10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2007; 3(4):225-236. doi:10.7150/ijbs.3.225 This issue Cite
Research Paper
MCEF is localized to the nucleus by protein sequences encoded within three distinct exons, where it represses HIV-1 Tat-transactivation of LTR-directed transcription
1. Ryerson University, Department of Chemistry & Biology, Toronto, Ontario, M5B2K3, Canada.
2. University of Guelph, Department of Chemistry, Guelph, Ontario, Canada.
Received 2006-12-16; Accepted 2007-2-27; Published 2007-3-1
Abstract
Translocations between the human Mixed Lineage Leukemia (MLL) and AF4 Family (AFF) member genes, are implicated in leukemia. Mutations to AFFs can disrupt lymphopoesis, CNS development and spermatogenesis. However, despite the growing list of pathologies linked to AFF members, their evolutionary relationship and the structure/function of individual members, remain to be elucidated. Here, we first report that database mining and phylogenetic analysis with AFF proteins from multiple species, revealed two monophyletic sister clades, suggesting a common Bilateria ancestor. We then examined the structure/function of the most recently discovered AFF member, MCEF (also known as AF5q31 or AFF4). In silico, the human MCEF gene was found to have 21 exons, and code for a protein with seven nuclear localization sequences (NLS). In HeLa cells, an MCEF-EGFP fusion protein, localized exclusively to the nucleus. Consequently, we made twenty constructs, expressing MCEF deletion mutants fused to EGFP and/or DsRed fluorescent proteins. Three distinct protein sequences, encoded by three separate MCEF exons, were found to mediate nuclear localization, only two of which were predicted in silico. Importantly, we also found that ectopic expression of MCEF, repressed HIV-1 LTR-directed RNA Polymerase II transcription, at the level of Tat-transactivation. We suggest that portions of MCEF could be exploited for chimeric transcription factor repression (CTFR) of HIV-1.
Keywords: Transcription, MCEF, AF5q31, AFF4, NLS, HIV, Tat
1. Introduction
Polycomb/trithorax proteins are epigenetic regulators of transcription [1]. The mixed lineage leukemia (MLL) protein is a mammalian homologue of Drosophila trithorax, functioning as a transcriptional maintenance factor during morphogenesis [2]. In fact, MLL normally regulates transcription initiation and elongation of a specific set of HOX genes. The human MLL gene on chromosome 11 is frequently translocated in acute lymphoblastic leukemia (ALL), generating chimeric proteins composed of amino-terminal MLL and carboxyl-terminal partners [3]. The reciprocal chimera can also occur [4]. There is evidence implicating these chimeras in dys-regulation of normal MLL-mediated HOX gene expression, leading to ALL [5]. Importantly, three AFF members (AF4, LAF4, MCEF, see below), are partners for MLL translocations, suggesting a role as transcription factors [6-12].
The human AF4 gene (designated HUMAN AFF1 by the Human Genome Nomenclature Committee (HGNC) [13]), on chromosome 4, activates transcription when fused to the DNA binding domain of yeast Gal 4 (GAL4), and tethered to a promoter with Gal 4 DNA binding sites (G4DBS) [14, 15]. In mice, AF4 is expressed in lymphoid tissues and brain [15]. Although knockout AF4 mice appear affected only in lymphoid tissues, it has recently been established that AF4 plays a role in the CNS, where a single aa mutation, modifying a Siah site, is associated with Purkinje cell loss; highlighting that even for the best studied AFF member, we still know little about its normal function in vivo [16].
The human FMR2 gene (designated HUMAN AFF2 by HGNC [13]) on chromosome X, has not been implicated in leukemia. Silencing FMR2 is involved in mild mental retardation (MMR) [17]. FMR2 is highly expressed in brain [17]. As for AF4, GAL4-FMR2 activates transcription [17].
The human LAF4 gene (designated HUMAN AFF3 by HGNC [13]) on chromosome 2, parallels AF4 expression, and is implicated in ALL and breast cancer [18]. Mouse LAF4 likely acts in lymphoid and CNS development [19]. As for AF4 and FMR2, GAL4-LAF4, activates transcription [17].
The human MCEF gene (designated HUMAN AFF4 by the HGNC [13] and also known as AF5q31) on chromosome 5, codes for a 1,163 aa protein with regions of homology to other AFF members, designated NHD, ALF, pSer, CHD [3, 9, 10, 12]. MCEF is expressed in adult heart, with lower levels in placenta, skeletal muscle, pancreas and brain [9, 10]. Higher expression occurs in fetal compared to adult lung and brain [9]. Unexpectedly, MCEF knockout mice were recently shown to be azoospermic [20].
Despite the growing list of pathologies implicating AFF members, little is known about their structure/function relationships. For this paper, we first mined databases for AFF sequences from multiple species, and determined phylogenetically that AFFs form sister clades, suggesting they have a common Bilateria ancestor. We then characterized MCEF in silico, finding seven putative NLS, within 3 of 21 exons. In HeLa cells, we showed that a full-length MCEF-EGFP fusion protein was nuclear. We consequently made twenty constructs expressing MCEF deletions, to test for functional NLS. We found that distinct protein sequences, encoded by three separate exons could mediate nuclear localization, only two of which were predicted in silico. Importantly, we also found that MCEF can directly repress RNA Polymerase II transcription of HIV-1 LTR-directed Tat-transactivation, in HeLa cells. These results establish that MCEF is a bonafide nuclear transcription factor, and suggest that portions of MCEF may be useful in designing a protein to be used in chimeric transcription factor repression (CTFR) of HIV-1.
