Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2009; 5(1):64-73. doi:10.7150/ijbs.5.64 This issue Cite
Research Paper
SpolvlgA is a DDX3/PL10-related DEAD-box RNA helicase expressed in blastomeres and embryonic cells in planarian embryonic development
Departament de Genètica, Facultat de Biologia, Universitat de Barcelona, Av. Diagonal 645, Edifici Annex, Planta 1, 08028 Barcelona, Catalunya, Spain
Received 2008-10-28; Accepted 2009-1-2; Published 2009-1-6
Abstract
Planarian flatworms have an impressive regenerative power. Although their embryonic development is still poorly studied and is highly derived it still displays some simple characteristics. We have identified SpolvlgA, a Schmidtea polychroa homolog of the DDX3/PL10 DEAD-box RNA helicase DjvlgA from the planarian species Dugesia japonica. This gene has been previously described as being expressed in planarian adult stem cells (neoblasts), as well as the germ line. Here we present the expression pattern of SpolvlgA in developing embryos of S. polychroa and show that it is expressed from the first cleavage rounds in blastomere cells and blastomere-derived embryonic cells. These cells are undifferentiated cells that engage in a massive wave of differentiation during stage 5 of development. SpolvlgA expression highlights this wave of differentiation, where nearly all previous structures are substituted by blastomere-derived embryonic cells. In late stages of development SpolvlgA is expressed in most proliferating and differentiating cells. Thus, SpolvlgA is a gene expressed in planarian embryos from the first stages of development and a good marker for the zygote-derived cell lineage in these embryos. Expression in adult worms is also monitored and is found in the planarian germ line, where it is showed to be expressed in spermatogonia, spermatocytes and differentiating spermatids.
Keywords: planarian, embryogenesis, neoblasts, germ line, PL10, DDX3.
Introduction
The embryonic development of planarians has remained poorly studied for years. Planarian embryonic development is highly derived from the standard spiral cleavage of other lophotrochozoan lineages [1]. It has some simple characteristics consistent with their suggested phylogenetic position [2], such as the presence of nuage material in early blastomeres [3, 4]. In the last ten years there has been a renewed interest in planarians as a model for regeneration [5-8] and this in turn has led to renewed efforts to understand developmental processes in planarian embryos [3, 4, 9].
Sexual hermaphrodite planarians lay polyembryonic ectolecithal egg capsules, each of them containing typically two or three zygotes in the planarian species Schmidtea polychroa. Zygotes cleave into different blastomeres which freely spread through a syncytium formed by fused yolk cells around the embryo. During the early stages of development blastomeres and blastomere-derived embryonic cells divide in the syncytium. A dramatic transition takes place during stage 5, when a massive wave of differentiation occurs. Embryonic cells start to differentiate in all cell types. During stages 6-8 embryonic cells proliferate and differentiate to finally form a worm-shaped last stage embryo.
Planarian blastomeres and blastomere-derived embryonic cells possess chromatoid bodies, electron dense granules similar to the ones in planarian adult somatic stem cells, the neoblast cells [4, 10, 11]. These structures are assumed to be analogous to germ granules such as polar granules in Drosophila melanogaster, P-granules in C. elegans and chromatoid body in mouse, collectively called nuage [12, 13]. Nuage is believed to be a structure involved in post-transcriptional silencing, RNA editing, transposon silencing and maintenance of undifferentiated cell states. Its conservation through animal kingdom is also observed at the molecular level, with the expression of components such as Vasa [14, 15] and Tudor [16-19].
Dugesia japonica vasa-like gene A (DjvlgA) is a planarian gene belonging to the DDX3/PL10 DEAD-box RNA helicases [20, 21], a family closely related to Vasa. DjvlgA is expressed in the planarian germ line and in neoblasts. PL10 homologues are maternally deposited in the germ line of some animals [22-24] though their expression is not as restricted to the germ line as Vasa expression [25]. Here we present the characterization of Schmidtea polychroa vasa-like gene A (SpolvlgA), a homologue of DjvlgA, during the embryonic development of S. polychroa, and show that it is expressed in blastomeres and embryonic cells throughout development.
Materials and methods
Animals and egg capsules
Schmidtea polychroa (Schmidt, 1862) individuals and egg capsules were collected in Sot de Ferrer (Castelló, Spain). Animals were kept in glass containers with 1:1 tap water:distilled water and fed beef liver twice a week.
Fixation
Alive egg capsules were laid on a glass surface, superficially perforated with a 0.5-mm needle, and immediately fixed for 4 h at 4°C in 4% formaldehyde in PBS. After an overnight wash in PBS at 4°C, egg capsules were stored at 4°C in 70% alcohol. When needed, they were rehydrated with PBS and dissected under the scope, using tweezers.
