korzick DH, Parker JL, Laughlin MH. Effects of gradual coronary artery occlusion and exercise training on gene expression in swine heart. Mol Cell Biochem. 2007;294:87-96
29. Laske TG, Skadsberg ND, Iaizzo PA. A novel ex vivo heart model for the assessment of cardiac pacing systems. J Biomech Eng. 2005;127:894-8
30. Casas F, Alam H, Reeves A, Chen Z, Smith WA. A portable cardiopulmonary bypass/extracorporeal membrane oxygenation system for the induction and reversal of profound hypothermia: feasibility study in a Swine model of lethal injuries. Artif Organs. 2005;29:557-63
31. Geddes LA, Roeder RA, Rundell AE, Otlewski MP, Kemeny AE, Lottes AE. The natural biochemical changes during ventricular fibrillation with cardiopulmonary resuscitation and the onset of postdefibrillation pulseless electrical activity. Am J Emerg Med. 2006;24:577-81
32. Larsen MO, Rolin B. Use of the Gottingen minipig as a model of diabetes, with special focus on type 1 diabetes research. ILAR J. 2004;45:303-13
33. Street CN, Sipione S, Helms L, Binette T, Rajotte RV, Bleackley RC, Korbutt GS. Stem cell-based approaches to solving the problem of tissue supply for islet transplantation in type 1 diabetes. Int J Biochem Cell Biol. 2004;36:667-83
34. Cooper DK, Gollackner B, Sachs DH. Will the pig solve the transplantation backlog? Annu Rev Med. 2002;53:133-47
35. Tseng YL, Sachs DH, Cooper DK. Porcine hematopoietic progenitor cell transplantation in nonhuman primates: a review of progress. Transplantation. 2005;79:1-9
36. Simon GA, Maibach HI. The pig as an experimental animal model of percutaneous permeation in man: qualitative and quantitative observations--an overview. Skin Pharmacol Appl Skin Physiol. 2000;13:229-34
37. Dalton CH, Hattersley IJ, Rutter SJ, Chilcott RP. Absorption of the nerve agent VX (O-ethyl-S-[2(di-isopropylamino)ethyl] methyl phosphonothioate) through pig, human and guinea pig skin in vitro. Toxicol In Vitro. 2006;20:1532-6
38. Stuetz A, Baumann K, Grassberger M, Wolff K, Meingassner JG. Discovery of topical calcineurin inhibitors and pharmacological profile of pimecrolimus. Int Arch Allergy Immunol. 2006;141:199-212
39. Huang YC, Wang TW, Sun JS, Lin FH. Epidermal morphogenesis in an in-vitro model using a fibroblasts-embedded collagen scaffold. J Biomed Sci. 2006;12:855-67
40. Geffrotin C, Crechet F, Le Roy P, Le Chalony C, Leplat JJ, Iannuccelli N, Barbosa A, Renard C, Gruand J, Milan D, Horak V, Tricaud Y, Bouet S, Franck M, Frelat G, Vincent-Naulleau S. Identification of five chromosomal regions involved in predisposition to melanoma by genome-wide scan in the MeLiM swine model. Int J Cancer. 2004;110:39-50
41. Zhi-Qiang D, Silvia VN, Gilbert H, Vignoles F, Créchet F, Shimogiri T, Yasue H, Leplat JJ, Bouet S, Gruand J, Horak V, Milan D, Le Roy P, Geffrotin C. Detection of novel quantitative trait loci for cutaneous melanoma by genome-wide scan in the MeLiM swine model. Int J Cancer. 2007;120:303-20
42. Imai H, Konno K, Nakamura M, Shimizu T, Kubota C, Seki K, Honda F, Tomizawa S, Tanaka Y, Hata H, Saito N. A new model of focal cerebral ischemia in the miniature pig. J Neurosurg. 2006;104(2 Suppl):123-32
43. Tambuyzer BR, Nouwen EJ. Inhibition of microglia multinucleated giant cell formation and induction of differentiation by GM-CSF using a porcine in vitro model. Cytokine. 2005;31:270-9