2. Materials and Methods
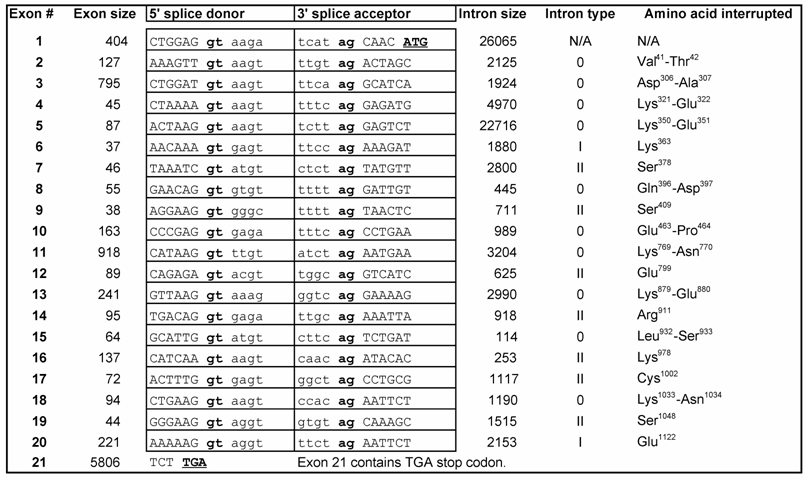
MCEF bioinformatics. The MCEF cDNA was re-sequenced with 3 fold redundancy, on a 310 Genetic Analyzer (ABI) (GenBank accession no. AF213987.2) [10]. The MCEF sequence represents the cDNA clone used in this paper, and generates the MCEF protein sequence (GenBank accession no. AAM00184.2). The AF5q31 sequence is a distinct cDNA, cloned by another group, that generates a protein sequence nearly identical to MCEF (GenBank accession no. AAF18981.1) [9]. The AFF4 consensus sequence is a distinct hypothetical reference sequence, also nearly identical to MCEF (GenBank accession no. NP_055238). Individual AFF1 (GenBank accession nos. P51825, 114594998, XP_001095779.1, AAU93698.1, XP_223161.4), AFF2 (nos. NP_002016.2, NP_001009042.1, BAE02338.1, NP_032058.1, XP_001054673.1), AFF3 (nos. AAH36895.1, XP_525831.1, XP_001104149.1, AAH52061.1, XP_343560.2), and AFF4 (nos. AAM00184.2, AAF18981.1, NP_055238, XP_517928.1, BAE00514.1, BAC35763.1, XP_220420.2), sequences were used for alignments. The out-group was Lilliputian (no. NP_722863.1) [21]. ClustalW (v1.4) in MacVector 2.22 (Accelerys), generated alignments. Best tree was derived by Neighbor Joining, and node values obtained from 32,000 bootstraps. BLAST (NCBI) searches identified the longest AFF4 mRNA (no: NM_014423.3), used to identify intron/exon junctions using the chromosome 5 contig (no: NT_034772.5) [22]. Junctions were predicted by Spidey (NCBI) [23]. The NHD, ALF, pSer, CHD, and a putative MCEF Transactivation domain (TAD), were derived from previous publications, then BLAST (NCBI) was used to locate homologous MCEF sequences [3, 24]. The 7 PEST sites and 7 NLS, were predicted by PESTFIND and PSORTII [16, 25].
Molecular constructs. The -pcMCEF construct expresses full-length MCEF from a CMV promoter and +pcMCEF contains an antisense MCEF sequence [10]. The antisense construct was used as a control, instead of empty vector, since it is of the exact same size as the MCEF-expressing construct. The MCEF ORF was amplified using forward primers with XhoI, and reverse primers with BamHI, sites incorporated for cloning into pEGFP-N1 or pDsRed (Clontech). Primers used to generate constructs in Fig. 3 were: c - MN-F2 (5'-CCGCTCGAGCGGATGAACCGTGAAGACCGG-3') and MN-R2 (5'-CGCGGATCCCTAGATATCAACTTGGCATCCTG-3'); d - MN-F2 and MN-R3 (5'-CGCGGATCCTGTCTGCTGGAGGGCATGC-3'); e - MN-F13a (5'-CCGCTCGAGATGAGTGGATCTGAAAGCAGCTCTGG-3') and MN-R7a (5'-CGCTGGATCCTCGGGAGATGCCTGGGATGG-3'); f - MN-F7 (5'-CCGCTCGAGATGCCTCGTGGAGGCCTGAAGATAG-3') and MN-R2; g - MN-F7 and MN-R2; h - MN-F20 (5'-CCGCTCGAGGCCACCATGAATATAAAGAAGGAGTCTAAG-3') and MN-R20 (5'-CGCTGGATCCGAAGATTTACTTGTTGACTTATATTTC-3'); i - MN-F21 (5'-CCGCTCGAGGCCACCATGCCTCGACCTACAGCAGAG-3') and MN-R20; j - MN-F5 (5'-CCGCTCGAGATGGAAGAGAAGGAACTTC-3') and MN-R2; k - MN-F5 and MN-R2; l - MN-F12 (5'-CCGCTCGAGATGAGGGAAATCATAGAAACAG-3') and MN-R11 (5'-CGCTGGATCCCCACTGCTTGACTTTAAGGA-3'); m - MN-F12 and MN-R6a (5'-CGCTGGATCCTCACTTTCATCTGAATCTGA-3'); n - MN-F16 (5'-CCGCTCGAGGCCACCATGGTTTCCAACAAAGGCAAGAGG-3') and MN-R9 (5'-CGCTGGATCCTTATGCTTCCTCTTGCC-3'); o - MN-F9 (5'-CCGCTCGAGATGTACAAAGAAACAGAGCCGCCC-3') and MN-R9; p - MN-F5 and MN-R9; q - MN-F12 and MN-R9; r - MN-F14 (5'-CCGCTCGAGATGAAGATTGACCTGAATCTT-3') and MN-R9; s - MN-F16 and MN-R10 (5'-CGCTGGATCCTTGTCCTCCGTTTTGGG-3'); t - MN-F15 (5'-CCGCTCGAGATGAGTGCTGCAAAAGAAAAGG-3') and MN-R2; u - MN-F11 (5'-CCGCTCGAGATGAACAGCAACAAGGAGACGAGTGG-3') and MN-R2; v - MN-F22 (5'-CCGCTCGAGGCCACCATGCCAACTCTTGATTCTTCTAAG-3') and MN-R22 (5'-CGCTGGATCCGCTGAATAATTTCTGTCATCAAAG-3'); w - MN-F15 and MN-R11. PCR, cloning and sequencing of constructs was by standard techniques. Constructs were tested using the T7 TNT-Coupled reticulocyte lysate system (Promega), per manufacturer's recommendations, using Redivue L-[35S] methionine (GE Healthcare).
Tissue culture and transfection. HeLa S3 cells (ATCC) were maintained in F-12K media, with 10% fetal bovine serum and 10% penicillin-streptomycin (Multicell), at 37ºC and 5% CO2. HeLa S3 cells were transfected using DreamFect (Oz Biosciences), with 1.0 μg of each construct, in 80% confluent 6-well plates. After 24 h, cells were observed under a confocal laser microscope or harvested for luciferase assays.
Fluorescent Microscopy. Cells were routinely examined under an AE31 microscope (Motic). For confocal laser microscopy, cells were grown on No. 1 glass cover slips (VWR International), inside 6-well plates, and transfected with DNA of interest. For observations under the LSM 510 microscope (Zeiss), cover slips were removed and cells covered with complete F-12K medium, containing 20 mM HEPES, to buffer against pH changes. The Plan-Neofluoar oil DIC 40x objective was used. Expression was scored as + or – , in the nucleus and cytoplasm.