Isolation of SpolvlgA and phylogenetic analysis
We used previously described degenerate primers [21] to amplify a fragment of the SpolvlgA gene from S. polychroa cDNA, made from total RNA (Trizol) using MMLV reverse transcriptase (Promega). Due to the high conservation of the central region of SpolvlgA we performed 3' RACE reactions amplifying the less conserved 3' region by means of specific primers and a poly-T primer linked to an anchor sequence. Phylogenetic analyses were performed with ClustalW aligments and MEGA4 software. A phylogenetic tree was inferred using Neighbour-Joining algorithms.
In situ hybridization
Antisense and sense digoxigenin RNA probes were synthesized in vitro from the 3' region of SpolvlgA with T7 and SP6 RNA polymerases (Roche). Whole mount in situ hybridizations and in situ hybridization on paraffin sections were carried out as described previously [9, 26]. Fluorescent in situ hybridizations were incubated overnight with antidigoxigenin-POD (Roche) at a 1:100 dilution and developed with the TSA system (Perkin Elmer).
Imaging
Whole mount animals were observed with a Leica MZ16F stereomicroscope and sections were observed with a Zeiss Axiophot microscope. Images were captured with a Leica DFC300FX camera. Fluorescent slides were counterstained with DAPI (Molecular Probes), mounted in Vectashield (Vector Labs) and imaged with a Leica TCS SP2 confocal microscope. Images were processed with Image J and Adobe Photoshop software.
Results and discussion
Isolation and phylogenetic analysis of SpolvlgA
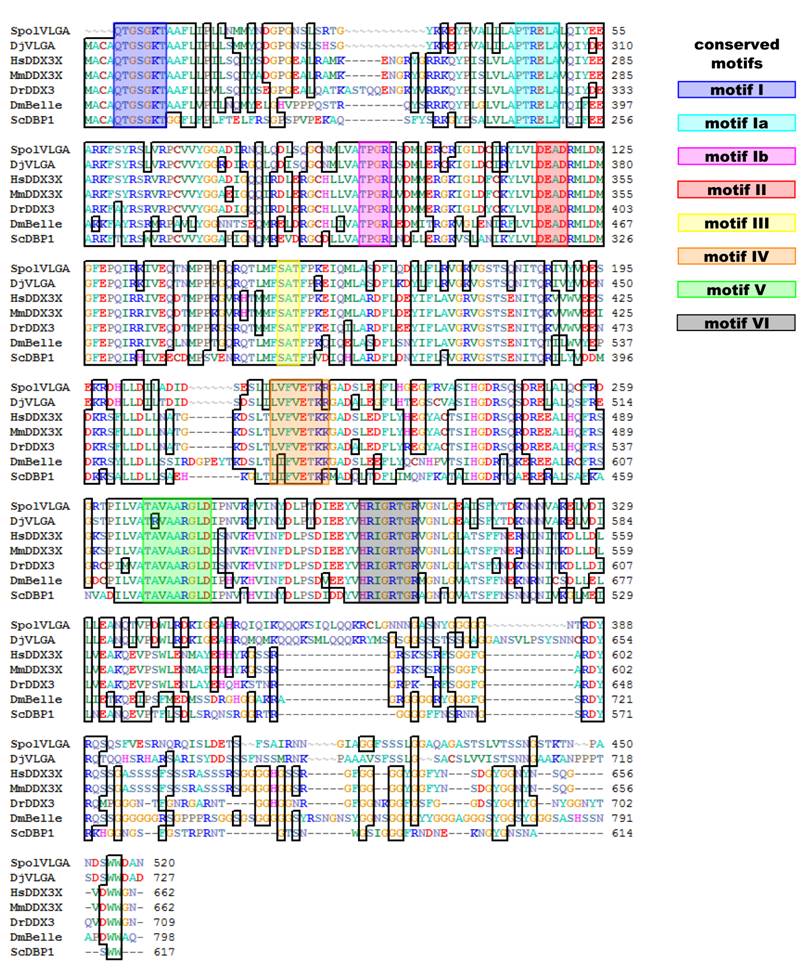
We have isolated a 1599 bp clone comprising the central region, the 3' region and the 3' UTR of SpolvlgA (Fig. 1). The isolated sequence contains all 8 conserved motifs typical of DEAD-box RNA helicases and presents a 78.9% amino acid identity to DjvlgA, with this value higher than 90% in the central region.
There has been controversy about the phylogenetic relationships of DjvlgA and its homologues [20, 21]. Our analyses indicated that SpolvlgA clusters together with the DDX3/PL10 RNA helicases rather than with Vasa and its homologues, with a 100% bootstrap value (Fig. 2). However, the position in this group is basal, clustering as an outgroup of the group formed by PL10 homologues and DBP1, a yeast homologue of the group. This result might be due to a long-branch effect and suggests rapid evolution of SpolvlgA.
