Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2009; 5(1):28-33. doi:10.7150/ijbs.5.28 This issue Cite
Short Research Communication
Differential display of expressed genes reveals a novel function of SFRS18 in regulation of intramuscular fat deposition
1. College of Animal Science & Technology, Nanjing Agricultural University, Nanjing 210095, China
2. Key Laboratory of Animal Physiology and Biochemistry, Nanjing Agricultural University, Nanjing 210095, China
3. Department of Animal Sciences, Washington State University, Pullman, WA 99164-6351, USA
Received 2008-10-29; Accepted 2008-12-15; Published 2008-12-17
Abstract
Intramuscular fat (IMF) content plays a key role in establishing pork quality. In the present study, differential-display reverse transcription-polymerase chain reaction (DDRT-PCR) was used to identify differentially expressed (DE) genes between longissimus dorsi (LD) muscles with extremely different IMF content. A major DE gene associated with IMF content was identified as splicing factor serine-arginine rich protein (SFRS18) gene, also known as SRrp130. The gene exhibited relatively higher expression levels in LD muscles with higher IMF content. A full-length cDNA sequence of pig SFRS18 gene was obtained by in silico comparative cloning coupled with PCR target sequencing, while the current EST (expressed sequence tag) database supported two transcript variants of the pig gene. Differential expression of the SFRS18 gene was further confirmed using quantitative PCR. The mRNA levels of SFRS18 gene showed significant and positive correlation with IMF content in LD muscle (r = 0.54, P < 0.01). Collectively, these results suggest that the SFRS18 gene is involved in the regulation of IMF deposition in pig and that it may be a useful tool in selecting animals for desired amounts of fatness for high quality pork.
Keywords: SFRS18, differential display, IMF, muscle, expression level
1. Introduction
The quality of red meats is partially determined by the type and amount of fat contained within the intramuscular fat (IMF) depot (1), and the IMF depot in red meat animals is one of the body fat depots that is desired. However, in an effort over the years to decrease the overall inefficient use of energy fed to red meat animals, cattle and pig production regimens have surprisingly reduced the IMF depot (1). Consequently, cattle and pigs must be fed even longer in order to obtain the correct amount of IMF for the quality that consumers prefer, and are willing to purchase (1). Knowledge gained regarding regulation of adipogenesis or lipid metabolism of the IMF depot will facilitate increasing the characteristics of desired meat quality without the need for increased feeding of production animals.
Splicing factor, serine-arginine rich protein 18 (SFRS18), also known as SRrp130, is a newly identified serine-arginine (SR) protein in human that co-purifies with pinin, which is also a SR-related protein (2). The SR-rich C-terminal portion of SFRS18 contains two well-conserved, and seven degenerated RRSRSXSX repeats that are the characteristic of proteins of SR family. The C-terminus of SFRS18 also shows similarity to splicing factors belonging to the SR family. The SR domains are necessary for specific protein-protein interactions among SR proteins, which play key roles in the early steps of spliceosome assembly. These interactions aid in the recruitment and stabilization of snRNPs to splice pre-mRNA. In spite of possessing evolutionally conserved functional domains, SR proteins behave differently with each other. It has been shown that preincubation of a labeled pre-mRNA with a specific purified SR protein could result in commitment of the RNA to undergo splicing. For instance, a β-globin pre-mRNA was committed by preincubation with SC35 but not ASF/SF2 or other SR proteins, whereas HIV tat pre-mRNA was committed specifically by ASF/SF2 and not SC35 (3). However, little information is currently available to understand the particular physiological pathway in which SFRS18 might be involved. As such, the hypothesis examined by this study was that SFRS18 expression is positively correlated to the amount of IMF.