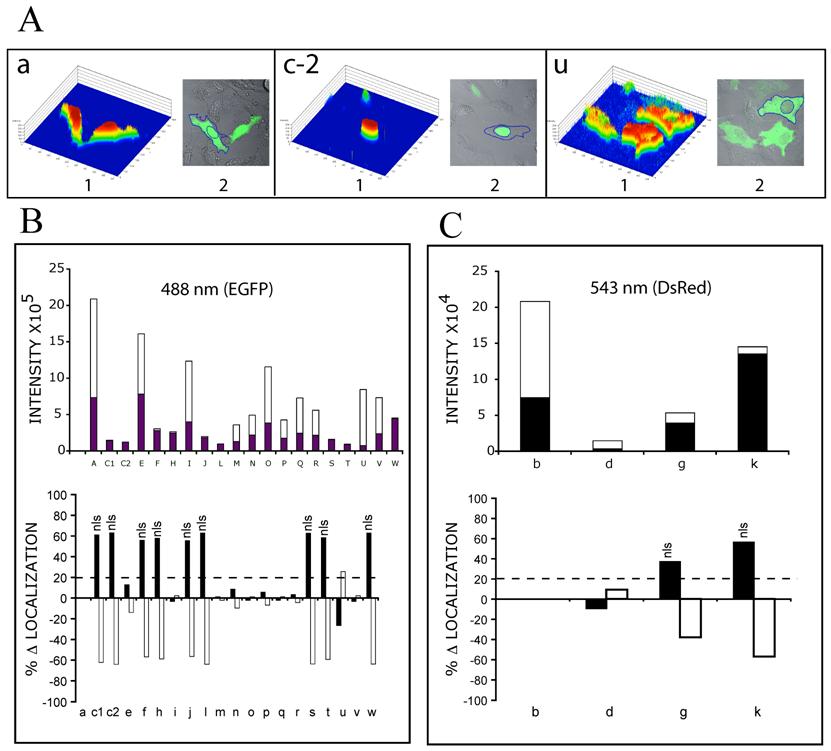
Single cell expression levels. Individual cells were analysed with LSM 510 Image Analysis Software (Zeiss). Histogram data were derived from individually outlined cells and nuclei. Total Expression Level (TEL) for each cell, was calculated as the [(Σ (1-250) pixel intensities in outlined cell) x (# of pixels at each intensity)]. Nuclear Expression Level (NEL) for each cell, was calculated as ([Σ (1-250) pixel intensities in outlined nucleus) x (# of pixels at each intensity)]. Cytoplasmic Expression Level (CEL) for each cell, was calculated as [TEL – NEL]. Data was normalized to EGFP (Fig. 3, 4, 5: a) settings of laser intensity and pinhole aperture (5% and 135 μm), but are presented separately in graphs B and C, since the intensity was approximately 10 fold less for DsRed constructs (543 nm) than for EGFP constructs (488 nm). The value of percentage change in localization (% Δ Nuclear Localization), was calculated as the [(% NEL for each construct) – (% NEL for EGFP alone)]. The % Δ Cytoplasmic Localization, was calculated as the [(% CEL for each construct) – (% CEL for EGFP alone)].
Luciferase assays. The HIV-1 subtype B LTR, clone pMCE36.1, was previously described and subcloned into a luciferase reporter construct by standard techniques [26]. The HIV-1 Tat expression construct (pCMV-Tat), –pcMCEF and +pcMCEF constructs, have been previously described [10, 26, 27]. Luciferase assays were performed as recommended by the manufacturer (Promega) and activity quantitated using a 20/20 Luminometer (Turner). Triplicate results, were adjusted for protein content via the Bradford assay (Bio-Rad) and for transfection efficiencies using a Renilla luciferase control (Promega). The CMV-RL construct (Promega) was used to analyze the effects of MCEF on the CMV promoter, with results represented as fold repression, compared to fold repression of LTR, both in the presence or absence of Tat. A fold activity of 1, was considered zero fold repression (see figure 6).
3. Results
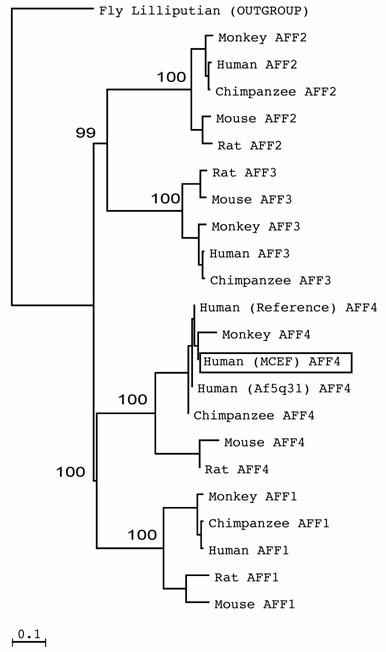
AFFs form sister clades. Few AFF sequences have been published. As a result, we mined databases to determine which species have AFF sequences. We found complete or partial AFF sequences in Fly, Zebra fish, Puffer fish, Frog, Chicken, Cow, Rat, Mouse, Orangutan, Macac, Chimpanzee and Humans (Fig. 1 and not shown). To establish which AFF members most resemble MCEF, we phylogenetically analyzed AFF members from various species, and found that AFF1 and AFF4 are sister clades, as are AFF2 and AFF3, with bootstrap values of 100% and 99% (Fig. 1). The distinct sequences for MCEF and Af5q31, result from the fact that they represent nearly identical but distinct clones. The AFF4 sequence is also distinct, but nearly identical to MCEF, since it is a theoretical consensus sequence.
AFF members form two sister clades. The tree, derived through phylogenetic analysis using ClustalW and Neighbor Joining, was created as described in Materials and Methods. Bootstrap values from 32,000 re-samplings, are given in percentage at the nodes. The outgroup sequence was from Drosophila melanogaster (Fly Lilliputian). Human MCEF is boxed in a rectangle.