Alignment of SpolvlgA central and C-terminal region with other DDX3/PL10 genes. Conserved amino acids are indicated with boxes. Known conserved motifs are indicated with color boxes. Sequences used for the alignment indicating organism and accession number: SpolvlgA, Schmidtea polychroa, FJ599746; DjVLGA, Dugesia japonica, AB017002; HsDDX3X, Homo sapiens, NM_001356; MmDDX3X, Mus musculus, NM_010028; DrDDX3, Danio rerio, BC133162; DmBelle, Drosophila melanogaster, NM_080522; ScDBP1, Saccharomyces cerevisiae, NP_015206.
Neighbor-joining phylogenetyc tree of DDX3/PL10 RNA helicases and DDX4/Vasa RNA helicases. The two groups of RNA helicases are indicated by color boxes. Bootstrap values are given in each branch. Sequences used for the phylogenetical analyses indicating organism and accession number: see Fig. 1 for DDX3/PL10 sequences; DmVasa, Drosophila melanogaster, NM_165103; DrVasa, Danio rerio, NM_131057; HsDDX4, Homo sapiens, NM_024415; MmDDX4, Mus musculus, NM_010029.
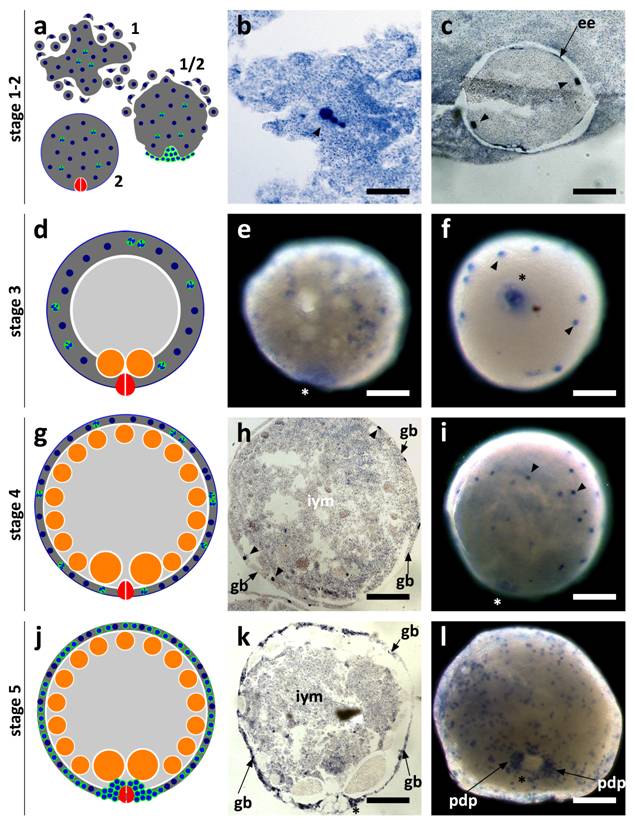
SpolvlgA is expressed in blastomeres and embryonic cells in early stages of planarian development
SpolvlgA expression was monitored by in situ hybridization both on whole mount embryos and on paraffin sections. We found that the expression of SpolvlgA could be detected as early as in stage 1 embryos (Fig. 3a-b, arrowhead). At this stage the zygote has cleaved into several blastomeres that do not stay close to each other, in a phenomenon called “blastomere anarchy”. At the same time a syncytial yolk mass is forming around them by means of cell fusion. In stage 2 the whole syncytial mass is covered by an embryonic epidermis that separates the syncytium containing a few SpolvlgA-labeled blastomeres from the rest of unfused yolk cells (Fig. 3c). By stage 3 (Fig. 3d) the newly formed embryonic pharynx ingests unfused yolk cells and as a consequence of this phenomenon a cavity is formed on the inner part of the embryo. Blastomere-derived embryonic cells located in the inner part of the embryo at early stage 3 (Fig. 3e) migrate to the periphery and are afterwards compressed to it by the ingested yolk mass (Fig. 3f), displaying a more peripheral and defined labeling. The signal displayed in the embryonic pharynx (Fig. 3e-f, asterisk) is likely a non-specific effect and it is also observed in negative controls performed with sense probes (data not shown).
As stage 4 proceeds (Fig. 3g) there is an increase in size and cell number in the germ band. The embryo also grows in size due to ingestion of yolk cells by the embryonic pharynx. Embryonic cells by stage 4 and previous stages are labeled throughout the germ band and display a regular cellular shape and size (Fig. 3h-i). In stage 5 (Fig. 3j) the number of SpolvlgA-positive embryonic cells is still growing (Fig. 3k-l). A cluster of embryonic cells accumulates on the area surrounding the embryonic pharynx (Fig. 3l), which corresponds with the primordium of the definitive pharynx.