44. Minuzzi L, Nomikos GG, Wade MR, Jensen SB, Olsen AK, Cumming P. Interaction between LSD and dopamine D2/3 binding sites in pig brain. Synapse. 2005;56:198-204
45. Eubanks DL, Cooper R, Boring JG. Surgical technique for long-term cecal cannulation in the Yucatan minipig (Sus scrofa domestica). J Am Assoc Lab Anim Sci. 2006;45:52-6
46. Qiu H, Xia T, Chen X, Zhao X, Gan L, Feng S, Lei T, Yang Z. Cloning, comparative characterization of porcine SCAP gene, and identification of its two splice variants. Mol Genet Genomics. 2006;276:187-96
47. Brambilla G, Cantafora A. Metabolic and cardiovascular disorders in highly inbred lines for intensive pig farming: how animal welfare evaluation could improve the basic knowledge of human obesity. Ann Ist Super Sanita. 2004;40:241-4
48. Reid G, Sanders ME, Gaskins HR, Gibson GR, Mercenier A, Rastall R, Roberfroid M, Rowland I, Cherbut C, Klaenhammer TR. New scientific paradigms for probiotics and prebiotics. J Clin Gastroenterol. 2003;37:105-18
49. Domeneghini C, Di Giancamillo A, Arrighi S, Bosi G. Gut-trophic feed additives and their effects upon the gut structure and intestinal metabolism. State of the art in the pig, and perspectives towards humans. Histol Histopathol. 2006;21:273-83
50. Bailey M, Haverson K, Inman C, Harris C, Jones P, Corfield G, Miller B, Stokes C. The development of the mucosal immune system pre- and post-weaning: balancing regulatory and effector function. Proc Nutr Soc. 2005;64:451-7
51. McClain S, Bannon GA. Animal models of food allergy: opportunities and barriers. Curr Allergy Asthma Rep. 2006;6:141-4
52. Schmitt KU, Snedeker JG. Analysis of the biomechanical response of kidneys under blunt impact. Traffic Inj Prev. 2006;7:171-81
53. Ellner SJ, Mendez J, Vera DR, Hoh CK, Ashburn WL, Wallace AM. Sentinel lymph node mapping of the colon and stomach using lymphoseek in a pig model. Ann Surg Oncol. 2004;11:674-81
54. Goldberg BB, Merton DA, Liu JB, Thakur M, Murphy GF, Needleman L, Tornes A, Forsberg F. Sentinel lymph nodes in a swine model with melanoma: contrast-enhanced lymphatic ultrasonography [US]. Radiology. 2004;230:727-34
55. Teo JC, Si-Hoe KM, Keh JE, Teoh SH. Relationship between CT intensity, micro-architecture and mechanical properties of porcine vertebral cancellous bone. Clin Biomech (Bristol, Avon). 2006;21:235-44
56. Chang CH, Kuo TF, Lin CC, Chou CH, Chen KH, Lin FH, Liu HC. Tissue engineering-based cartilage repair with allogenous chondrocytes and gelatin-chondroitin-hyaluronan tri-copolymer scaffold: A porcine model assessed at 18, 24, and 36 weeks. Biomaterials. 2006;27:1876-88
57. Drespe IH, Polzhofer GK, Turner AS, Grauer JN. Animal models for spinal fusion. Spine J. 2005;5(6 Suppl):209S-216S
58. Kawashita Y, Fujioka H, Ohtsuru A, Kaneda Y, Kamohara Y, Kawazoe Y, Yamashita S, Kanematsu T. The efficacy and safety of gene transfer into the porcine liver in vivo by HVJ (Sendai virus) liposome. Transplantation. 2005;80:1623-9
59. Lassota N, Kiilgaard JF, Prause JU, la Cour M. Correlation between clinical and histological features in a pig model of choroidal neovascularization. Graefes Arch Clin Exp Ophthalmol. 2006;244:394-8