2. Materials and Methods
2.1 Samples collection and IMF quantification
The animal experiments and protocols used in this study were approved by the Nanjing Agricultural University Institutional Animal Care and Use Committee. Longissimus dorsi (LD) muscle samples were collected from 90 castrated Sutai pigs. Sutai is a newly developed pig breed which contains 50% Erhualian and 50% Duroc. All animals were raised in the Sutai Breeding Center located in Suzhou, Jiangsu Province. Sutai pigs were slaughtered at approximately 180 d of age. Samples were taken immediately (within 5 to 10 min of death) from the LD muscle at the last rib. A portion of the sample was put immediately into liquid nitrogen and then stored at −80 °C, whereas the remainder was used to determine IMF content. The IMF content was measured in triplicate via the Soxhlet extraction method using petroleum ether as the solvent (4).
The average IMF content in these 90 castrated Sutai pigs was 2.72 ± 0.13 %. Twelve animals, six with highest IMF and six with lowest IMF were selected as H and L groups. The IMF for L and H groups was 0.98 ± 0.09 % and 5.79 ± 0.35 %, respectively (P < 0.01).
2.2 RNA extraction and first strand synthesis
Total RNA was extracted from LD muscle using Trizol (Invitrogen, California, US). RNA pool was established for each group by mixing equal amounts of individual RNA samples. RNA was treated using DNase I (Takara, Dalian, China) before the first-strand cDNA was synthesized. The quality and fidelity were analyzed by agarose gel electrophoresis. Two μg of RNA were reverse-transcribed in a 25 μl reaction mixture at 42 ºC for 60 min with M-MLV reverse transcriptase (Promega, San Luis Obispo, US) using anchor primer: 5′ -AAG CTT TTT TTT TTT (A, G, C)-3′. The product was stored at -20 °C until use.
2.3 DDRT-PCR
DDRT-PCR was performed according to Liang and Pardee (5) with minor modifications. Reverse transcription reaction products (2 μl) were amplified (MyCyclerTM Thermal cycler, BIO-RAD, California, US) in the presence of one anchored primer and one arbitrary 13nt primer. Sample reactions were amplified by PCR in 36 primer combinations, using 3 anchored primers coupled to 12 arbitrary primers. Amplifications were performed by 4 min at 94 ºC, 40 cycles of 30 s at 94 ºC, 2 min at 42 ºC, 30 s at 72 ºC, and final extension for 10 min at 72 ºC. For each individual, differential display PCR reactions were performed in triplicate and PCR products were then separated on an 8% non-denaturing polyacrylamide gel in adjacent lanes and visualized by silver staining (6).
2.4 Cloning
Bands of interest were recovered from the gel and extracted using polyacrylamide Gel DNA Extraction Kit (TIANGEN, Beijing, China). The fragments were re-amplified with the same primer combination and PCR conditions described above. The re-amplified DNA was ligated to a PTZ57R/T plasmid (insT/AcloneTM PCR Product Cloning Kit, MBI, US), and JM109 Escherichia coli bacterium strain (Invitrogen, California, US) allowing blue-white selection to be transformed with the ligated constructs. Inserted DNA sequences of the plasmid clones were determined by automated sequencing (ABI3730, InVitrogen, Shanghai, China). All sequences were compared with the non-redundant GenBank database using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
The complete coding sequence was obtained for the pig SFRS18 gene by using a combination of an in silico comparative cloning with a PCR target cloning approach described previously by Jiang and colleagues (7). Basically, a BLAST search against the public databases using full-length cDNA sequences of human SFRS18 gene as a reference was performed to retrieve all pig orthologous sequences, particularly ESTs. These ESTs were then assembled and the gaps were filled with target amplification and sequencing.
2.5 RT- PCR
Reverse transcript polymerase chain reaction (RT-PCR) was used to verify the differential expression of the SFRS18 gene between two groups. First-strand cDNA synthesis parameters were described as above. PCR amplification was performed by 4 min at 94 ºC, 33 cycles of 30 s at 94 ºC, 30 s at 60 ºC, 30 s at 72 ºC, and final extension for 10 min at 72 ºC (MyCyclerTM Thermal cycler, BIO-RAD, California, US). The primer sets were designed according to the cloned sequences as follows: forward 5'-TTT CAT CCT CCT TAT TGG C-3' and reverse 5'-TGG AAG AGT CCT GCG TTT-3'. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the invariant control. The primers for GAPDH (GenBank accession number AF017079) are: forward 5'-CAC CAT CTT CCA GGA GC-3' and reverse 5'-CTT CAA GTG AGC CCC AG-3'.