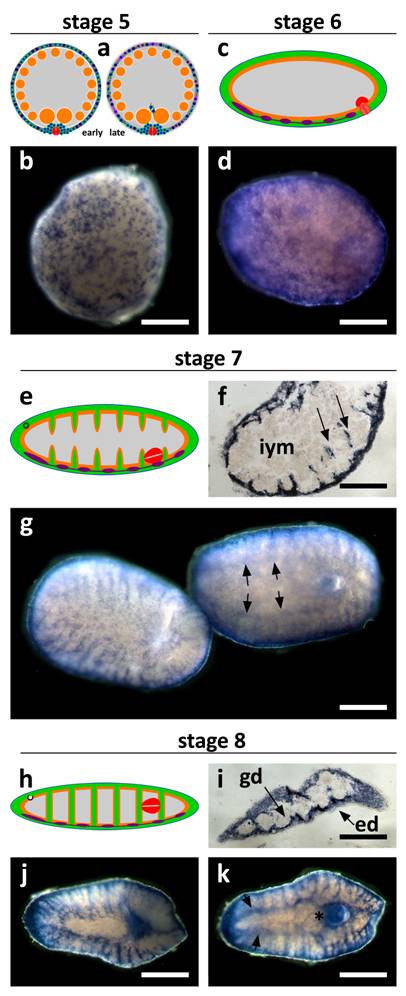
SpolvlgA is expressed in embryonic cells during stage 5 of planarian development and highlights massive embryonic cell differentiation
In early stage 5 embryonic cells are regular in shape and size and are dispersed in the germ band (Fig. 3k-l). However, as massive differentiation takes place during stage 5, embryonic cells start to vary in shape and size (Fig. 4a-b). This differentiation occurs in a wave fashion, some embryos display an early stage 5 pattern on one side and a late stage 5 pattern on the other (data not shown). SpolvlgA is expressed in most, if not all, embryonic cells during this wave. Its labeling pattern shifts from a pattern of discrete regular cells of around 20 µm, to a pattern were cells with different sizes are observed (Fig. 4b).
During stage 6 (Fig. 4c) this massive differentiation proceeds and causes the disruption of the syncytium. Nearly all the space previously occupied by the syncytium is filled with embryonic differentiating cells, and the expression of SpolvlgA is found in all the cells on the surface of the embryo (Fig. 4d).
It has been suggested that early stages of planarian development might constitute a cryptic larval stage that represents either the remnants of a vestigial larva or the evolutionary result of the competition between multiple embryos for the maternal yolk resources in the polyembryonic ectolecithal egg capsule [4]. SpolvlgA expression highlights the end of this cryptic larval stage, with the complete substitution of all maternal and transitory tissues of the cryptic larva (negative for SpolvlgA expression) with differentiating cells derived from undifferentiated embryonic cells (positive for SpolvlgA expression). This substitution might represent a cryptic metamorphic stage in which embryonic cell lineage can be traced by SpolvlgA expression. The presence of SpolvlgA signals in embryonic cells during this wave of differentiation suggest that SpolvlgA is expressed in undifferentiated blastomeres and embryonic cells as well as in differentiating cells.
SpolvlgA expression in early developmental stages of S. polychroa. a,d,g,j Schematic representations of S. polychroa developmental stages 1 to 5. e-f, i, l SpolvlgA whole mount in situ hybridizations. b-c, h, k SpolvlgA in situ hybridization on paraffin sections. a Schematic of stages 1 and 2. The zygote-derived blastomeres (blue) cleave in the syncytial yolk (dark grey) during stage 1 (top). Some blastomeres differentiate and form an embryonic epidermis and an embryonic pharynx (right). By stage 2 (bottom) these structures are formed and the embryonic epidermis covers the whole syncytium, containing blastomeres (blue, with a white cleft) and syncytial yolk nuclei (blue). b Three signal positive cleaving blastomeres in the syncytial mass (arrowhead). c A stage 2 embryo with two SpolvlgA-positive blastomeres (arrowheads) attached to the embryonic epidermis (arrow). d Schematic of stage 3. A cavity filled with unfused yolk cells (light grey) is formed due to the activity of the embryonic pharynx. e Early stage 3 embryo. SpolvlgA signal is found deep in the embryo. f By late stage 3 SpolvlgA-positive embryonic cells (two of them indicated by arrowheads) have migrated completely to the periphery and are compressed by the inner cavity. g Schematic of a stage 4 embryo. Embryonic cells and the syncytium are restricted to the outer layer, called germ band. Gastrodermal cells have differentiated from embryonic cells (orange). h Section of a stage 4 embryo. The germ band is in the periphery (indicated by arrows) and embryonic cells (three of them indicated by arrowheads) are located in the germ band. The cavity is full of inner yolk mass, made of ingested yolk cells. i Stage 4 embryo, displaying SpolvlgA-positive embryonic cells in the germ band (two of them indicated by arrowheads). j Schematic of stage 5. All kinds of tissue start to differentiate massively within the germ band. A subset of embryonic cells forms the primordium of the definitive pharynx beneath the embryonic pharynx. k Section of a stage 5 embryo. The number of SpolvlgA-positive embryonic cells in the germ band (indicated by arrows) has increased. l Early stage 5 embryo. SpolvlgA-positive embryonic cells are located in the peripheral germ band. A cluster of SpolvlgA-positive cells forms the primordium of the definitive pharynx (indicated by arrows). Asterisks indicate the position of the embryonic pharynx. ee, embryonic epidermis; gb, germ band; iym, inner yolk mass; pdp, primordium of the definitive pharynx. Scale bars: b, 60 µm; c, e-f, 150 µm; h-i, 200 µm; k-l, 250 µm.
SpolvlgA expression in late developmental stages of S. polychroa. a, c, e, h Schematic representations of S. polychroa developmental stages 5 to 8. b, d, g, j, k SpolvlgA whole mount in situ hybridizations. f, i SpolvlgA in situ hybridization on paraffin sections. a Schematic of stage 5. A massive wave of differentiation takes place in the germ band. Most embryonic cells start to differentiate to all cell types simultaneously. The syncytium is disrupted. b Late stage 5 embryo. The pattern of regular SpolvlgA-positive embryonic cells of the same size approximately has shifted to a more diffuse pattern with different types of SpolvlgA-positive cells with different shapes and sizes, due to massive cell differentiation. c Schematic of stage 6. Muscle tissue, nervous system and gastrodermal tissue are represented in green, orange and violet, respectively. The syncytium has been disrupted and completely filled with embryonic differentiating cells. d Stage 6 embryo. SpolvlgA signal is seen throughout the periphery of the embryo, comprising embryonic cells proliferating and differentiating to all cell types. e Schematic of stage 7. Embryonic cells grow towards the inner side of the embryo (muscular in green) to form body walls and shape muscular system, gut and parenchyma. f Section of stage 7 embryo. Ingrows are positive for SpolvlgA expression (two of them indicated by arrows). g Two stage 7 embryos. Left, dorsal view. Right, ventral view. Anterior is to the left. The ingrows are seen in both the dorsal and the ventral view. Expression in the CNS is indicated by arrows. h Schematic of stage 8. Ingrows have finally shaped muscular system and gut (green, orange) and the parenchyma. i Section of a stage 8 embryo. Nearly all the space between the epidermis and the gastrodermis is positive for SpolvlgA expression. This space is full of growing cells and differentiating cells, as well as putative differentiated cells. j Dorsal view of a stage 8 embryo. All the parenchyma of the embryo is positive for SpolvlgA expression. k Ventral view of a stage 8 embryo. Expression in the brain is indicated by arrows. A prominent specific labeling is also seen in the growing definitive pharynx (asterisk). iym, inner yolk mass; gd, gastrodermis; ed, epidermis. Scale bars: b,d, f-h, j-k, 300 µm; I, 500 µm.
SpolvlgA is expressed in differentiating embryonic cells during late stages of development
During stage 6 all the remnants of the disrupted syncytium of the germ band disappear. The embryo becomes worm-shaped due to muscular differentiation and by stage 7 (Fig. 4e) the embryonic cells that occupied their place begin to form integuments made of muscular and gastrodermal tissues. These septa are responsible for the final patterning of the gut and the muscular system. SpolvlgA is expressed in cells responsible for the formation of these ingrowths (Fig. 4f-g). Expression in the developing central nervous system (CNS) is also observed, though it becomes faint in later stages. During stage 8 (Fig. 4h) SpolvlgA is observed in most of the developing parenchyma, as well as in the growing pharynx. Expression is observed in most of the tissue in between the epidermis and the gastrodermis (Fig. 4i), which will eventually become the parenchyma of the adult. Most differentiating tissues display SpolvlgA signals, such as muscular system (Fig. 4j), pharynx (Fig. 4k) and brain (Fig. 4k, arrows). These results suggest that SpolvlgA is expressed in differentiating cells as well as in the cellular subset which will eventually become the neoblast cells of the adult worm. Taking together all these results we suggest that SpolvlgA is expressed in both undifferentiated blastomeres and embryonic cells and continues to be expressed in these cells as proliferation and differentiation occurs. During Dugesia japonica regeneration, DjvlgA is expressed in the regeneration blastema [21], which contains neoblast-derived differentiating cells. Consistently, our experiments indicate that SpolvlgA is expressed both in undifferentiated cells and differentiating cells during embryonic development. The presence of SpolvlgA signals in differentiated cells cannot be ruled out during late stages of development, since most of the space between the epidermis and the gastrodermis is positive for SpolvlgA signals and this space might include some differentiated cells. However, it is important to state that the end of differentiation is difficult to identify in this tissue and that fully differentiated tissues such us the epidermis and the gastrodermis lack SpolvlgA signals.
SpolvlgA is expressed in testes and parenchymatic cells in adult flatworms
SpolvlgA expression in adult organisms was monitored by in situ hybridization and fluorescent in situ hybridization on paraffin sections. We found that the expression of SpolvlgA could be dectected in testis easily (Fig. 5a-e). However, the expression in ovaries was found to be weaker than in testis, consistent with what was previously shown [21].The expression is localized in most of the testis tissue and not only in the peripheral layer where the spermatogonia are, consistently with previous data [21]. This latter case is typical of neoblast-specific genes like Djpcna [27] and Spoltud-1 [28] when monitored in the planarian germ line. Nevertheless, SpolvlgA is expressed not only in the undifferentiated germ stem cells, but also in differentiating spermatids (Fig. 5b-e). The previously described expression in cells scattered in the parenchyma [21] was found to be highly variable and faint. Scattered cells within the parenchyma are visible in fluorescent in situ hybridization (Fig. 5c-e), although we cannot confirm if they are neoblast cells or differentiating cells or both.
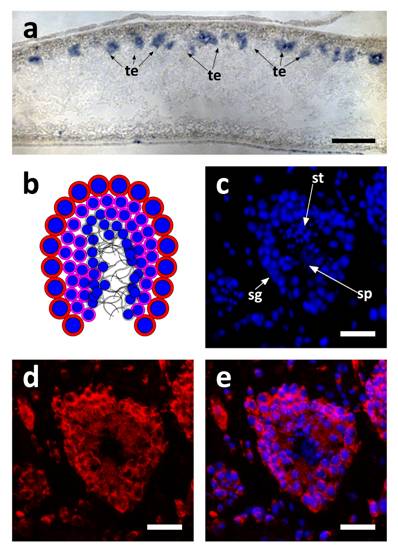
SpolvlgA expression in testis of adult organisms. a SpolvlgA in situ hybridization on paraffin sections. b-d SpolvlgA fluorescent in situ hybridization on paraffin sections of a testes. a Sagital section of an adult organism. SpolvlgA expression is observed in testis, located in the dorsal side of the animal, indicated by arrows. b Schematic of a planarian testis. Spermatogonia and spermatocytes are located in the outer layer (red cells with blue nuclei). Post-meiotic spermatids are located inside (pink cells with smaller blue nuclei). Fully differentiated sperm cells are located in the inner part (grey). c DAPI-stained nuclei of a planarian testes. Spermatogonia are located on the outer layer. Post-meiotic spermatids appear like small nuclei and sperm cells appear like fibers in the inner side of the testes, indicated by arrows. d SpolvlgA is expressed throughout the testes, except in sperm cells. Some of the SpolvlgA-positive cells outside the testes correspond to neighboring testis, but some of them are parenchymatic SpolvlgA-positive cells. e overlay of c and e. te, testis; sg, spermatogonia, st, spermatids; sp, sperm cells. Scale bars: a, 200 µm; c-e, 40 µm.
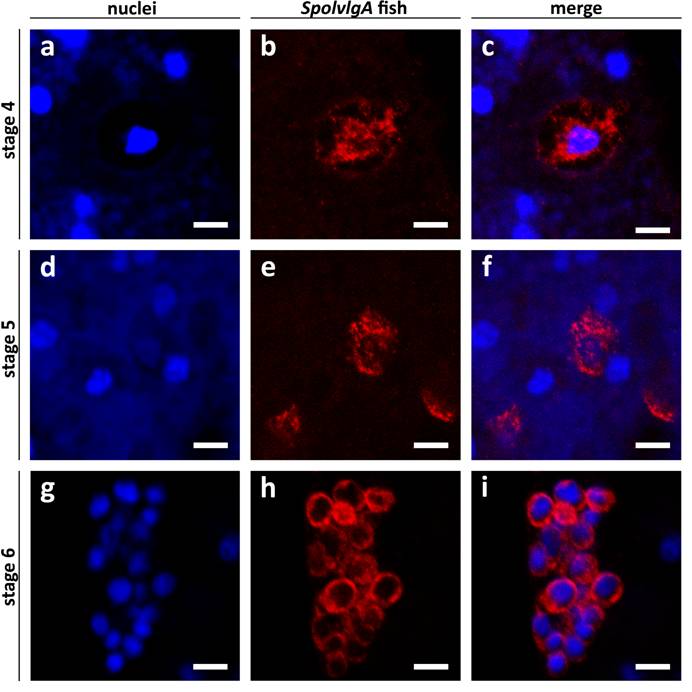
SpolvlgA mRNA subcellular localization in early and late developmental stages of S. polychroa. a-i Confocal sections of SpolvlgA fluorescent in situ hybridizations. a-f Whole mount fluorescent in situ hybridization. g-i Fluorescent in situ hybridization on paraffin sections. a, d, g Nuclei stained with DAPI. b, e, h SpolvlgA mRNA. c, f, i overlay images. a nuclei of a stage 4 embryo. Bright blue nuclei are syncytial yolk nuclei, dark blue nucleus is a embryonic cell nucleus, less stained due to its high content in uncondensed chromatin. Syncytial plasm is faintly stained by DAPI due to its high content in RNA, contrasting the embryonic cell shape. b SpolvlgA mRNA is localized attached to the multilobed nucleus and in an outer rim. c overlay of a and b. The outer rim of expression colocalizes with the embryonic cell membrane contrasted by DAPI. d nuclei of a stage 5 embryo. Bright blue nuclei are syncytial yolk nuclei, embryonic cell nuclei appear as darker blue nuclei surrounded by a dark area. e SpolvlgA mRNA is localized in most embryonic cells and occupies most of the cytoplasm. f Overlay of d and e. g nuclei of a stage 6 embryo, the inner side of the embryo is to the right, where visible nuclei correspond to inner yolk mass cells nuclei. h SpolvlgA mRNA is localized in all embryonic cells and occupies all the cytoplasm. I Overlay of g and h. Scale bars: 10 µm
