Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2009; 5(4):366-376. doi:10.7150/ijbs.5.366 This issue Cite
Review
Targeting Promyelocytic Leukemia Protein: A Means to Regulating PML Nuclear Bodies
Department of Biochemistry, School of Medicine, Case Western Reserve University (CWRU) and the Comprehensive Cancer Center of CWRU. 10900 Euclid Avenue, Cleveland, Ohio 44106, USA
Received 2009-4-1; Accepted 2009-5-6; Published 2009-5-22
Abstract
The promyelocytic leukemia protein (PML) is involved in many cellular processes including cell cycle progression, DNA damage response, transcriptional regulation, viral infection, and apoptosis. These cellular activities often rely on the localization of PML to unique subnuclear structures known as PML nuclear bodies (NBs). More than 50 cellular proteins are known to traffic in and out of PML NBs, either transiently or constitutively. In order to understand the dynamics of these NBs, it is important to delineate the regulation of PML itself. PML is subject to extensive regulation at transcriptional, post-transcriptional, and post-translational levels. Many of these modes of regulation depend on the cellular context and the presence of extracellular signals. This review focuses on the current knowledge of regulation of PML under normal cellular conditions as well as the role for regulation of PML in viral infection and cancer.
Keywords: PML, nuclear body, tumor suppressor, cell signaling, virus
Discovery of PML
PML was discovered due to its role in the oncogenesis of Acute Promyelocytic Leukemia (APL). The PML gene is involved in a chromosomal translocation with the gene for retinoic acid receptor α (RARα). This translocation results in the expression of the fusion proteins PML-RARα and RARα-PML (1). The expression of these proteins is the driving force in the development of APL. The two best treatments for APL are with the natural RARα ligand, all-trans retinoic acid (ATRA), or with arsenic trioxide (As2O3) (2). Interestingly, in APL patients where PML-RARα is expressed, the normal localization of PML into PML NBs in the cell is disrupted (1; 3; 4). The use of either of these treatments leads to restoration of PML NBs (5). While these are secondary effects of ATRA treatment, As2O3 targets the PML portion of PML-RARα directly (6). This activity will be investigated later in this review.
PML Nuclear Bodies
PML NBs (also known as PODs and ND10) are discrete subnuclear structures suggested to act as cellular organizing centers for the coordinated regulation of different processes including transcriptional regulation, DNA damage response, apoptosis, and senescence. PML NBs range in size from 0.2 to 1 μm and in number from 1 to 30 bodies per cell (7). More than 50 proteins are known to localize in PML NBs either constitutively or transiently, including p53, CBP/p300, Daxx, BLM, Pin1, HDAC7, and pRB (8-13). Furthermore, while some of these proteins, such as Daxx, have been shown to bind to PML directly, many are recruited to PML NBs via indirect interaction with another NB component. PML NBs are absent in PML-/- primary cells, but can be reconstituted by the expression of exogenous PML (8; 14), indicating that PML is essential for the formation and integrity of PML NBs.
The accumulation of PML can be regulated in response to specific cellular stimuli at multiple steps, namely at transcriptional, post-transcriptional and post-translational events. These regulatory events not only control PML protein levels, but also modifications of PML that are important for both NB formation and interactions with other proteins.
Transcriptional Control of PML Expression
Transcriptional induction of PML is an important mechanism by which extracellular signals can orchestrate a response involving PML NBs. Interferons (IFN) have been shown to activate PML transcription, leading to increased PML protein levels, nuclear body size and number in a variety of cell types (15-19). Both type I and type II IFNs (α, β, and γ) are able to induce expression of PML transcripts. This is mediated through binding of IFN-stimulated transcription factors, known as signal transducers and activators of transcription (STATs), to both an IFN-α/β stimulated response element (ISRE) and an IFN-γ activation site (GAS) in the first exon of PML (16). Not surprisingly, interferons are also able to induce expression of the oncogenic fusion protein PML-RARα (20; 21). Furthermore, IFN-regulatory factor 3 (IRF3) directly regulates PML levels by binding to the PML promoter. This increased PML transcription is a key regulatory event in the ability of IRF3 to promote p53-dependent cellular senescence and inhibition of cell growth in both normal and U87MG astrocyte cancer cells (22). Moreover, in activated macrophages, the myeloid cell transcription factor, IFN-regulatory factor 8 (IRF8), is required for IFN-γ-induced up-regulation of PML (23). These observations indicate that different cell types may have evolved distinct mechanisms mediating IFN-induced stimulation of PML transcription. Therefore, up-regulation of PML and thus PML NBs is an important mediator of the IFN response. In addition, the STATs have been shown to negatively regulate PML expression during mammary ductal morphogenesis. Disruption of the expression of PML by either gene knockout of Stat6 (leads to increased PML expression) or gene knockout of PML disrupted proper mammary gland development in mice (24). This work highlights the importance of proper regulation of PML for maintaining normal cellular responses.
In a parallel mechanism, there are other intrinsic pathways that can up-regulate PML at the transcriptional level. PML can be transcriptionally up-regulated by p53. The first intron of PML harbors a p53 binding site and p53 is capable of associating with this site both in vitro and in vivo. Induction of p53 can up-regulate PML expression and increase PML NB number and size (25). Furthermore, oncogenic Ras, whose downstream effects include up-regulation of p53, is also able to induce PML transcription (26; 25). Since various cell stimuli can induce interferon signaling or control p53 activity, it is likely that PML levels are controlled transcriptionally in a wide range of cellular conditions. Together, these data indicate that an increase in PML transcription is part of the response for a distinct set of pathways that are involved in genome stability, oncogenesis, and viral responses. Furthermore, they suggest that PML likely plays a role in other intracellular signaling events in response to various extracellular stimuli.
Control of PML through alternative splicing
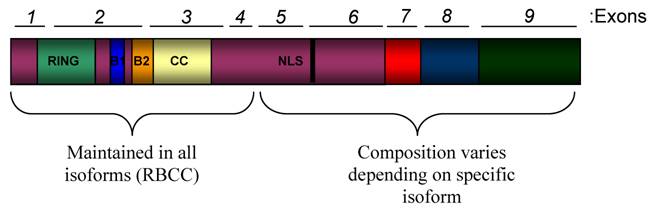
PML expression is regulated by the cell or tissue type and the differentiation stage of the cell (27-29). One of the main contributing factors to this varied expression is alternative splicing which results in the expression of at least 11 different isoforms (30). The human promyelocytic leukemia (PML) gene is located across 53,000 bases that compose 9 exons at chromosomal location 15q22. Different PML isoforms are derived from alternative splicing of the C-terminal exons (5-9) (Figure 1). All PML isoforms maintain the RBCC motif in the N-terminal half of the protein, comprised of a zinc-finger RING domain followed by two zinc finger B Boxes, followed by a coiled-coil domain (30). This motif is an important mediator of homo-oligomerization of PML as well as interactions of PML with other proteins. Since not all PML isoforms retain the nuclear localization sequence in exon 6, there are both nuclear and cytoplasmic PML isoforms (30-33).
Cytoplasmic PML
One of the major distinctions among PML isoforms is their subcellular localization (Figure 2). This can be one of the main determinants of their role in the cell. It has become clear that the cytoplasmic forms are an important area of investigation for both their ability to regulate nuclear PML as well as their own (cytoplasmic) functions. Cytoplasmic PML can redistribute nuclear PML to the cytoplasm, thus decreasing nuclear body formation and size (34; 35). For example, this sequestration was shown to lead to inhibition of the growth suppressive function of p53, which is normally mediated by nuclear PML (34; 35). In this manner, the subcellular distribution of certain PML isoforms is able to regulate the overall activity of PML. There is also crosstalk between cytoplasmic forms of PML and the TGFβ signaling pathway through interactions with its downstream regulators Smad2/3 and SARA (36). Furthermore, the homeodomain protein TGIF, which is a negative regulator of TGFβ signaling, is able to sequester cytoplasmic PML to the nucleus (37). Interestingly, herpes simplex virus -1 (HSV-1) infection leads to a change in splicing of PML pre-mRNA resulting in an increased ratio of cytoplasmic to nuclear isoforms (38). Overall, controlling the subcellular localization of PML appears to be an important regulatory mechanism for PML. Research into the mechanisms that determine PML localization may provide one of the most effective ways by which to therapeutically manipulate PML.
Domain Structure of PML. The schematic shows all functional domains that are present in PML proteins. This schematic is not representative of any specific naturally occurring isoform. The human PML gene is comprised of 9 exons that are alternatively spliced into transcripts that encode at least 11 isoforms. All of the isoforms contain exons 1-4, but alternative splicing of exons 5-9 leads to variation at the C-terminal ends of the PML isoforms. All isoforms contain the N-terminal RBCC (RING/B-Box/Coiled coil) motif, which is important for protein-protein interactions and contains many regulatory sites for PML. Most isoforms also contain a nuclear localization sequence (NLS) found in exon 6.
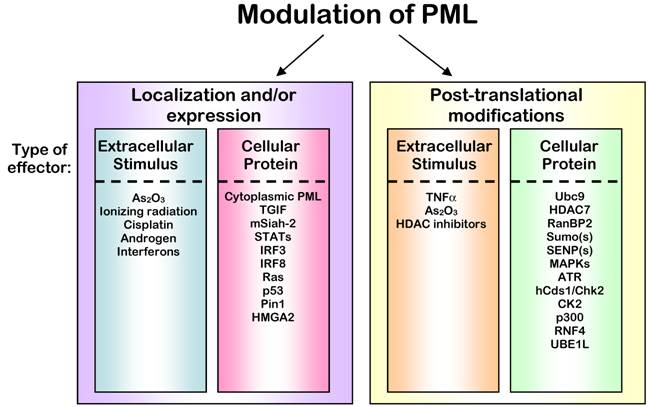
Regulation of PML Protein. The activity of PML is ultimately controlled through its localization and expression levels. The control of these characteristics of PML can be accomplished through altering binding partners or stability of PML that lead to immediate changes (left panel) or by post-translationally modifying PML, which leads to subsequent changes in localization and stability indirectly (right panel). Signals to control PML often result from extracellular stimuli, but can also originate from interactions with other proteins within the cell. In some cases, a single agent or protein can affect all of these modes of regulation; however, in all cases the outcome is an alteration of PML activity.
Post-Translational Modifications of PML
PML is subject to multiple posttranslational modifications including sumoylation, phosphorylation, ubiquitination, acetylation, and ISGylation. These modifications lead to alteration of PML protein levels and activity (Figure 2). One of the best characterized of these modifications is sumoylation of PML. PML can be modified by Sumo-1 and Sumo-2/3 at lysines 65, 160 and 490 (39-41). It has recently been shown that these different Sumo modifications can have distinct consequences for the sumoylated protein based on the Sumo used and the length of the polysumo chain (42; 43). For example, one report suggests that modification of PML specifically by Sumo-3 is integral to its nuclear localization (42). Sumoylation of PML is stimulated by the only known E2 sumo ligase, Ubc9 (44). Similar to other sumo-conjugated proteins, PML can be sumoylated in vitro in the absence of E3 ligases. Using in vitro translated PML as a substrate, the E3 sumo ligase RanBP2 is capable of promoting PML sumoylation, but whether RanBP2 sumoylates PML in vivo remains unclear (45). Furthermore, we have recently shown that the TNFα-stimulated association of histone deactylase 7 (HDAC7) with PML and its subsequent Sumo-1 modification, leads to increased PML NB formation (46). Using purified recombinant proteins, we demonstrated that HDAC7 potently stimulates PML sumoylation with Sumo-1 in a reconstituted system. More importantly, knockdown of HDAC7 abrogates basal and TNFα-induced PML sumoylation and reduces PML protein levels and PML NB formation (12). These data provide a mechanistic link between PML, its Sumo-1 modification and its stability. In support, Sumo1-/- mice express reduced levels of PML protein and fewer PML NBs (47). Lastly, as mentioned previously, the sumoylation status of PML is regulated in a cell cycle-dependent manner. During interphase, Sumo-1 is conjugated to PML. PML is de-sumoylated once mitosis begins (48).
De-sumoylation of PML may be an important mechanism for the regulation of PML activity, but has not been widely investigated. SENP-1, a member of the Sumo-specific protease family, appears to decrease PML sumoylation after treatment of cells with interleukin-6 (IL-6) (49). Similarly, SENP-5 can remove Sumo-2 or Sumo-3 from lysines 160 or 490 or Sumo-1 or Sumo-3 from lysine 65 in transient transfection assays (50). Surprisingly, despite retention of two of the three PML sumoylation sites, sumoylation of PML-RARα has only been observed in vitro when Sumo1 is overexpressed (39; 44). Differences in PML and PML-RARα sumoylation may account for the ability of As2O3 to induce degradation of PML-RARα more rapidly than PML (51).
Recent studies indicate that the RING-domain-containing ubiquitin E3 ligase, RNF4, catalyzes ubiquitination of sumoylated PML and PML-RARα. This activity is attributed to its Sumo-interacting motifs (SIMs), domains in its N-terminus known to interact with the Sumo moeity. This ubiquitinylation is required for the As2O3-induced PML degradation (52-54). Since PML sumoylation plays an important role in regulation of PML accumulation and NB dynamics, it is important to further understand the mechanisms and control of sumoylation of PML in order to alter PML NB formation under disease situations.
Phosphorylation is another important post-translational modification of PML. PML is phosphorylated at multiple sites in response to various stimuli, with each phosphorylation having a distinct effect on PML activity. Interestingly, phosphorylation of PML can be directly linked to sumoylation in response to As2O3. Using an in vitro kinase assay, it was shown that PML could be phosphorylated by the mitogen-activated protein kinase ERK2, at threonine 28 and serines 36, 38, 40, 527, and 530. Furthermore, As2O3-induced PML phosphorylation at some of these sites is ERK2-dependent. Mutation of these residues not only prevents phosphorylation but also blocks sumoylation of PML. These observations led to a model in which the phosphorylation of PML is essential for PML sumoylation and As2O3-induced apoptosis (6). Although the detailed mechanisms by which PML phosphorylation promotes or inhibits sumoylation are currently unknown, there is likely crosstalk between these two modifications in response to extracellular signals.
Phosphorylation of PML can also regulate PML activity independently of sumoylation. In response to the DNA-damaging agent doxorubicin, PML accumulates in the nucleolus as part of the S-phase cell cycle checkpoint. This nucleolar localization is dependent on phosphorylation of PML by the checkpoint kinase ataxia telangiectasia Rad-3 related (ATR) kinase, although the site(s) of this phosphorylation are unknown (55). In addition, in response to γ-irradiation, hCds1/Chk2 phosphorylates PML and this phosphorylation is required for γ-irradation-induced apoptotsis (56).
Aside from its roles in the DNA damage response and apoptosis, PML is also widely studied as a tumor suppressor. This activity can be regulated in a phosphorylation-dependent manner. The oncogenic kinase, casein kinase 2 (CK2), is able to phosphorylate PML on serine 565 and promote proteosome-dependent degradation of PML. Consequently, PML mutants that cannot be phosphorylated by CK2 have more potent tumor suppressor properties, providing a strong link between regulation of PML by CK2 and tumorigenesis (57).
In addition to sumoylation and phosphorylation, other PML modifications have been reported. PML and PML-RARα are subject to ISGylation by interferon-stimulated gene- 15 (ISG15). Like Sumo, ISG15 is a small ubiquitin-like peptide that can be conjugated to proteins in an E1/E2/E3-dependent manner. Expression of the ISG15 conjugation system can be stimulated by type-I interferons, bacterial lipopolysaccharides, and viral infection. Once expressed, ISG15 can be secreted or conjugated to proteins (58). Protein ISGylation can affect protein-protein interactions (59; 60) and block other modifications such as neddylation (61). In one case, conjugation of ISG15 to the eukaryotic translation initiation factor 4E (eIF4E) family member 4E-HP (or eIF4E-2) increases the affinity of 4E-HP to the mRNA 5' cap structure, thus affecting control of cap-dependent translation (59). Interestingly, PML has been shown to interact with family member eIF4E (or eIF4E-1) and decrease its affinity for mRNA, thus also controlling mammalian protein expression. It is not known whether ISGylation plays a role in this pathway (62). ISGylation of the PML domain in PML-RARα has been proposed to decrease its accumulation (63). The mechanism by which PML ISGylation is regulated is completely unknown. Recent work also indicates that PML is subject to acetylation. This acetylation appears to correlate with increased PML sumoylation and may be important in PML-dependent cell death pathways (64). Lastly, as previously mentioned, PML is known to be ubiquitinylated by RNF4 (52-54). This ubiquitinylation is induced in response to As2O3, which increases sumoylation of PML. Furthermore, since many other extracellular signals regulate PML sumoylation and or PML stability, it is likely that RNF4 acts downstream of other signals, and that there are likely other undiscovered E3 ubiquitin ligases that act on PML.
Post-translational modifications of PML are important for the regulation of PML protein levels, localization, and activity and it is likely that there are other PML modifications yet to be identified. This intricate network of modifications of PML exemplifies the elaborate regulation of PML activity in the cell.
Control of PML Protein Stability
Several distinct mechanisms have been shown to regulate PML protein levels (Figure 2). The mammalian homologues of the Drosophila Seven in Absentia (SIAHs) are known to target proteins involved in cell growth and tumorigenesis for degradation. Specifically, mSiah-2 targets PML for proteosome-dependent degradation. The decrease in PML protein levels correlates with a decrease in PML NB number. It is possible that this pathway is part of a regulatory feedback loop involving the p53 protein, since p53 can activate SIAH-1 transcription (65). PML-RARα, is also subject to mSiah-2-mediated degradation; however, ectopic overexpression of mSiah-2 only partially rescues the differentiation block that is characteristic of APL cells, possibly due to residual PML-RARα that is not degraded (66). Another p53-dependent pathway, the DNA damage response, leads to increased PML protein levels. Treatment of cells with ionizing radiation or cisplatin increases PML protein levels and PML NB number and size; however PML mRNA levels are unchanged (67). Though the exact mechanism underlying DNA damage-induced increases in PML protein is unknown, it may be dependent on a kinase activated by ionizing radiation, inhibition of a negative kinase regulator, or through another cell cycle checkpoint regulator such as ATM/Chk2 (68; 69). By contrast, androgen leads to down-regulation of PML protein levels in the prostate cancer cell line CWR22R. Though the detailed mechanism of this degradation has yet to be delineated, it has been suggested that the decrease in PML protein levels is post-translational because little change in the PML mRNA levels was observed. Functionally, this data suggests that PML may suppress the cell growth of androgen-dependent prostate cancers (70). While little is known about the regulation of steady-state levels of PML in the absence of exogenous signals, work in our lab has shown that PML interacts with the peptidyl-prolyl isomerase Pin1 (13). Interaction with Pin1 results in decreased stability of PML. Pin1-mediated PML degradation is likely to be blocked by Sumo1 modification since Sumo1-modified PML no longer binds to Pin1. Furthermore, while the interaction requires PML phosphorylation, the kinases responsible for mediating the interaction, as well as the downstream mechanism of degradation of PML, are unknown (13). Similarly, overexpression of the architectural protein HMGA2 leads to ubiquitin-dependent degradation of PML protein, but whether this is due to a direct interaction is currently unknown. The effects of HMGA2 are responsive to As2O3 and dependent on HMGA2 sumoylation (71). In summary, these data indicate that certain signals and protein-protein interactions can promote degradation of PML.
Viral Regulation of PML
Findings by several labs suggest that many viruses target PML to modify its levels, localization or activity (Table 1). Viruses can regulate PML through multiple mechanisms to promote viral replication. For example, several viruses target PML to alter its sumoylation including human cytomegalovirus and herpes simplex virus type I (HSV-1) (72; 73). In many cases viral proteins induce decreases in sumoylation of PML resulting in disruption of PML stability and overall decreases in PML protein levels. Interestingly, the targeting of PML is required for the infection and survival of HSV-1 (74), a result that implicates an inhibitory role for PML in viral replication. Similarly, other viral-induced changes in PML modifications control PML activity in response to viral infection such during adenovirus type 5 infection where slower migrating PML species increase while Sumo-1 modified forms of PML decreases (75; 76).
Viral Proteins that Regulate PML. Many viruses have mechanisms that target and control PML. Several of these controls occur post-translationally to effect PML stability and/or localization. The viral protein components that regulate PML are listed in the first column. The second column indicates the effect that these proteins have on PML.
| Viral Proteins | Effect on PML | Reference |
|---|---|---|
| IE1, human cytomegalovirus | Promotes de-sumoylation of PML | (72; 73) |
| ICP0, herpes simplex virus, type I | Promotes PML degradation, alternative splicing, enriches cytoplasmic PML | (77; 78; 38) |
| ORF75c, murine gammaherpesvirus 68 | Promotes PML degradation | (79) |
| E4orf3, adenovirus type 5 | Disrupts PML NBs | (75; 76) |
| Core protein, hepatitis C virus | Interacts with and sequesters PML | (80) |
| E7, human papilloma virus | Interacts with PML | (81) |
| ?, human papilloma virus | Increases PML NB number | (82) |
| ?, human herpes virus 6B | Increases PML expression, decreases PML NB number | (83) |
| Pre-integration complex, human immunodeficiency virus | Promotes redistribution of PML to the cytoplasm | (84) |
| Z protein, lymphocytic choriomeningitis virus | Promotes redistribution of PML to the cytoplasm | (85) |
| EBNA-1, Epstein-barr virus | Promotes PML degradation, disrupts PML NBs | (86) |
Viruses also directly affect PML stability or activity via mechanisms that have not been shown to include post-translational modification of PML. These include proteosome-dependent degradation of PML during infection by murine gammaherpesvirus 68 (gammaHV68) and direct interaction with PML by the hepatitis C virus (HCV) core (80; 79). In both cases the functional outcome of modulation of PML is to promote viral replication. Furthermore, as discussed earlier, changes in the subcellular localization of PML can be an important regulator of its activity. Components of both human immunodeficiency virus (HIV) and lymphocytic choriomeningitis virus (LCMV) can induce redistribution of nuclear PML to the cytoplasm (85; 84).
In certain cases, some viruses appear to stabilize or promote PML levels. The human papilloma oncoprotein E7 directly interacts with PML and blocks PML-mediated cellular senescence. It is currently unknown whether interaction of E7 with PML also plays a role in the HPV infection (81). However, recent work indicates that the number of PML NBs increases during HPV infection of human keratinocytes (82), which would indicate a mechanism opposite to that of other viruses. Similarly, infection of cells with human herpesvirus 6B (HHV-6B) led to increased levels of PML protein and mRNA (83).
Clearly the mechanisms governing these observations are more complex than can be explained with the current understanding of PML NB regulation. However, it is clear that targeting PML is a common mechanism of viral infection and appears to be required for viral survival in many cases.
Regulation of PML in Cancer
PML is known to act as a potent tumor suppressor not only in APL, but also in other cancers. Although PML-/- mice develop normally, they are prone to develop tumors in chemical and physical models of carcinogenesis (87). The ability of PML to suppress oncogenesis may be due to its ability to promote apoptosis by both intrinsic and extrinsic pathways (9; 10). Changes in PML localization, levels, and activity are likely to be equally important factors in the anti-tumorigenic function of PML. PML protein expression is reduced or greatly diminished in tumor cell lines derived from prostate adenocarcinomas, colon adenocarcinomas, breast carcinomas, lung carcinomas, lymphomas, CNS tumors, and germ cell tumors; however, there is no change in PML transcript levels in these cells compared to their normal counterparts (88). There are several known mechanisms by which PML levels are decreased in these cancers. First, as previously mentioned, decreased PML levels in cancer can be altered by phosphorylation of PML by CK2 (57). There is an inverse correlation between PML protein levels and CK2 activity in human lung-cancer derived cell lines. Decreased PML levels are also associated with increased tumorigenesis in a mouse model of lung cancer.
Conversely, as previously mentioned IRF3 has been shown to directly regulate PML levels by binding to the PML promoter. This IRF3-induced transcription of PML is a key regulatory event in the promotion of p53-dependent cellular senescence and inhibition of cell growth in U87MG astrocyte cancer cells (22).
Interestingly, PML was recently shown to be transcriptionally up-regulated during haematopoiesis which correlated with increased PML expression in patients with the haematopoietic malignancy chronic myeloid leukemia (CML). Furthermore, down-regulation of PML levels led to a decrease in quiescent leukemia-initiating cells (89). These data suggest that up-regulation of PML is a critical event in CML. However, it remains to be seen whether this is unique to CML or more general to other haematopoietic malignancies. Nonetheless, these data highlight the importance for regulated control of PML expression.
These examples support the notion that PML is a key regulatory factor in tumorigenesis. It will be important to elucidate the regulatory mechanisms that lead to decreased PML levels in cancer cells and what role PML plays in cancer progression. Furthermore, understanding the normal regulation of PML is central to elucidating the mechanisms by which PML is de-regulated in many cancers including in APL.
Conclusions and Perspective
It appears that PML is targeted by numerous extracellular signals both in physiological and pathological conditions that result in modifications and changes in PML levels and function (Figure 3). These signals may each control PML independently; however, it is likely that some act in a collaborative fashion. There are likely several undiscovered enzymes such as kinases, phosphatases, E3 ubiquitin ligases, E3 sumo ligases, etc. that modify and modulate PML activity. The recent findings that sumo modifications play a role in the stability control of PML protein and the newly identified PML modifications such as acetylation and ISGylation suggest that PML activity is controlled by complex regulatory networks. It is probable that these modifications affect PML regulation in a combinatorial manner.
Even after more than 15 years of investigation, there are still many unanswered questions. For example, little is known about regulation of the alternative splicing of the PML transcript or even the unique function of each of the spliced isoforms. Similarly, while sumoylation of PML is known to be important for NB formation and dynamics, little is known about steady-state regulation of PML sumoylation. Even more apparent is the lack of mechanistic details on the specific role of PML in processes such as the interferon response, DNA damage response, and cell cycle.
Controlling PML protein levels and activity is an important mechanism determining cell fate. Extracellular stimuli and other events such as viral infection and oncogenesis are known to affect PML activity. This regulation occurs at five distinct levels: transcriptional, alternative mRNA splicing, post-translational modification, subcellular localization, and stability control. The exact mechanisms of these changes are complex, and often include multiple modes of regulating PML. The result is a net increase or decrease in PML activity in the cell. (S= sumoylation, A= acetylation, P= phosphorylation, I = ISGylation, U= ubiquitinylation)
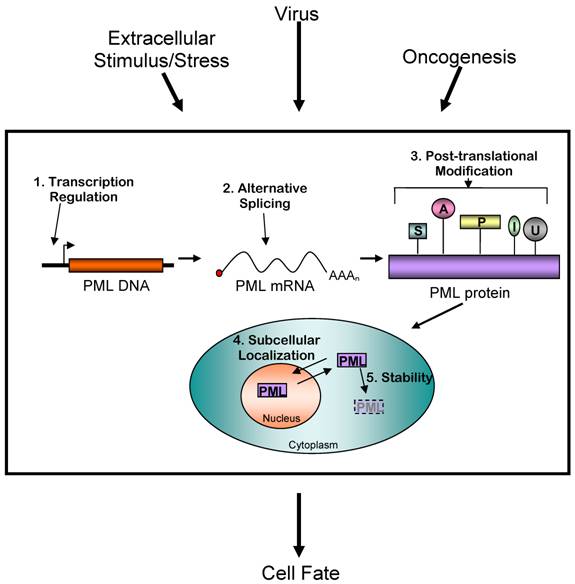
PML appears to play a role in many cellular pathways. Disruption of PML NBs will affect these pathways and therefore may have profound effects on normal cellular function. PML has been implicated in several illnesses, most notably viral infection and cancer. Interestingly, PML is a common target of viruses in order to promote viral infection. The reasons behind this are likely intimately linked with the ability of PML to promote apoptosis and arrest cell cycle progression. A similar effect is observed in cancer, though the causes of the decreased PML expression are currently unknown. Other diseases involving PML and NBs include those with impaired DNA damage responses such as Rothmund-Thomson syndrome and Bloom's Syndrome or several neurodegenerative diseases including ataxia, Huntington's disease and dentatorubral pallidoluysian atrophy (90).
It is of interest that PML-/- mice appear to develop and survive relatively normally. However, recent reports demonstrate that perturbing PML does, in fact, affect normal cellular functioning. For example, alteration of PML expression resulted in abnormal mammary gland development in mice (24). Furthermore, PML expression is found exclusively in the neural progenitor cells of the developing mouse neocortex. Loss of this expression led to defects in the ratio of progenitor types and ultimately effected adult brain development (91). It is likely that introduction of PML expression into other cells in the developing brain may also have deleterious effects. Many progresses from other labs and ours have shown that changes in PML activity or expression levels have been found occur in response to extracellular signals or can be modified by such. Furthermore, the diseases in which PML plays a role are diseases that have defects in “response-driven” pathways, such as those impaired in DNA damage, inflammation, or in conditions such as cancer or viral infection where normal cellular signaling is disrupted by other means. Under these circumstances, changes in PML levels or activity are found to be important. Therefore, by modulating the PML activities or expression levels during cellular stress caused by aberrant cellular signaling, one may be able to partially alleviate some of the symptoms of these diseases. Armed with new understanding of the mechanisms underlying PML regulation, we are one step closer to effectively manipulating NBs for therapeutic purposes or disease prevention.
Acknowledgements
We thank Drs. Samols, Snider, and Stanya for their comments on the manuscript. Hung-Ying Kao is supported by NIH (DK078965), the Pardee Foundation, and the American Cancer Society.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675-684
2. Ohno R, Asou N, Ohnishi K. Treatment of acute promyelocytic leukemia: strategy toward further increase of cure rate. Leukemia. 2003;17:1454-1463
3. Kakizuka A, Miller WHJr, Umesono K, Warrell RPJr, Frankel SR, Murty VV, Dmitrovsky E, Evans RM. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663-674
4. Dyck JA, Maul GG, Miller WHJr, Chen JD, Kakizuka A, Evans RM. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333-343
5. Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345-356
6. Hayakawa F, Privalsky ML. Phosphorylation of PML by mitogen-activated protein kinases plays a key role in arsenic trioxide-mediated apoptosis. Cancer Cell. 2004;5:389-401
7. Dellaire G, Bazett-Jones DP. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays. 2004;26:963-977
8. Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748-2752
9. Hofmann TG, Will H. Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ. 2003;10:1290-1299
10. Takahashi Y, Lallemand-Breitenbach V, Zhu J, de The H. PML nuclear bodies and apoptosis. Oncogene. 2004;23:2819-2824
11. Buschbeck M, Uribesalgo I, Ledl A, Gutierrez A, Minucci S, Muller S, Di Croce L. PML4 induces differentiation by Myc destabilization. Oncogene. 2007;26:3415-3422
12. Gao C, Ho CC, Reineke E, Lam M, Cheng X, Stanya KJ, Liu Y, Chakraborty S, Shih HM, Kao HY. Histone deacetylase 7 promotes PML sumoylation and is essential for PML nuclear body formation. Mol Cell Biol. 2008;28:5658-5667
13. Reineke EL, Lam M, Liu Q, Liu Y, Stanya KJ, Chang KS, Means AR, Kao HY. Degradation of the tumor suppressor PML by Pin1 contributes to the cancer phenotype of breast cancer MDA-MB-231 cells. Mol Cell Biol. 2008;28:997-1006
14. Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honore N, Doubeikovsky A, Duprez E, Pandolfi PP, Puvion E, Freemont P, de The H. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med. 2001;193:1361-1371
15. Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi PP, Pelicci PG, Dejean A. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene. 1995;11:871-876
16. Stadler M, Chelbi-Alix MK, Koken MH, Venturini L, Lee C, Saib A, Quignon F, Pelicano L, Guillemin MC, Schindler C. et al. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene. 1995;11:2565-2573
17. Grotzinger T, Sternsdorf T, Jensen K, Will H. Interferon-modulated expression of genes encoding the nuclear-dot-associated proteins Sp100 and promyelocytic leukemia protein (PML). Eur J Biochem. 1996;238:554-560
18. Heuser M, van der Kuip H, Falini B, Peschel C, Huber C, Fischer T. Induction of the pro-myelocytic leukaemia gene by type I and type II interferons. Mediators Inflamm. 1998;7:319-325
19. Vannucchi S, Percario ZA, Chiantore MV, Matarrese P, Chelbi-Alix MK, Fagioli M, Pelicci PG, Malorni W, Fiorucci G, Romeo G, Affabris E. Interferon-beta induces S phase slowing via up-regulated expression of PML in squamous carcinoma cells. Oncogene. 2000;19:5041-5053
20. Chelbi-Alix MK, Pelicano L, Quignon F, Koken MH, Venturini L, Stadler M, Pavlovic J, Degos L, de The H. Induction of the PML protein by interferons in normal and APL cells. Leukemia. 1995;9:2027-2033
21. Nason-Burchenal K, Gandini D, Botto M, Allopenna J, Seale JR, Cross NC, Goldman JM, Dmitrovsky E, Pandolfi PP. Interferon augments PML and PML/RAR alpha expression in normal myeloid and acute promyelocytic cells and cooperates with all-trans retinoic acid to induce maturation of a retinoid-resistant promyelocytic cell line. Blood. 1996;88:3926-3936
22. Kim TK, Lee JS, Oh SY, Jin X, Choi YJ, Lee TH, Lee E, Choi YK, You S, Chung YG, Lee JB, DePinho RA, Chin L, Kim H. Direct transcriptional activation of promyelocytic leukemia protein by IFN regulatory factor 3 induces the p53-dependent growth inhibition of cancer cells. Cancer Res. 2007;67:11133-11140
23. Dror N, Rave-Harel N, Burchert A, Azriel A, Tamura T, Tailor P, Neubauer A, Ozato K, Levi BZ. Interferon regulatory factor-8 is indispensable for the expression of promyelocytic leukemia and the formation of nuclear bodies in myeloid cells. J Biol Chem. 2007;282:5633-5640
24. Li W, Ferguson BJ, Khaled WT, Tevendale M, