10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2007; 3(7):471-476. doi:10.7150/ijbs.3.471 This issue Cite
Research Paper
The Growth-hormone inducible transmembrane protein (Ghitm) belongs to the Bax inhibitory protein-like family
Department for Plastic, Hand and Reconstructive Surgery, Medical School Hannover, Podbielskistraße 380, D-30659 Hannover, Germany
*These authors contributed equally.
Received 2007-7-23; Accepted 2007-11-20; Published 2007-11-22
Abstract
The conserved protein domain UPF0005 is a protein family signature distributed among many species including fungi and bacteria. Although of unknown functionality this motif has been found in newly identified antiapoptotic proteins comprising the BI-1 family, namely Bax-inhibitory Protein-1 (BI-1), Lifeguard (LFG), and h-GAAP. In a search for vertebrate proteins presumably belonging to the BI-1 family, we found that Growth-hormone inducible transmembrane protein (Ghitm) is another prospective member of the BI-1 family. Here we characterise Ghitm in a first analysis regarding its phylogeny, expression in cancer cell lines, and proteomical properties.
Keywords: protein family, Bax-inhibitor, programmed cell death, bioinformatics, apoptosis inhibitors, transmembrane protein
1. Introduction
The evolutionarily conserved motif UPF0005, although functionally uncharacterised so far, is found in a protein family wildly distributed among all kingdoms. Many proteins bearing this motif have been identified and incorporated into the public databases including three novel antiapoptotic proteins, Bax Inhibitor-1 (BI-1), Lifeguard (LFG), and h-GAAP. It has been proposed that BI-1 and LFG and probably other closely related proteins represent a new family of cytoprotective proteins called BI-1 family [1].
Due to the hydrophobic nature of the UPF0005 motif, it has been predicted that it involves six to eight probable transmembrane spanners. Experimental evidence as well as bioinformatic analysis indicate that proteins belonging to this family might reside in the membrane of the endoplasmatic reticulum and are involved in the regulation of cell death control by the Bcl-2 family [2-4].
BI-1 is the far most best characterised member of the BI-1 family. The antiapoptotic function of BI-1 was detected by a screen for functional repressors of the proapoptotic properties of Bax in yeast [4]. LFG has been shown to protect against Fas-induced apoptosis in mammalian cells [5]. H-GAAP is an inhibitor against intrinsic and extrinsic stimuli with a highly conserved counterpart found in vaccinia virus [6].
BI-1 is up-regulated in several malignant tumors including pulmonary adenocarcinoma, breast cancer, lymphoma and prostate cancer, whilst Ghitm was found to be over-expressed in breast cancers [7-10].
For a better characterisation of the BI-1 family, we searched the database for proteins containing the UPF0005 family signature. We emphasized that the proteins should have homologues in higher vertebrate species and expression data might indicate a functional role in the regulation of crucial cellular processes. One of the proteins found was the Growth hormone-inducible transmembrane protein (Ghitm). Ghitm was first identified in the brown adipose tissue from mouse [9]. Ghitm is ubiquitously expressed in mammalian cells with relatively low expression in intestine and thymus [11]. It was proposed that it functions in tumorigenesis and in adipose tissues [9], although a functional mechanism has not been described.
We analysed the relation between LFG, BI-1, and Ghitm in a phylogenetic analysis and used bioinformatical tools to summarise the molecular characteristics of Ghitm. An expression analysis for ghitm transcripts was performed for some cancer cell lines.
2. Material and Methods
Phylogenetic tree construction
Amino acid sequences used in CLUSTAL W analysis and phylogenetic analysis with their accession numbers are given in brackets: C. Elegans F40F9 (CAA94766), chicken LFG (XP_424507), human LFG (NP_036438), mouse LFG (NP_082500), rat LFG (NP_653357), Xenopus BI-1 (AAH47131), human BI-1 (AAH36203), rat BI-1 (P55062), mouse BI-1 (AAH05588), macaque BI-1 (AAV98554), Xenopus Ghitm (NP_001017195), Xenopus Ghitm-prov (AAH41226), human Ghitm (CAH72661), human Ghitm (CAG38550), rat Ghitm (NP_001005908), mouse Ghitm (NP_510963), dog Ghitm (XP_536408), cow Ghtim (NP_001029224), zebrafish Ghitm (NP_956885), chicken Ghitm (NP_001026388), dog GAAP (XP_531662), human GAAP (AAF14868), mouse glutamate receptor (NP_075657), putative MAPK activating protein (BAC77379), human RECS1 (Q969X1). Sequences were aligned using ClustalW. The software package PHYLIP 3.64 was used for phylogenetic analysis. The Protdist program with Jones-Taylor-Thornton matrix was used to generate a distance matrix. The neighbor-joining algorithm [12] and the maximum likelihood method intergrated in the software tools Neighbor-joining and PROTML, respectively were used for generation of phylogenetic trees. Seqboot and Consense programs were used to statistically assess the strength of the trees using bootstrap resampling. A consensus tree was viewed with TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). ClustalW was used to determine the percentage of sequence identity between the amino acid sequences of Ghitm from different vertebrate species.