We have shown that SpolvlgA expression in planarian developing embryos is localized in blastomeres and blastomere-derivative embryonic cells during early stages. During stage 5 a massive wave of differentiation takes place within the developing embryo. Most embryonic cells, previously undifferentiated, are positive for SpolvlgA and undergo differentiation from this stage on. During late stages, SpolvlgA is expressed in most embryonic tissues, both in undifferentiated cells and differentiating cells, although we cannot rule out if some of these cells are fully differentiated. In adult tissues SpolvlgA is expressed in ovaries and testis and in cells distributed throughout the parenchyma. This pattern of expression is consistent to that of DjvlgA, the orthologous gene in the planarian species Dugesia japonica [21]. DjvlgA is expressed in cells distributed throughout the parenchyma of asexual worms which resemble neoblasts and in ovaries and testes of the sexualized individuals. Within the testis, expression is not only restricted to the peripheral layer where spermatogonial cells reside, but is also expressed in differentiating spermatid cells. However, expression of DjvlgA is not observed in mature fully differentiated sperm cells. These results are consistently seen in our in situ hybridizations in adult tissues. Taken together all these observations we conclude that SpolvlgA is expressed in undifferentiated and differentiating cells but not in differentiated cells.
SpolvlgA mRNA subcellular localization changes during planarian development
In order to investigate the subcellular localization of SpolvlgA mRNA during the different stages of development we performed fluorescent in situ hybridization experiments on paraffin sections and whole mount embryos. These experiments showed three main patterns of mRNA subcellular localization for SpolvlgA: a pattern displayed in early stages, a pattern displayed in late stages and a transitional pattern observed in stage 5.
SpolvlgA mRNA at early stages of development (2-4) is localized in granules closely attached to the nucleus of the embryonic cells at those stages of development. Furthermore, a thin rim is observed closely attached to the cell membrane. The high content of mRNA in the yolk syncytium contrasts the perimeter of the blastomere and embryonic cell membrane in a DAPI staining (Fig. 6a,c). This fact allowed us to map localisation of the mRNA to the periphery of embryonic nuclei and to close association with the cell membrane (Fig. 6b-c). This subcellular localization is extremely similar to the subcellular localization of Spoltud-1 protein at these stages of planarian development [28]. Spoltud-1 is a component of chromatoid bodies in adult neoblasts and embryonic cells. These results suggest that the subcellular localization of SpolvlgA might correspond to localization in chromatoid bodies of these cells. Nevertheless, it has been described that embryonic cells in early stages of planarian development typically contain large vesicles which can occupy a substantial part of the cytoplasm. We cannot rule out that the subcellular localization of SpolvlgA in early stages is caused by these large vesicles, although the pattern observed for other genes expressed in blastomeres and embryonic cells differs from the one displayed by SpolvlgA (not shown).
In stage 5 embryos the subcellular localization pattern changes progressively to a cytoplasmic distribution (Fig. 6d-f). As stage 5 proceeds to stage 6 the subcellular distribution becomes more prominent and occupies all the cytoplasm of embryonic cells (Fig. 6g-i). Taken together all these results we conclude that the pattern of subcellular localization of SpolvlgA changes from early to late stages of development and suggest that this change might represent a localization of SpolvlgA in chromatoid bodies in early stages of development.
Acknowledgements
We are grateful to: Francesc Cebrià for critical review of the manuscript, Jose María Martín Duran, Francesc Monjo and Miquel Vila-Farré for kindly sharing reagents and for helpful discussion, and the anonymous reviewers for highly improving the quality of this manuscript. J.S is supported by a BRD grant from the University of Barcelona. This work was supported by a BFU07-63209 grant to R.R.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Le Moigne A. Etude du développement embryonnaire de Polycelis nigra (Turbellarié, Triclade). Bull Soc Zool Fr. 1963;88:403-422
2. Baguna J, Riutort M. Molecular phylogeny of the Platyhelminthes. Canadian Journal of Zoology-Revue Canadienne De Zoologie. 2004;82(2):168-193
3. Cardona A, Hartenstein V, Romero R. The embryonic development of the triclad Schmidtea polychroa. Dev Genes Evol. 2005;215(3):109-31
4. Cardona A, Hartenstein V, Romero R. Early embryogenesis of planaria: a cryptic larva feeding on maternal resources. Dev Genes Evol. 2006;216(11):667-81
5. Handberg-Thorsager M, Fernandez E, Salo E. Stem cells and regeneration in planarians. Front Biosci. 2008;13:6374-94
6. Newmark P.A, Sanchez Alvarado A. Not your father's planarian: a classic model enters the era of functional genomics. Nat Rev Genet. 2002;3(3):210-9
7. Rossi L, Salvetti A, Batistoni R. et al. Planarians, a tale of stem cells. Cell Mol Life Sci. 2008;65(1):16-23
8. Salo E. The power of regeneration and the stem-cell kingdom: freshwater planarians (Platyhelminthes). Bioessays. 2006;28(5):546-59
9. Cardona A, Fernandez J, Solana J. et al. An in situ hybridiza-tion protocol for planarian embryos: monitoring myosin heavy chain gene expression. Dev Genes Evol. 2005;215(9):482-88
10. Coward S.J. Chromatoid bodies in somatic cells of the planarian: observations on their behavior during mitosis. Anat Rec. 1974;180(3):533-45
11. Hori I. An Ultrastructural Study of the Chromatoid Body in Planarian Regenerative Cells. J Electron Microsc (Tokyo). 1982;31(1):63-72
12. Eddy E.M. Germ plasm and the differentiation of the germ cell line. Int Rev Cytol. 1975;43:229-80
13. Saffman E.E, Lasko P. Germline development in vertebrates and invertebrates. Cell Mol Life Sci. 1999;55(8-9):1141-63
14. Lasko P.F, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335(6191):611-7
15. Raz E. The function and regulation of vasa-like genes in germ-cell development. Genome Biol. 2000;1(3):REVIEWS1017
16. Boswell R.E, Mahowald A.P. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43(1):97-104
17. Chuma S, Hosokawa M, Kitamura K. et al. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci U S A. 2006;103(43):15894-9
18. Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40(3):164-70
19. Thomson T, Lasko P. Tudor and its domains: germ cell formation from a Tudor perspective. Cell Res. 2005;15(4):281-91
20. Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T. Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev Genes Evol. 2001;211(6):299-308
21. Shibata N, Umesono Y, Orii H. et al. Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev Biol. 1999;206(1):73-87
22. Gururajan R, Mathews L, Longo F.J. et al. An3 mRNA encodes an RNA helicase that colocalizes with nucleoli in Xenopus oocytes in a stage-specific manner. Proc Natl Acad Sci U S A. 1994;91(6):2056-60
23. Gururajan R, Weeks D.L. An3 protein encoded by a localized maternal mRNA in Xenopus laevis is an ATPase with substrate-specific RNA helicase activity. Biochim Biophys Acta. 1997;1350(2):169-82
24. Johnstone O, Deuring R, Bock R. et al. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev Biol. 2005;277(1):92-101
25. Rosner A, Rinkevich B. The DDX3 subfamily of the DEAD box helicases: divergent roles as unveiled by studying different organisms and in vitro assays. Curr Med Chem. 2007;14(23):2517-25
26. Umesono Y, Watanabe K, Agata K. A planarian orthopedia homolog is specifically expressed in the branch region of both the mature and regenerating brain. Dev Growth Differ. 1997;39(6):723-7
27. Orii H, Sakurai T, Watanabe K. Distribution of the stem cells (neoblasts) in the planarian Dugesia japonica. Dev Genes Evol. 2005;215(3):143-57
28. Solana J, Lasko P, Romero R. Spoltud-1 is a chromatoid body component required for planarian long-term self-renewal. Dev Biol. in press
Author contact
![]() Correspondence to: Dr. Jordi Solana, jsolanaedu
Correspondence to: Dr. Jordi Solana, jsolanaedu

 Global reach, higher impact
Global reach, higher impact