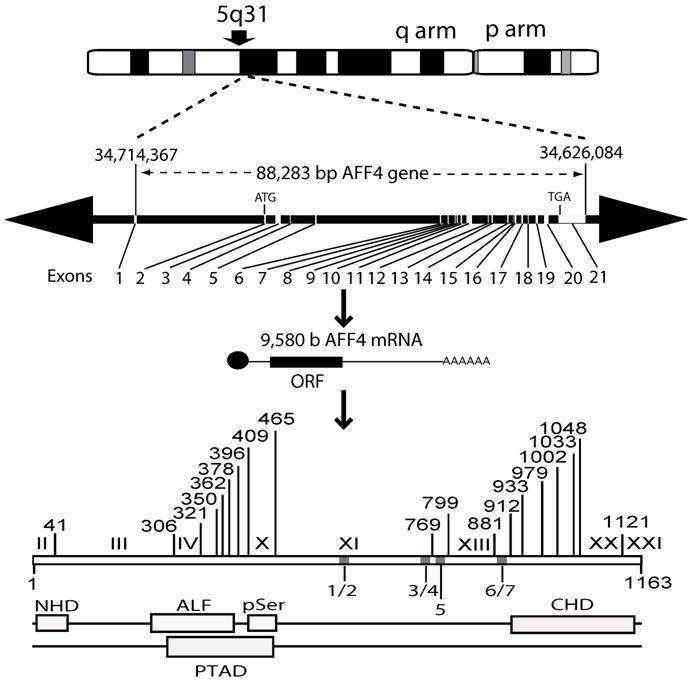
MCEF has seven putative NLS. In silico, the 21 exon MCEF gene on chromosome 5 was found to span 88,283 bp, to be transcribed into a 9,580 b mRNA and to code for a 1,163 aa protein, with seven putative NLS, distributed over 4 regions, designated 1/2, 3/4, 5, 6/7 (Fig. 2 and Table 1). The putative NLS are outside the NHD, ALF, pSer, CHD and a putative transcription activation domain (TAD) regions [3, 24]. In silico, we also found seven high scoring putative PEST (PP1 to PP7) sequences, in addition to a previously reported Siah site (Fig. 2) [16, 25]. PEST and Siah1 sites are involved in proteosome protein turnover [16, 25].
MCEF gene, mRNA and protein overview. Schematic representation of the MCEF protein in relation to the AFF4 mRNA, the 21-exon gene, and the corresponding chromosomal locus. The long (q) and short (p) arms of chromosome 5 are labeled and the AFF4 gene location is indicated as 5q31. The 88,283 bp AFF4 gene is represented with Exons and Introns positioned on a 41,199,371 bp human chromosome 5 genomic contig. The positions of 34,714,367 and 34,626,084 correspond to the first 5' base of AFF4 mRNA and the last 3' position at 9,580 b on the contig, respectively. The 9,580 b reference AFF4 mRNA sequence was used to determine intron/exon boundaries (see table 1) and for the relative position of exons 1-21. The ATG start codon in Exon 2 and the TGA stop codon in Exon 21 are indicated. The relative positions of the MCEF open reading frame (ORF), poly-adenylation bases (AAA), and cap (l), are indicated. The bottom of the figure is a representation of the MCEF protein derived from the 1,163 aa full length ORF translation of the MCEF cDNA. NHD, ALF, pSer CHD and the putative TAD, are indicated as open boxes (see text) in their relative positions. Five regions (1/2, 3/4, 5, 6/7) are designated containing the seven NLS. See materials and methods section for further details including Genbank Accession numbers for sequences used.
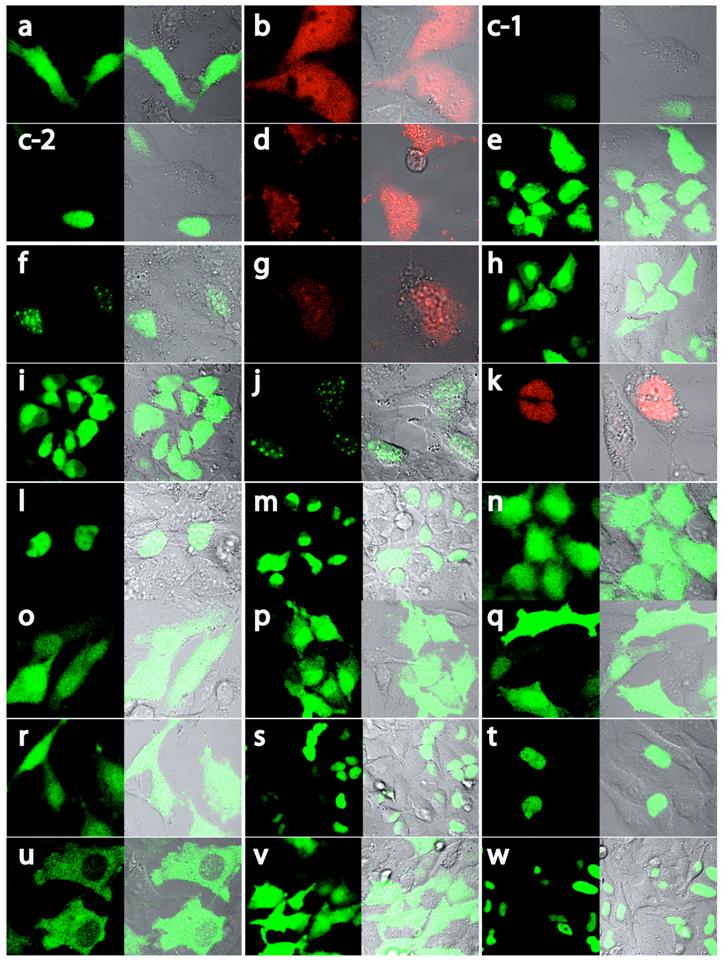
MCEF localizes to the nucleus. Although T-cells are a preferred target for HIV-1, the HeLa cell was selected for our studies because: (i) our MCEF clone was derived from HeLa cells, and (ii) we have previously shown that MCEF represses HIV-1 transcription in HeLa cells [10]. To determine MCEF localization, we transfected HeLa cells with a construct over-expressing a full-length MCEF-EGFP fusion protein. In contrast to EGFP or DsRed alone, green fluorescence was strictly nuclear (Fig. 3 and 4; a and b, versus c). Despite the compartmentalized restriction, we estimated the expression at 1/20 that of EGFP (Fig. 4, c-1 versus a, and Fig 5B upper graph). The cells in panel c-2 (Fig. 4) and all subsequent panels (Fig. 4, d to w), were photographed after laser power was adjusted to approximately match EGFP or DsRed expression (Fig. 4a and b). However, the data in Figure 5B and C upper graphs, is quantitative. Using this data, we calculated the % change in nuclear or cytoplasmic localization of EGFP, as a result of MCEF fusion, using a 20% change as indicative of localization signals. Clearly, there is at least one NLS in MCEF, since its fusion to EGFP results in an approximately 60% change in nuclear localization, accompanied by a corresponding -60% cytoplasmic localization (Fig. 5B lower graph).