Sequence analysis
NCBI RPS-BLAST was used to search for conserved domain database screening (http://www.ncbi.nlm.nih.gov/structure/cdd/wrpsb.cgi).
Sequence analysis was performed with the primary amino acid sequence of human and mouse Ghitm. A Kyte and Doolittle hydropathic plot [13] was generated with ProtScale (http://www.expasy.org/cgi-bin/protscale.pl) with Kyte and Doolittle option. Protein topology was predicted with [14, 15] TOPPRED2 (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html), TMPRED (http://www.ch.embnet.org/software/TMPRED_form.html), TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) HMMTOP (http://www.enzim.hu/hmmtop/html/adv_submit.html), and SMART (http://smart.embl-heidelberg.de).
SignalP was used for the prediction of signal peptides and the possible localisation was calculated with PSORT II [16]: http://psort.nibb.ac.jp/ SignalP: (http://www.cbs.dtu.dk/services/SignalP/) Target P: http://www.cbs.dtu.dk/services/TargetP/). Sequence pattern and motif search was performed with the various tools collected at the EXPASY proteomics server (http://www.expasy.ch/).
Gene expression analysis
Total RNA was isolated from the cancer cell lines and reverse transcribed in a reaction containing 0.25 mM dNTP-mix, 1µg random hexamers, 20U recombinant StratascriptII (Stratagene) with 1x Stratascript buffer supplied by the manufacturer following instructions. 1µl of the reaction mixture was used in polymerase chain reaction with the specific primers (sense 5'-GGG CCT GGG TCT CGT CTT TG-3'; antisense 5'-ATC CAC CGT ACA TTG CCA CTG AG-3'). Cycling conditions were 35 cycles of 94°C for 30s, 65°C for 30s and 72°C for 1min 30s. As a reaction control primers for 18S rRNA were used. The amplification products were analysed on a 2% agarose gel supplemented with EtBr.
3. Results and Discussion
Ghitm is a new member of the BI-1 family
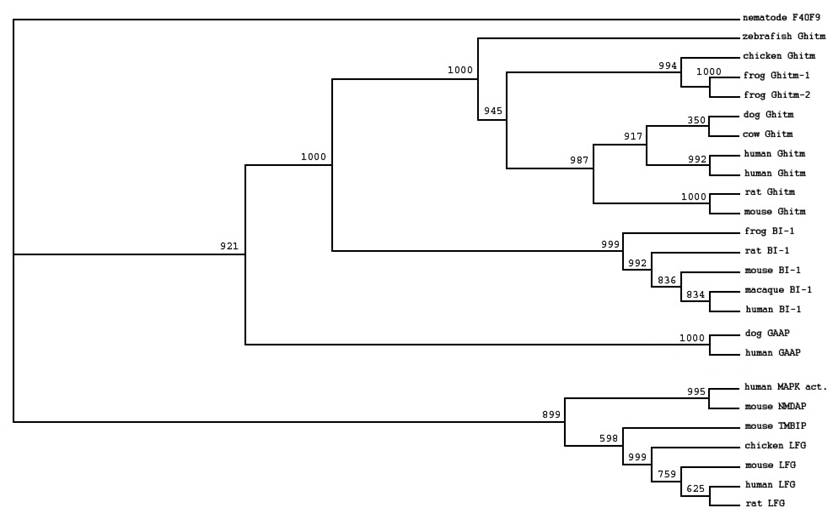
We identified members of the BI-1 family by searching the public databases using RPS-BLAST. Like all other family members Ghitm has the UPF0005 family signature at its C-terminal end. To characterise the relation between Ghitm, BI-1 and other members of the BI-1 family we selected sequences of different vertebrate species and arranged them in multiple alignments. Based on ClustalW alignment phylogenetic trees were constructed as described in the Materials and Methods section. The data are presented in a phylogenetic tree using the UPF0005 motif containing hypothetical protein F40F9 from C. Elegans as an outgroup (figure 1). Ghitm is found in a clade together with BI-1, while the recently described apoptosis inhibitory protein h-GAAP is more distantly related. Another group is build by the antiapoptotic protein LFG and the human so far uncharacterised putative MAPK activating protein and the related murine N-methyl D-asparate-associated protein 1.