2.6 Quantitative reverse transcriptional PCR (qRT-PCR)
To investigate the relation of the SFRS18 gene with the IMF content, 15 individuals with highest IMF content and 15 with lowest IMF content were exploited for mRNA expression analysis by real-time quantitative PCR system (Continuous fluorescence detector, MJ Research. Inc., BIO-RAD, San Francisco, US). A total of 20 μl reaction volume contained 200 nM primers, 2 μl cDNA and 10 μl SYBR Premix Ex TaqTM (Takara, Dalian, China). The PCR primers and amplified conditions are the same as RT-PCR. Amplification specificity was assessed by 8 % polyacrylamide gel electrophoresis. Experiments were performed in triplicate for each data point. The relative amount of mRNA was calculated with GAPDH mRNA as the invariant control (8). The independent T-test procedure (SPSS Inc., Chicago, IL) was used to compare the average relative expression level between two groups.
3 Results
3.1 Identification and Cloning of Pig SFRS18 gene
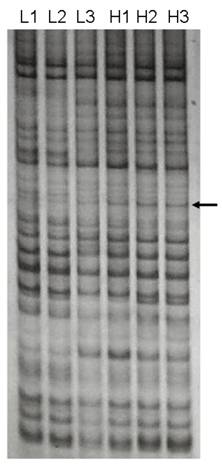
We identified a 691-bp cDNA fragment that appeared to be a visual DE gene between two LD muscle groups with significantly different IMF content (Figure 1). The cDNA fragment showed a higher expression in the high IMF group. Sequence analysis of the cDNA revealed that it is 99 % homologous to the human SFRS18 (GenBank Accession Number AF 314184) (2).
Although the SFRS18 complete coding sequence has been identified for humans, the corresponding sequence in pigs remains unknown. We initially used the human SFRS18 sequence to obtain pig ESTs from GenBank. Two contigs were established for 5' and 3' ends, but a gap was left between them. A pair of primers (forward, 5'-AGT AAA ACG CAG GAC TCT TC-3' and reverse, 5'-TTG CTT CTC TTC TTC CAT CT-3') was then designed to target the gap region. Finally, a sequence of 3227 bp was generated for the pig SFRS18 gene (GenBank Accession Number EU308569), including 215 bp of sequence for 5' untranslated region, 2,418 bp for opening reading frame and 594 bp for 3'untranslated region, respectively. The open reading frame encodes 805 deduced amino acids, which is 98, 98, 95 and 95 % identical to those of humans, monkeys, mice and rats, respectively.
DDRT-PCR revealed a differentially expressed cDNA. The cDNA was amplified from two total RNA pools with significantly different IMF contents in LD muscle by primer combination of 5' AAG CTT TTT TTT TTT C-3' (anchor primer) and 5'-AAG CTT GAT TGC C-3' (arbitrary primer). H1, H2, H3 were amplified products from the high IMF group, while L1, L2, L3 were from the low IMF group. Arrow indicates the differentially expressed cDNA between two groups.
3.2 An unusual splicing in the pig SFRS18 gene
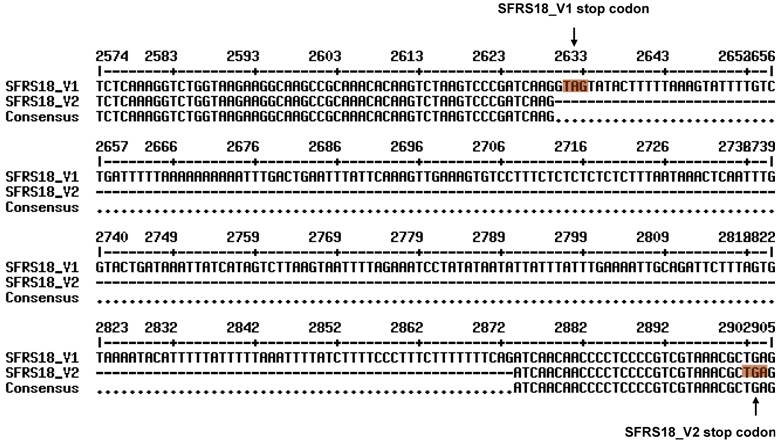
In order to further characterize the porcine SFRS18 gene, we used the cDNA sequence described above as a query and performed a BLAST search against the porcine EST database. Interestingly, we found this porcine gene might have two splicing forms, which were illustrated in Figure 2.
The transcript variant 2 turned the translation end region and partial 3' untranslated region of transcript variant 1 into a novel intron, which has a regular exon-intron boundary with GT (2,630 - 2,631) at the 5' splice site and AG (2,872 - 2,873) at the 3' splice site (Figure 2). Such a change also shifted the stop codon from positions 2,631 - 2,633 in variant 1 to the position 2,902 - 2,904 bp in the variant 2 (see masked nucleotides in Figure 2). As a consequence, transcript variant 2 is 244-bp shorter than variant 1 in length. However, both variants share a common protein with 805 amino acids, but the transcript variant 2 contains extra nine amino acids (STTPPRRKR) at its C terminus.
Two transcript variants detected by ESTs retrieved from pig EST database using our cloned SFRS18 mRNA as a reference sequence. Alternative stop codons are used in the translation of two transcript variants, which are marked in dark background. Our cloned sequence EU308569 represents pig SFRS18 transcript variant 1 (SFRS18_V1). It is supported by 15 additional ESTs in the GenBank database, including AJ948023, CK464951, CN164095, AJ938658, CK463016, CK463348, EW328287, EW423619, EW353476, BQ597517, AJ944568, AJ946990, EW509565, EW613877 and EW556555. Eight ESTs - AJ951908, AJ950607, AJ958378, AJ950217, AJ948941, AJ659463, AJ659329 and EW232314 contributed to the formation of transcript variant 2 of the pig SFRS18 (SFRS18_V2).
3.3 Confirmation of expression pattern by RT-PCR
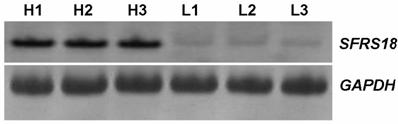
The expression levels of the SFRS18 gene were confirmed between two pools of animals by RT-PCR (Figure 3). The result of RT-PCR was clearly consistent with that of DDRT-PCR, supporting higher expression of SFRS18 gene in animals with high IMF content.
3.4 Real-time quantitative PCR
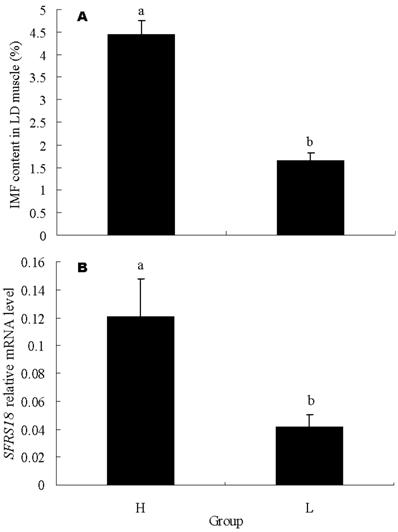
To further confirm the relation of SFRS18 gene with IMF content, 15 individuals with highest IMF content and 15 with lowest IMF content were exploited for mRNA expression analysis by real-time quantitative PCR. The IMF contents of these two groups averaged 1.65 ± 0.16 % vs 4.46 ± 0.29 %, and were significantly different (P < 0.05) (Figure 4A). The result showed that the SFRS18 expression level is positively correlated with IMF contents (r = 0.54, P < 0.01). The average mRNA levels are significantly different between two groups (P < 0.05) (Figure 4B).
RT-PCR analysis of SFRS18 gene. H1, H2, H3 were amplified products from the high IMF group and L1, L2, L3 were those from the low group. GAPDH was used as the invariant control.
Real-time PCR analysis of SFRS18 gene. (A) IMF contents in LD muscle of the two groups used for mRNA expression analysis by real-time quantitative PCR. Each group consisted of 15 individuals with highest IMF content or 15 with lowest IMF content. (B) SFRS18 gene relative mRNA level of the two groups. H and L stand for groups with high and low IMF content, respectively. Averages with different letters indicate significant difference (P<0.05).
4. Discussion
In this study, we found that differential expression of SFRS18 gene in muscle is correlated with IMF content in pigs. SFRS18 is a serine-arginine (SR) rich splicing factor, which exhibits speckled nuclear distribution that aligned with components of the pre-mRNA splicing machinery (2, 9). A working hypothesis of our result was that the SFRS18 gene is implicated in the pre-mRNA splicing of key genes regulating IMF deposition.
Within the mammalian cell nucleus, most splicing factors are concentrated in 20-40 distinct domains called speckles, which are highly dynamic structures that respond specifically to activate nearby genes and the dynamic events are dependent on RNA polymerase II transcription. Dynamic events of speckles are sensitive to inhibitors of protein kinases and Ser/Thr phosphatases. When single gene transcription is activated in living cells, splicing factors leave speckles and accumulate at the new site of transcription (10). Although having similar structures and conserved domains, SR proteins are specific in their behaviors (3).
A number of studies have demonstrated that pre-mRNA splicing occurs co-transcriptionally (11-14). For instance, Dellaire et al. (2002) propose that pre-mRNA processing protein 4 kinase (PRP4K) links transcriptional regulation and pre-mRNA splicing by modulating the coordination of both processes via phosphorylation of proteins within the spliceosome and the co-regulator complex (15).
SFRS18 is one of the components in pre-mRNA spliceosome (2). Although no direct evidence indicates that PRP4K directly modulates SFRS18, PRP4K does interact with pinin protein (15), which is localized to spliceosome speckles in mammals (16) with interactions with SFRS18 (2). Moreover, PRP4K can co-immunoprecipitate with nuclear receptor co-activator complex switching defective/sucrose nonfermenting (SWI/SNF) component of Brahma-related gene 1 (BRG1) (15). The expression of a dominant negative BRG1 blocks adipogenesis in adipocytes and inhibits the expression of PPARγ by suppressing the recruitment of general transcription factors and RNA polymerase II at the PPARγ promoter and affects the expression level of downstream genes, such as fatty acid binding protein (AP2) (17). Despite its extremely low level, mouse muscle PPARγ is critical for the maintenance of normal adiposity by modulating muscle lipid metabolism (18, 19). Collectively, we proposed that SFRS18 may participate in the splicing of some key genes that regulate muscle lipid metabolism in pigs.
As an extension of our original study, we devised a comparative map between what is known in humans to the pig. This map shows that the SFRS18 gene should be located around 54-55 cM on Sus scrofa chromosome 1 (SSC1). Interestingly, this location harbors a QTL region for marbling in pigs, which flanks a region from 52-65 cM with a peak at 52 cM (20). The current draft pig genome assembly (http://www.ncbi.nlm.nih.gov/genome/guide/pig/) also indicates that the pig SFRS18 gene is located at 39.85 Mb on SSC1 (see NW_001886145). Therefore, future research with both the SFRS18 gene product and the QTL locus might provide a direct link between the two and serve as sites with which to target for manipulation of intramuscular adiposity in red meat animals.
In conclusion, the collective results from this study provide knowledge about a potentially novel function of SFRS18, whose expression level in pigs is positively correlated with IMF content in LD muscle. Therefore, we suggest that SFRS18 might be a promising candidate gene for improving pig IMF deposition.
Acknowledgements
This study was supported by National Natural Science Foundation of China (No. 30800780). This study was also supported by hi-tech research and development program of China (2007AA10Z100, 2006AA10Z136) and National Basic Research Program of China (2004CB117505). This activity was also funded, in part, with an Emerging Research Issues Internal Competitive Grant from the Washington State University, College of Agricultural, Human, and Natural Resource Sciences, Agricultural Research Center to Z.J.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Hausman GJ, Dodson MV, Ajuwon K. et al. Board Sponsored Invited Review: The biology and regulation of preadipocytes and adipocytes in meat animals. J Anim Sci. 2008 [Epub ahead of print]
2. Zimowska G, Shi J, Munguba G. et al. Pinin/DRS/memA Interacts with SRp75, SRm300 and SRrp130 in Corneal Epithelial Cells. Invest Ophthalmol Vis Sci. 2003;44:4715-4723
3. Fu XD. Specific commitment of different pre-mRNA to splicing by single SR proteins. Nature. 1993;365:82-85
4. AOAC. Official Methods of Analysis of the Association of Analytical Chemists, 17th rev ed. Washington, DC: AOAC Int. 2000
5. Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967-971
6. Jiang Z, Jouquand S. Galibert F. Identification and characterization of a SSCP marker at the canine signal sequence receptor beta subunit gene. Anim Genet. 1997;28:460-461
7. Jiang Z, Kunej T, Michal JJ. et al. Significant associations of the mitochondrial transcription factor A (TFAM) promoter polymorphisms with marbling and subcutaneous fat depth in Wagyu x Limousin F2 crosses. Biochem Biophys Res Commun. 2005;334:516-523
8. Louet JF, Coste A, Amazit L. et al. Oncogenic steroid receptor coactivator-3 is a key regulator of the white adipogenic program. Proc Natl Acad Sci U S A. 2006;103:17868-17873
9. Brandner JM, Reidenbach S, Kuhn C. et al. Identification and characterization of a novel kind of nuclear protein occurring free in the nucleoplasm and in ribonucleoprotein structures of the “speckle” type. Eur J Cell Biol. 1998;75:295-308
10. Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523-527
11. Ge H, Si Y, Wolffe AP. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol Cell. 1998;2:751-759
12. Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415-1429
13. Monsalve M, Wu Z, Adelmant G. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell. 2000;6:307-316
14. Martinez E, Palhan VB, Tjernberg A. et al. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with premRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782-95
15. Dellaire G, Makarov EM, Cowger JJ. et al. Mammalian PRP4 kinase copurifies and interacts with components of both the U5 snRNP and the N-CoR deacetylase complexes. Mol Cell Biol. 2002;22:5141-5156
16. Brandner JM, Reidenbach S, Kuhn C. et al. Identification and characterization of a novel kind of nuclear protein occurring free in the nucleoplasm and in ribonucleoprotein structures of the “speckle” type. Eur J Cell Biol. 1998;75:295-308
17. Salma N, Xiao H, Mueller E. et al. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor g nuclear hormone receptor. Mol Cell Biol. 2004;24:4651-4663
18. Norris AW, Chen L, Fisher SJ. et al. Muscle-specific PPAR gamma deficient mice develop increased adiposity and insulin resistance but respond to thiazolidinediones. J Clin Invest. 2003;112:608-618
19. Kokta TA, Dodson MV, Gertler A, Hill RA. Review: Intercellular signaling between adipose tissue and muscle tissue. Domest Anim Endocrinol. 2004;27:303-331
20. Kim JJ, Zhao H, Thomsen H, Rothschild MF, Dekkers JC. Combined line-cross and half-sib QTL analysis of crosses between outbred lines. Genet Res. 2005;85:235-248
Author contact
![]() Correspondence to: Dr. Jie Chen, Tel: +86 25 84395278; Fax: +86 25 84395314; E-mail: jiech1119edu.cn
Correspondence to: Dr. Jie Chen, Tel: +86 25 84395278; Fax: +86 25 84395314; E-mail: jiech1119edu.cn

 Global reach, higher impact
Global reach, higher impact