Exon-intron structure of AFF4 gene.
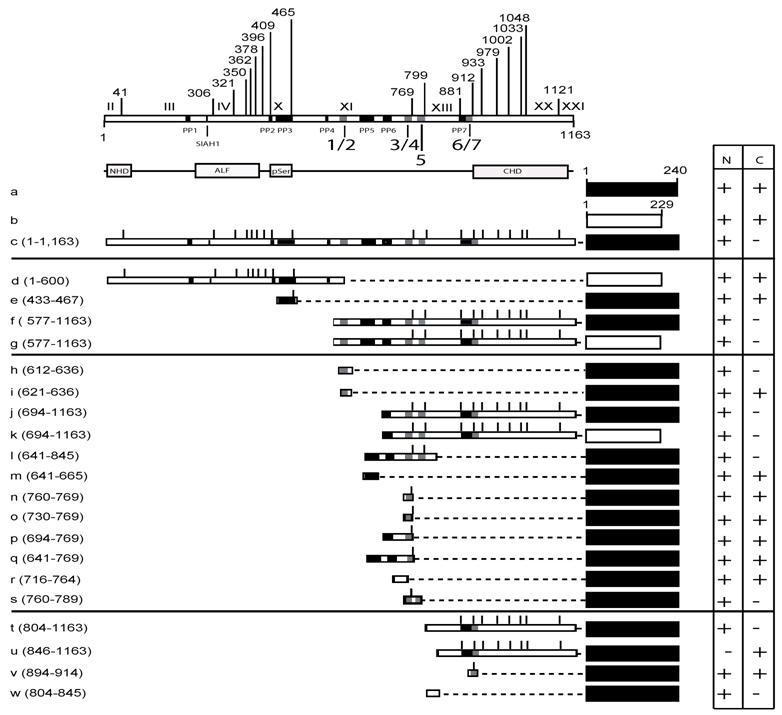
The C-terminal half of MCEF mediates nuclear localization. To map nuclear localization sequences in HeLa cells, we transfected constructs expressing MCEF deletion mutants, fused to EGFP and/or DsRed proteins (Fig. 3, 4 and 5). N-terminal MCEF localized to both the cytoplasm and the nucleus, whereas C-terminal MCEF localized only to the nucleus (Fig. 3, 4 and 5; d, versus f and g). This is consistent with in silico C-terminal prediction of NLS (see above). Fusion of PP3 alone to EGFP, had no effect on localization (Fig. 3, 4 and 5; e versus a).
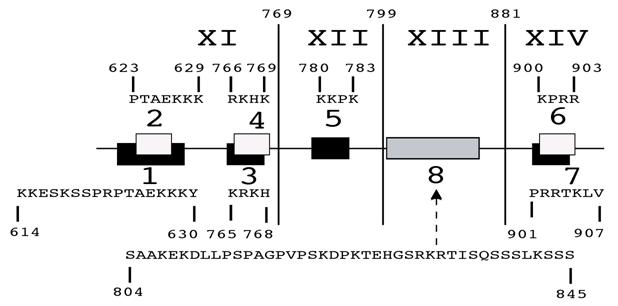
NLS 1, and 5, within exons XI and XII, mediate nuclear localization. From the C-terminal portion of MCEF, a construct containing NLS 1 and 2, fused to EGFP, resulted in nuclear localization, whereas a construct with only NLS 2, did not (Fig. 3, 4 and 5; h versus i). This indicates NLS 1 is functional. However, constructs containing NLS 3, 4, 5, 6 and 7, also localized to the nucleus (Fig. 3, 4 and 5; j, k). A construct with only NLS 3, 4 and 5, also localized to the nucleus (Fig. 3, 4 and 5; l). We also tested PP6 fused to EGFP, but found no effect on EGFP localization (Fig. 3, 4 and 5; m versus a). Five additional constructs containing either NLS 3, 4, or sequences between NLS 3, 4 and NLS 1, 2, all failed to localize fused EGFP to the nucleus (Fig. 3, 4 and 5; n, o, p, q, r). In contrast, a construct containing NLS 3, 4 and 5, localized exclusively to the nucleus (Fig. 3, 4 and 5; s). This indicates that in addition to NLS 1, in exon XI, NLS 5 in exon XII, is functional.
A non-NLS sequence within exon XIII, mediates nuclear localization. To test NLS 6 and 7, we made a construct encompassing the C-terminal 804-1,163 aa and found that it localized to the nucleus (Fig. 3, 4 and 5; t). Surprisingly, a further deletion to aa 846, ablated nuclear localization, despite leaving NLS 6 and 7 intact (Fig. 3, 4 and 5; u). We further tested NLS 6 and 7 directly and confirmed no nuclear localization (Fig. 3, 4 and 5; v). This suggests NLS 6 and 7 are not functional, but another sequence, containing aa 804-845, can mediate nuclear localization. In fact, sequences from 804-845, were able to mediate nuclear localization (Fig. 3, 4 and 5; w). These results establish that a non-NLS sequence, within exon XIII mediates nuclear localization, in addition to the sequences in exons XI and XII, summarized in Fig. 6.
MCEF-EGFP and MCEF-DsRed constructs and localization. Schematic representation of the MCEF-EGFP and MCEF-DsRed constructs transfected into HeLa cells. The constructs are labelled a to w. The upper part of the diagram has the exons I to XXI indicated, with the aa numbers above. Beneath the upper representation are the seven putative pest domains (PP1 to PP7), the Siah1 site, and the seven putative NLS sequences (1/2, 3/4, 5, 6/7). Below this, the conserved NHD, pSer, ALF, CHD regions are indicated, for reference below and with Fig. 1. The representation of the constructs includes the C-terminal EGFP (black rectangles) from aa 1 to 240, and the C-terminal DsRed (white rectangles) from aa 1 to 229. The MCEF aa numbers fused to the respective fluorescent protein, for each construct, are indicated beside the letter designation, in brackets. The MCEF deletions are represented with dashed lines, to simplify comparison. Vertical lines represent the exon boundaries. To the right, the expression in the nucleus (N) and the cytoplasm (C) are indicated, as either negative (-), or positive (+). Construct letters correspond to the respective panels in Fig. 4.
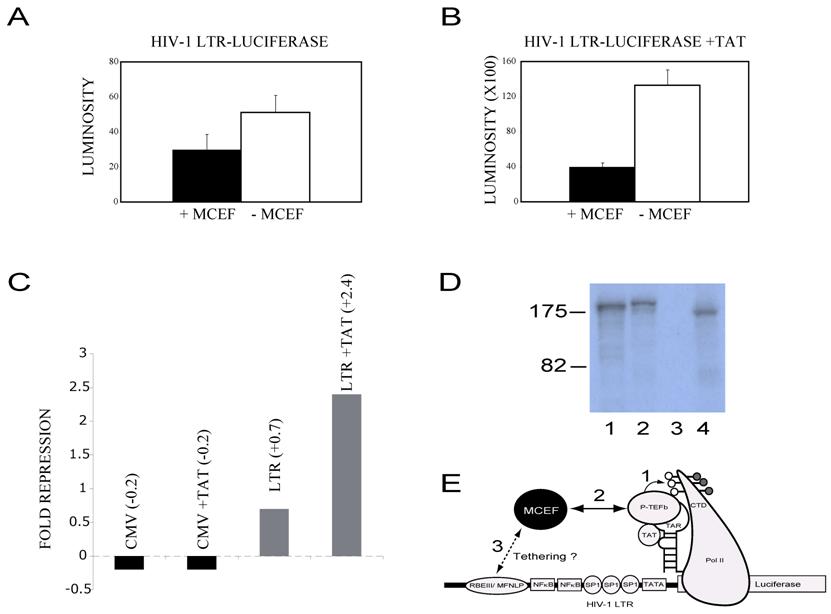
MCEF represses HIV-1 Tat Transactivation. Because MCEF was isolated as a co-immunoprecipitating protein with P-TEFb (see discussion), we directly tested its ability to impinge Tat-transactivation [10]. In order to ensure that our construct was in fact expressing MCEF, we tested the expression, in a coupled transcription-translation reaction, and a band of the expected size was detected (Fig. 7D, lane 4) [10]. In the absence of Tat, MCEF had a minimal effect upon HIV-1 LTR-directed transcription, 0.7 fold (Fig. 7A and 7C). However, in the presence of Tat, it repressed Tat-transactivation, 2.4 fold (Fig. 7B). In order to test for promoter specificity of the repression, we tested the CMV promoter in the presence or absence of Tat and in the presence or absence of MCEF. These were the exact same conditions used for the LTR experiments. Under these conditions, MCEF had a minimal activation effect, independent of the presence or absence of Tat (Fig. 7C). Similarly, we found that the TK promoter was also un-affected by MCEF expression (not shown).
Localization of MCEF-EGFP and MCEF-DsRed proteins. Confocal laser microscopy of HeLa cells transfected with EGFP-expressing (a), DsRed-expressing (b), full-length MCEF-EGFP-expressing (c) and deletion mutants of MCEF-EGFP (e, f, h, i, j, l to w) or deletion mutants of MCEF-DsRed (d, g, k). The constructs correspond with the schematic representations in Fig. 3. Panels are in sets of two. The first, identified with the letter corresponding to the construct transfected, was taken under fluorescent microscopy. The second, to the right, is an overlay of fluorescence and visible light microscopy images.
Single cell expression levels of MCEF-EGFP and MCEF-DsRed, fusion proteins. Individual single cells were analyzed with Image Analysis Software (Zeiss) and intensities derived from outlined cells. Total Expression Levels (TEL), Nuclear Expression Levels (NEL) and Cytoplasmic Expression Levels (CEL) were derived as described in Materials and Methods. The letters (a to w) correspond to those in Fig. 3 and 4. (A) Examples of 3 cells, transfected with the respective constructs (a, c-2 and u), used for expression level analysis (the color 3-D histogram is shown in 1, and the outlined cell and nucleus are shown in 2). (B) Bar graph of the Intensity of EGFP signal (collected at 488nm wavelength), in the nucleus (black bars) and the cytoplasm (white bars), for the respective MCEF-EGFP constructs (upper graph); and bar graph of the % change (D) in EGFP intensity (with respect to EGFP alone), in the nucleus (black bars) and the cytoplasm (white bars). A cut-off of 20% was selected as indicative of specific localization (dashed line), with those above 20% indicated (nls). (C) As for B, but for DsRed constructs (collected at 543 nm wavelength).
4. Discussion
All AFF sequences we mined, came from Deuterostomia, except the Fly sequence (Arthropoda). This suggests the ancestor to Deuterostome AFFs, is at least as old as the common ancestor to Deuterostome and Ecdysozoa (from which Arthropoda developed), which is Bilateria [28]. As more AFF-like sequences are discovered in species distant to humans, they should be classified according to where they fall on a robust tree, with nodes supported by high bootstrap values. In Fly, there appears to be only one distantly related family member, Drosophila lilliputian [21]. Since three of the four AFFs are known to be involved in translocations, it is entirely possible that the four AFFs developed from ancestral duplications of a single sequence, as the Bilateria evolved, approximately 600 million years ago [28].
Precedent for differentially spliced AFF isoforms, was set by OX19, an FMR2 variant [17]. PEST sequences are involved in proteosome pathway degradation of proteins, and interestingly, a Siah1 site, conserved in AFF members, is involved in proteosome pathway degradation of AFF1[16, 25]. Therefore, our in silico results initially suggested MCEF, or differentially spliced MCEF isoforms, containing exons XI, and/or XII and/or XIV, would be nuclear, with expression levels of at least some isoforms, regulated by proteosomal turnover. The experimental data however, indicated that only the bipartite NLS 1, and NLS 5, in exons XI and XII were functional in HeLa cells [29]. Interestingly, we also discovered that a non-NLS containing sequence, in exon XIII, mediates nuclear localization. One possibility is that this sequence is a binding region involved in co-transport via association with another nuclear localized factor. In this regards, MCEF is known to interact with such a factor, P-TEFb [10]. Further experiments will be required to test if this same interaction is involved in the nuclear localization through exon XIII.
Our results suggest that the fusion of full-length MCEF to EGFP, reduces expression levels by a factor of 20 (Fig. 5B, upper graph; a versus c-1 or c-2). However, we could not map this reduced expression level to specific Pest sites. Further experiments will be required to determine if the Pest sites are indeed functional or not, in mediating expression levels. Alternatively, Kozak sequences affect expression levels in eukaryotes [30]. In fact, the naturally-occurring MCEF Kozak sequence is weak [9, 10]. However, the weak Kozak sequence in our original MCEF cDNA clone, was replaced in the MCEF-EGFP fusion constructs. At any rate, our results show that MCEF isoforms with exons XI, XII and XIII, will be nuclear, and suggest translational or post-translational control of MCEF expression levels may be occurring.
MCEF was initially cloned as a protein immunoprecipitating with P-TEFb, itself composed of CDK9 and Cyclin T [10]. Importantly, HIV-1 Tat, P-TEFb and a nascent RNA stem-loop structure called TAR, form a ternary complex, permitting CDK9 to hyperphosphorylate the RNA Polymerase II Carboxy Terminal Domain (Pol II CTD) [10]. This hyperphosphorylation is essential for transcription elongation to occur at the HIV-1 LTR promoter (Fig. 7 E). Previous tethering of GAL4-AF5q31 or GAL4-MCEF, did not activate a minimal promoter with G4DBS [10, 17]. However, tethering of GAL4-MCEF repressed an enhancer containing promoter, and ectopic MCEF expression has been previously shown to repress HIV-1 replication [10]. It is therefore significant that MCEF represses Tat-transactivation (this paper), and we propose the model illustrated in Fig. 7 E. In this model, MCEF interactions with P-TEFb would interfere with the proper formation of the ternary complex (see above), possibly inhibiting CDK9 hyperphosphorylation of the RNA Pol II CTD. This mechanism may appear to be at odds with another groups data, where they failed to inhibit P-TEFb phosphorylation of the RNA Pol II CTD, with AF5q31 [20]. However, their results were derived outside the cellular environment, and they did not look at the assembled RNA Pol II complex on a promoter, in the presence of Tat and TAR, as would be the case in our assays inside HeLa cells. Further experiments will be required to test our model, and to determine if MCEF can interfere with other well-known treatments that activate the HIV-1 LTR, independent of Tat, such as ionomycin/PMA.
MCEF exons XI, XII and XIII, can mediate nuclear localization. Schematic representation of nuclear localization results for MCEF sequences transfected into HeLa cells. The exons XI to XIV are indicated, with the relative position of NLS identified in silico, indicated by either black or white rectangles. The sequences for these regions are indicated above (2, 4, 5, 6) or below (1, 3, 7). The non-NLS sequence, found to mediate nuclear localization, is also indicated (8).
MCEF represses HIV-1 Tat Transactivation. (A) Bar graph showing HIV-1 LTR-Luciferase activity (LUMINOSITY), in the absence of HIV-1 Tat (Y axis), with the co-transfected MCEF-expressing construct, -pcMCEF (+), or in the presence of the MCEF antisense construct, +pcMCEF (-) (X axis). (B) Exactly as in A, with the co-tranfection of a construct expressing HIV-1 Tat. (C) Fold repression of CMV promoter and LTR promoters, in the presence or absence of a co-transfected Tat-expressing vector. Fold repression value of 1 was considered no repression. (D) In vitro transcription-translation of pGAL4-MCEF (Lane 1), pMN-MCEF (1-1,163)-EGFP (Lane 2), +pcMCEF (Lane 3) and -pcMCEF (Lane 4). The size of two standards is given to the left of the autoradiogram, in KD. The -pcMCEF, expresses MCEF, whereas the +pcMCEF clone, expresses an MCEF antisense mRNA. (E) Cartoon of proposed mechanism for MCEF repression of HIV-1 replication and Tat Transactivation. The HIV-1 LTR is depicted with the Luciferase reporter gene fused (right side of cartoon) and several DNA binding sites depicted (RBEIII/ MFNLP, NFkB, SP1, TATA). RNA Pol II is depicted after promoter clearance and transcription of the RNA stem-loop structure TAR. The ternary complex between Tat-Tar and P-TEFb is indicated. The carboxy Terminal Domain (CTD) of RNA Pol II is shown phosphorylated, with hyperphosphorylation occurring through P-TEFb (1). In (2) the interaction between MCEF (Black circle) and P-TEFb would inhibit and/or disrupt the ternary complex from hyperphosphorylating the CTD. In (3), the cartoon suggests that if tethered to the highly conserved RBEIII/MFNLP site, this could increase the repressive effects of MCEF. This strategy of making a chimeric transcription factor, between MCEF and a component of RBF2 (USF or TFIIi), we dub Chimeric Transcription Factor repression (CTFR) of HIV.
Chimeric transcription factors, encompassing a portion of a protein encoded by one gene, fused to a portion of another protein encoded by a second gene, are naturally-occurring inside cells, for example, in the case of MLL-AFF member translocations (see introduction). These chimeric proteins appear to be able to change expression patterns of sets of genes. Similarly, fusions between GAL4 and a long list of proteins, can be used to artificially tether these proteins to promoters with appropriate binding sites. Engineered tethering of transcription repressors to the HIV-1 LTR has also been shown to be possible [31]. Extending this line of thinking, despite the extreme sequence variability of HIV-1, there is a highly conserved DNA binding site, proximal to its minimal promoter, termed RBEIII/ MFNLP, which binds the transcription factor RBF2. We have recently shown that RBF2 consist of USF and TFIIi [26, 27, 32, 33, and M.C.E. unpublished data]. Given that MCEF represses HIV-1 replication, that it represses transcription from an enhancer containing promoter with GAL4 DBS, that it interacts with P-TEFb, and that it represses Tat-transactivation (this paper) - we propose that an RBF2-MCEF chimeric protein, may result in potent Chimeric Transcription Factor Repression (CTFR) of HIV-1 (Fig. 7E).
Acknowledgements
This work was supported by CFI, OIT, NSERC, Canfar and Ryerson University grants, to MCE. MCE conceived the project, designed the experiments, interpreted the data, made the figures and wrote the manuscript. MFN did the experiments with assistance from RH and ZI. The authors thank John Marshall for access to confocal laser microscopy. MFN thanks MCE and Mario Monteiro, for co-supervising MFN at the University of Guelph, in the Department of Chemistry graduate program.
Conflict of interest
The authors have declared that no conflict of interest exists.