60. van Kooten TG, Koopmans S, Terwee T, Norrby S, Hooymans JM, Busscher HJ. Development of an accommodating intra-ocular lens--in vitro prevention of re-growth of pig and rabbit lens capsule epithelial cells. Biomaterials. 2006;27:5554-60
61. Moroni L, Poort G, Van Keulen F, de Wijn JR, van Blitterswijk CA. Dynamic mechanical properties of 3D fiber-deposited PEOT/PBT scaffolds: an experimental and numerical analysis. J Biomed Mater Res A. 2006;78:605-14
62. Brown AL, Srokowski EM, Shu XZ, Prestwich GD, Woodhouse KA. Development of a model bladder extracellular matrix combining disulfide cross-linked hyaluronan with decellularized bladder tissue. Macromol Biosci. 2006;6:648-57
63. Hu JC, Yamakoshi Y, Yamakoshi F, Krebsbach PH, Simmer JP. Proteomics and genetics of dental enamel. Cells Tissues Organs. 2005;181:219-31
64. Miller TL, Touch SM, Shaffer TH. Matrix metalloproteinase and tissue inhibitor of matrix metalloproteinase expression profiles in tracheal aspirates do not adequately reflect tracheal or lung tissue profiles in neonatal respiratory distress: observations from an animal model. Pediatr Crit Care Med. 2006;7:63-9
65. Perlman CE, Cook KE, Seipelt JR, Mavroudis JC, Backer JC, Mockros LF. In vivo hemodynamic responses to thoracic artificial lung attachment. ASAIO J. 2005;51:412-25
66. Turner DJ, Noble PB, Lucas MP, Mitchell HW. Decreased airway narrowing and smooth muscle contraction in hyperresponsive pigs. J Appl Physiol. 2002;93:1296-300
67. Watremez C, Roeseler J, De Kock M, Clerbaux T, Detry B, Veriter C, Reynaert M, Gianello P, Jolliet P, Liistro G. An improved porcine model of stable methacholine-induced bronchospasm. Intensive Care Med. 2003;29:119-25
68. Cheetham S, Souza M, Meulia T, Grimes S, Han MG, Saif LJ. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J Virol. 2006;80:10372-81
69. Gonzalez AM, Nguyen TV, Azevedo MS, Jeong K, Agarib F, Iosef C, Chang K, Lovgren-Bengtsson K, Morein B, Saif LJ. Antibody responses to human rotavirus (HRV) in gnotobiotic pigs following a new prime/boost vaccine strategy using oral attenuated HRV priming and intranasal VP2/6 rotavirus-like particle (VLP) boosting with ISCOM. Clin Exp Immunol. 2004;135:361-72
70. Hasslung F, Wallgren P, Ladekjaer-Hansen AS. et al. Experimental reproduction of postweaning multisystemic wasting syndrome (PMWS) in pigs in Sweden and Denmark with a Swedish isolate of porcine circovirus type 2. Vet Microbiol. 2005;106:49-60
71. Butler JE, Sinkora M, Wertz N, Holtmeier W, Lemke CD. Development of the neonatal B and T cell repertoire in swine: implications for comparative and veterinary immunology. Vet Res. 2006;37:417-41
72. Elahi S, Brownlie R, Korzeniowski J. et al. Infection of newborn piglets with Bordetella pertussis: a new model for pertussis. Infect Immun. 2005;73:3636-45
73. Pomeranz LE, Reynolds AE, Hengartner CJ. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev. 2005;69:462-500
74. Dawson HD, Beshah E, Nishi S, Solano-Aguilar G, Morimoto M, Zhao A, Madden KB, Ledbetter TK, Dubey JP, Shea-Donohue T, Lunney JK, Urban JF Jr. Localized multi-gene expression patterns support an evolving Th1/Th2-like paradigm in response to infections with Toxoplasma gondii and Ascaris suum in pigs. Infection and Immunity. 2005;73:1116-1128
75. Dvorak CM, Hirsch GN, Hyland KA, Hendrickson JA, Thompson BS, Rutherford MS, Murtaugh MP. Genomic dissection of mucosal immunobiology in the porcine small intestine. Physiol Genomics. 2006;28:5-14
76. Houdebine LM. Use of transgenic animals to improve human health and animal production. Reprod Domest Anim. 2005;40:269-81
77. Hammamieh R, Bi S, Das R, Neill R, Jett M. Modeling of SEB-induced host gene expression to correlate in vitro to in vivo responses. Biosens Bioelectron. 2004;20:719-27
78. Perez J, Garcia PM, Bautista MJ, Millan Y, Ordas J, Martin de las Mulas J. Immunohistochemical characterization of tumor cells and inflammatory infiltrate associated with cutaneous melanocytic tumors of Duroc and Iberian swine. Vet Pathol. 2002;39:445-51
79. Apiou F, Vincent-Naulleau S, Spatz A, Vielh P, Geffrotin C, Frelat G, Dutrillaux B, Le Chalony C. Comparative genomic hybridization analysis of hereditary swine cutaneous melanoma revealed loss of the swine 13q36-49 chromosomal region in the nodular melanoma subtype. Int J Cancer. 2004;110:232-8
80. Tissot RG, Beattie CW, Amoss MS Jr. The swine leucocyte antigen (SLA) complex and Sinclair swine cutaneous malignant melanoma. Anim Genet. 1989;20:51-7
81. Blangero J, Tissot RG, Beattie CW, Amoss MS Jr. Genetic determinants of cutaneous malignant melanoma in Sinclair swine. Br J Cancer. 1996;73:667-71
82. Zhao S-H, Kuhar D, Lunney JK, Dawson HD, Guidry C, Uthe J, Bearson S, Recknor J, Nettleton D, Tuggle CK. Gene expression profiling in Salmonella Choleraesuis infected porcine lung using a long oligonucleotide microarray. Mammalian Genome. 2006;17:777-789
83. Andersson L, Georges M. Domestic-animal genomics: deciphering the genetics of complex traits. Nat Rev Genet. 2004;5:202-12
84. Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the US. J Am Vet Med Assoc. 2005;227:385-92
85. Miller LC, Fox JM. Apoptosis and porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol. 2004;102:131-42
86. Wang C, Hawken RJ, Larson E, Zhang X, Alexander L, Rutherford MS. Generation and mapping of expressed sequence tags from virus-infected swine macrophages. Anim Biotechnol. 2001;12:51-67
87. Vincent AL, Thacker BJ, Halbur PG, Rothschild MF, Thacker EL. An investigation of susceptibility to porcine reproductive and respiratory syndrome virus between two genetically diverse commercial lines of pigs. J. Anim. Sci. 2006;84:49-57
88. Royaee AR, Husmann R, Dawson HD, Calzada-Nova G, Schnitzlein WM, Zuckermann F, Lunney JK. Deciphering the involvement of innate immune factors in the development of the host responses to PRRSV vaccination. Vet. Immunol. Immunopathol. 2004;102:199-216
89. Petry DB, Holl JW, Weber JS, Doster AR, Osorio FA, Johnson RK. Biological responses to porcine respiratory and reproductive syndrome virus in pigs of two genetic populations. J Anim Sci. 2005;83:1494-1502
Author contact
![]() Correspondence to: Joan K. Lunney, Ph.D., APDL, ANRI, ARS, USDA, Building 1040, Room 103, BARC-East, Beltsville, MD 20705 USA. PH: 301-504-9368 FAX: 301-504-5306 Current email: jlunneybarc.usda.gov
Correspondence to: Joan K. Lunney, Ph.D., APDL, ANRI, ARS, USDA, Building 1040, Room 103, BARC-East, Beltsville, MD 20705 USA. PH: 301-504-9368 FAX: 301-504-5306 Current email: jlunneybarc.usda.gov

 Global reach, higher impact
Global reach, higher impact