All species are found in accordance with the genealogical tree.
Homologues of Ghitm have been found in many different vertebrate species as depicted in figure 1. Sequence identity values for the vertebrate homologues range between 60.3% (zebrafish and rat) and 93.4% (dog and human). For human and the clawed frog two nearly identical sequences have been found (human: 93.2%; clawed frog: 93.1%). Additional analysis is necessary to comment on the nature of the similarities and differences between these sequences.
A high degree of sequence conservation among different species is a typical feature for the members of the BI-1 family. Bolduc et al. showed that BI-1 homologues from Brassica napus and Nicotiana tabacum suppress Bax-induced apoptosis in human 293 cells. BI-1 inhibits mammalian Bax-induced cell death in yeast cells, and plants implying functional conservation in an ancient mechanism of cytoprotection [4, 17]. For LFG and h-GAAP homologues have been identified for many different species [1, 6]. For Ghitm – apart from the vertebrate sequences described in this study – sequences for non-vertebrate species have also been found such as fruitfly and nematode [11].
Phylogenetic tree of selected members of the BI-1 family based on ClustalW alignment of the deduced amino acid sequences derived from the public databases. Accession numbers were stated in the Methods section. An unnamed protein with UPF0005 motif from C. elegans (CAA94766F4) was used for tree rooting. Bootstrap values are given at each node.
Bioinformatical characterisation of Ghitm
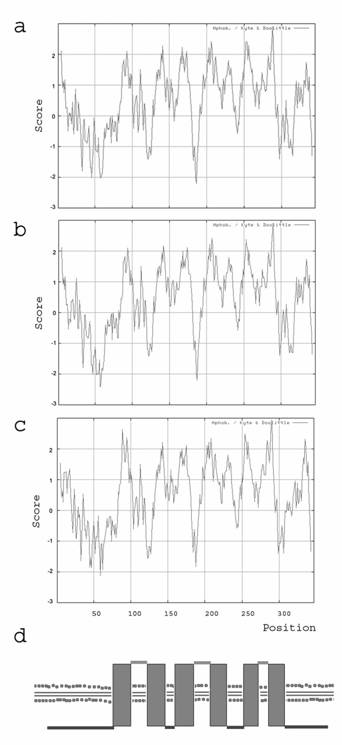
Kyte and Doolittle hydrophobicity plots have been carried out for the amino acid sequences of human, mouse and zebrafish. It shows a hydrophobic pattern for the examined species human, mouse and clawed frog with 6 dominant spikes (figure 2a-c). According to the Kyte and Doolittle pattern we assumed that Ghitm is a polytopic membrane protein with 6 transmembrane (TM) domains (figure 2d). The proposed topology had been arranged with the aid of various topology programs. Due to their predictions Ghitm belongs to the membrane proteins type 3a and probably resides either in the endoplasmatic reticulum (ER) or in the plasma membrane according to PSORT II, SignalP and TargetP. A subcellular localisation in the endoplasmatic reticulum has been assumed for BI-1 and LFG as well [1-3]. In contrast to the possible protein topology of LFG it was predicted for Ghitm that the N-terminus as well as the C-terminus are found on the cytoplasmatic side of the ER membrane, while the short N-terminus of LFG is probably found in the ER lumen.
Interestingly, it has been reported for Ghitm that it possesses an N-terminal signal peptide and that N-terminal cleavage is important for the regulation of protein expression. We carefully looked at the N-termini of selected sequences of vertebrate Ghitm. The signal peptide prediction program SignalP uses a combined neural network and hidden Markov models for the prediction of signal peptides and their cleavage sites. The data are expressed in score values expressing the output from the signal peptide/non signal peptide networks (S-score), the output from the cleavage site/non-cleavage site networks (C-score), and geometric average of the C-score and a smoothed derivative of the S-score, termed Y-score [18]. The prediction is summarised in a probability value. The N-terminal sequences of selected vertebrate Ghitm homologues are depicted in figure 3. The homologues of Ghitm found in the lower vertebrates are predicted to have a signal peptide, while the probability decreases during evolution. This is extremely interesting, taking into concern that Yoshida et al. found out that cleavage of the N-terminal signal peptide is essential for protein expression and function in the mouse [11]. A critical examination of the function of the signal peptide during the evolution of the Ghitm protein is needed.
In figure 4 an overview over the protein structure of four known members of the BI-1 family is given with the putative transmembrane domains underlined. It is clearly visible that, while all four proteins are highly similar, the two seven-span transmembrane proteins LFG and RECS1 are organized the same with a proline and glycine rich intracellular N-terminus containing several PXXP motifs and one potential PEST sequence [19]. The two six-span transmembrane proteins BI-1 and Ghitm differ more in their arrangement of putative transmembrane domains. N-terminus and C-terminus are predicted to be inside the cell and both are characterized by a number of basic residues: arginine 10% frequency, lysine 8.75% frequency at the N-terminus, arginine 5.66% frequency, lysine 9.34% frequency at the C-terminus. In contrast to RECS1 which has a proline content of 24% in the human sequence proline residues are found to a much lesser extent in Ghitm (N-terminus: 6.25%, C-terminus 3.77%). PXXP and PPXY motifs have not been found.
Ghitm expression in cancer cell lines
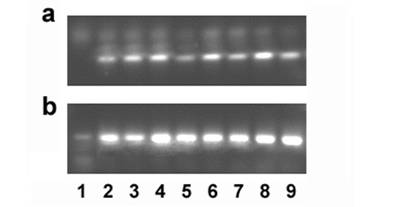
Ghitm gene expression in several cancer cell lines was determined by RT-PCR. Specific primers were designed to generate a 101bp fragment. Bands corresponding to the expected size are detected in the cDNAs derived from all cancer cell lines tested that is A549 lung carcinoma cells, ACTL lymphoma cells, HeLa cervix carcinoma, HepG2 hepatoma, SW872 liposarcoma, MCF-7 breast cancer, SW-620 colon carcinoma, and U2-OS osteosarcoma cells (figure 5).
A direct cytoprotective function of Ghitm has not been demonstrated so far, but observations published in the literature suggest a role in energy metabolism, and, with regard to its association to the growth hormone signalling, in aging and immunology [9, 11]. These functions might be linked to a cytoprotective role in the cellular accomplishment of ER stress but must be evaluated in corresponding experimental designs.
4. Conclusion
Although the precise function of the C-terminal domain UPF0005 of the BI-1 family is not known, a biological function is suggested by the high degree of sequence conservation between species. Computational analysis revealed that Ghitm shares the UPF0005 motif with the cytoprotective members of the BI-1 family members, and is closely related to BI-1, h-GAAP, and LFG. Although there is still a need for more experimental evidence these proteins have many molecular characteristics in common. The predicted protein topology indicates that they are transmembrane proteins of the type 3a with a variable number of transmembrane spanners from six to eight localized in the endoplasmatic reticulum. Variablity was found in the N-termini probably due to modifications in biological mechanisms which have yet to be defined. Further studies are needed to elucidate the physiological and probably pathological function of Ghitm.
Protein topology prediction based on the hydrophobicity of the deduced amino acid sequences. Kyte and Doolittle plots of the deduced amino acid sequences of human (a), mouse (b), and clawed frog (c). The putative TM segments are visible as hydrophobe spikes. (d) Calculated transmembrane domain organisation of Ghitm with six membrane passages and cytoplasmic N- and C-terminal ends.
Ghitm N-terminal sequences of selected vertebrate species aligned with ClustalW. The possible cleavage site as determined with SignalP is indicated by an apostrophe. The outcome of the probability score calculated with SignalP is given at the right.

Deduced amino acid sequences of the human BI-1, LFG, RECS1, and Ghitm aligned with Clustal W. Identical amino acids are illustrated by stars; points indicate conservative changes. The putative transmembrane domains calculated by TMHMM are indicated by vertical bars below the sequences.

Gene expression of Ghitm is found in several cancer cell lines (a). 18S rRNA amplification is used as a reaction control (b). 1 no template control, 2 A549, 3 ACTL, 4 HeLa, 5 HepG2, 6 MCF-7, 7 SW-620, 8 SW-872, 9 U2-OS
Acknowledgements
The work was funded by the Claudia-von-Schilling Breast Cancer Foundation. The authors are grateful to Christina Allmeling for helpful comments on manuscript and figures and thank Andrea Lazaridis for help with the gene expression analysis.
Conflict of interest
The authors have declared that no conflict of interest exists.
References
1. Reimers K, Choi CY, Mau-Thek E. et al. Sequence analysis shows that Lifeguard belongs to a new evolutionarily conserved cytoprotective family. Int J Mol Med. 2006;18:729-34
2. Chae HJ, Ke N, Kim HR. et al. Evolutionarily conserved cytoprotection provided by Bax Inhibitor-1 homologs from animals, plants, and yeast. Gene. 2003;323:101-13
3. Ihara-Ohori Y, Nagano M, Muto S. et al. Cell death suppressor Arabidopsis bax inhibitor-1 is associated with calmodulin binding and ion homeostasis. Plant Physiol. 2007;143:650-60
4. Xu Q Reed JC. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell. 1998;1:337-46
5. Somia NV, Schmitt MJ, Vetter DE. et al. LFG: an anti-apoptotic gene that provides protection from Fas-mediated cell death. Proc Natl Acad Sci U S A. 1999;96:12667-72
6. Gubser C, Bergamaschi D, Hollinshead M. et al. A new inhibitor of apoptosis from vaccinia virus and eukaryotes. PLoS Pathog. 2007;3:e17
7. Grzmil M, Thelen P, Hemmerlein B. et al. Bax inhibitor-1 is overexpressed in prostate cancer and its specific down-regulation by RNA interference leads to cell death in human prostate carcinoma cells. Am J Pathol. 2003;163:543-52
8. Grzmil M, Kaulfuss S, Thelen P. et al. Expression and functional analysis of Bax inhibitor-1 in human breast cancer cells. J Pathol. 2006;208:340-9
9. Li Y, Kelder B, Kopchick JJ. Identification, isolation, and cloning of growth hormone (GH)-inducible interscapular brown adipose complementary deoxyribonucleic acid from GH antagonist mice. Endocrinology. 2001;142:2937-45
10. Tanaka R, Ishiyama T, Uchihara T. et al. Expression of the Bax inhibitor-1 gene in pulmonary adenocarcinoma. Cancer. 2006;106:648-53
11. Yoshida T, Nagata S, Kataoka H. Ghitm is an ortholog of the Bombyx mori prothoracic gland-derived receptor (Pgdr) that is ubiquitously expressed in mammalian cells and requires an N-terminal signal sequence for expression. Biochem Biophys Res Commun. 2006;341:13-8
12. Saitou N Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406-25
13. Kyte J Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105-32
14. Kall L Sonnhammer EL. Reliability of transmembrane predictions in whole-genome data. FEBS Lett. 2002;532:415-8
15. Melen K, Krogh A, von Heijne G. Reliability measures for membrane protein topology prediction algorithms. J Mol Biol. 2003;327:735-44
16. Nakai K Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34-6
17. Bolduc N, Ouellet M, Pitre F. et al. Molecular characterization of two plant BI-1 homologues which suppress Bax-induced apoptosis in human 293 cells. Planta. 2003;216:377-86
18. Nielsen H, Engelbrecht J, Brunak S. et al. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1-6
19. Zhao H, Ito A, Kimura SH. et al. RECS1 deficiency in mice induces susceptibility to cystic medial degeneration. Genes Genet Syst. 2006;81:41-50
Author contact
![]() Correspondence to: Kerstin Reimers, Ph.D., Klinik für Plastische, Hand- und Wiederherstellungschirurgie, Medizinische Hochschule Hannover, Podbielskistraße 380, D-30659 Hannover. Phone: +49-511-906 3917; Fax: +49-511-906 3480; E-mail: reimers.kerstinde
Correspondence to: Kerstin Reimers, Ph.D., Klinik für Plastische, Hand- und Wiederherstellungschirurgie, Medizinische Hochschule Hannover, Podbielskistraße 380, D-30659 Hannover. Phone: +49-511-906 3917; Fax: +49-511-906 3480; E-mail: reimers.kerstinde

 Global reach, higher impact
Global reach, higher impact