References
1. Orlando V. Polycomb, epigenomes, and control of cell identity. Cell. 2003 (112):599-606
2. Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci U S A. 1998 (95):10632-10636
3. Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001 (20):5695-5707
4. Bursen A, Moritz S, Gaussmann A, Moritz S, Dingermann T, Marschalek R. Interaction of AF4 wild-type and AF4.MLL fusion protein with SIAH proteins: indication for t(4;11) pathobiology? Oncogene. 2004 (23):6237-6249
5. Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ, Look AT. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003 (102):262-268
6. Domer PH, Fakharzadeh SS, Chen CS, Jockel J, Johansen L, Silverman GA, Kersey JH, Korsmeyer SJ. Acute mixed-lineage leukemia t(4;11)(q21;q23) generates an MLL-AF4 fusion product. Proc Natl Acad Sci U S A. 1993 (90):7884-7888
7. Hiwatari M, Taki T, Taketani T, Taniwaki M, Sugita K, Okuya M, Eguchi M, Ida K, Hayashi Y. Fusion of an AF4-related gene, LAF4, to MLL in childhood acute lymphoblastic leukemia with t(2;11)(q11;q23). Oncogene. 2003 (22):2851-2855
8. von Bergh AR, Beverloo HB, Rombout P, van Wering ER, van Weel MH, Beverstock GC, Kluin PM, Slater RM, Schuuring E. LAF4, an AF4-related gene, is fused to MLL in infant acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2002 (35):92-96
9. Taki T, Kano H, Taniwaki M, Sako M, Yanagisawa M, Hayashi Y. AF5q31, a newly identified AF4-related gene, is fused to MLL in infant acute lymphoblastic leukemia with ins(5;11)(q31;q13q23). Proc Natl Acad Sci U S A. 1999 (96):14535-14540
10. Estable MC, Naghavi MH, Kato H, Xiao H, Qin J, Vahlne A, Roeder RG. MCEF, the newest member of the AF4 family of transcription factors involved in leukemia, is a positive transcription elongation factor-b-associated protein. J Biomed Sci. 2002 (9):234-245
11. Imamura T, Morimoto A, Ikushima S, Kakazu N, Hada S, Tabata Y, Yagi T, Inaba T, Hibi S, Sugimoto T, Imashuku S. A novel infant acute lymphoblastic leukemia cell line with MLL-AF5q31 fusion transcript. Leukemia. 2002 (16):2302-2308
12. Deveney R, Chervinsky DS, Jani-Sait SN, Grossi M, Aplan PD. Insertion of MLL sequences into chromosome band 5q31 results in an MLL-AF5Q31 fusion and is a rare but recurrent abnormality associated with infant leukemia. Genes Chromosomes Cancer. 2003 (37):326-331
13. Wain HM, Lush MJ, Ducluzeau F, Khodiyar VK, Povey S. Genew: the Human Gene Nomenclature Database. 2004 updates. Nucleic Acids Res. 2004 (32):D255-257
14. Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Leshkowitz D, Croce CM, Canaani E. Domains with transcriptional regulatory activity within the ALL1 and AF4 proteins involved in acute leukemia. Proc Natl Acad Sci U S A. 1995 (92):12160-12164
15. Baskaran K, Erfurth F, Taborn G, Copeland NG, Gilbert DJ, Jenkins NA, Iannaccone PM, Domer PH. Cloning and developmental expression of the murine homolog of the acute leukemia proto-oncogene AF4. Oncogene. 1997 (15):1967-1978
16. Bitoun E, Davies KE. The robotic mouse: unravelling the function of AF4 in the cerebellum. Cerebellum. 2005 (4):250-260
17. Hillman MA, Gecz J. Fragile XE-associated familial mental retardation protein 2 (FMR2) acts as a potent transcription activator. J Hum Genet. 2001 (46):251-259
18. To MD, Faseruk SA, Gokgoz N, Pinnaduwage D, Done SJ, Andrulis IL. LAF-4 is aberrantly expressed in human breast cancer. Int J Cancer. 2005 (115):568-574
19. Britanova O, Lukyanov S, Gruss P, Tarabykin V. The mouse Laf4 gene: exon/intron organization, cDNA sequence, alternative splicing, and expression during central nervous system development. Genomics. 2002 (80):31-37
20. Urano A, Endoh M, Wada T, Morikawa Y, Itoh M, Kataoka Y, Taki T, Akazawa H, Nakajima H, Komuro I, Yoshida N, Hayashi Y, Handa H, Kitamura T, Nosaka T. Infertility with defective spermiogenesis in mice lacking AF5q31, the target of chromosomal translocation in human infant leukemia. Mol Cell Biol. 2005 (25):6834-6845
21. Wittwer F, van der Straten A, Keleman K, Dickson BJ, Hafen E. Lilliputian: an AF4/FMR2-related protein that controls cell identity and cell growth. Development. 2001 (128):791-800
22. International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004 (431):931-945
23. Horowitz DS, Krainer AR. Mechanisms for selecting 5' splice sites in mammalian pre-mRNA splicing. Trends Genet. 1994 (10):100-106
24. Nilson I, Reichel M, Ennas MG, Greim R, Knorr C, Siegler G, Greil J, Fey GH, Marschalek R. Exon/intron structure of the human AF-4 gene, a member of the AF-4/LAF-4/FMR-2 gene family coding for a nuclear protein with structural alterations in acute leukaemia. Br J Haematol. 1997 (98):157-169
25. Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996 (21):267-271
26. Estable MC, Bell B, Merzouki A, Montaner JS, O'Shaughnessy MV, Sadowski IJ. Human immunodeficiency virus type 1 long terminal repeat variants from 42 patients representing all stages of infection display a wide range of sequence polymorphism and transcription activity. J Virol. 1996 (70):4053-4062
27. Estable MC, Bell B, Hirst M, Sadowski I. Naturally occurring human immunodeficiency virus type 1 long terminal repeats have a frequently observed duplication that binds RBF-2 and represses transcription. J Virol. 1998 (72):6465-6474
28. Peterson KJ, Lyons JB, Nowak KS, Takacs CM, Wargo MJ, McPeek MA. Estimating metazoan divergence times with a molecular clock. Proc Natl Acad Sci U S A. 2004 (101):6536-6541
29. Poon IK, Jans DA. Regulation of nuclear transport: central role in development and transformation? Traffic. 2005 (6):173-186
30. Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999 (234):187-208
31. Reynolds L, Ullman C, Moore M, Isalan M, West MJ, Clapham P, Klug A, Choo Y. Repression of the HIV-1 5' LTR promoter and inhibition of HIV-1 replication by using engineered zinc-finger transcription factors. Proc Natl Acad Sci U S A. 2003 (100):1615-1620
32. Chen J, Malcolm T, Estable MC, Roeder RG, Sadowski I. TFII-I regulates induction of chromosomally integrated human immunodeficiency virus type 1 long terminal repeat in cooperation with USF. J Virol. 2005 (79):4396-4406
33. Estable MC, Hirst M, Bell B, O'Shaughnessy MV, Sadowski I. Purification of RBF-2, a transcription factor with specificity for the most conserved cis-element of naturally occurring HIV-1 LTRs. J Biomed Sci. 1999 (6):320-332
Author contact
![]() Correspondence to: Dr. Mario C. Estable, E-mail: mestableca; Telephone: 416-979-5000 extension 4517; Fax: 416-979-5044
Correspondence to: Dr. Mario C. Estable, E-mail: mestableca; Telephone: 416-979-5000 extension 4517; Fax: 416-979-5044

 Global reach, higher impact
Global reach, higher impact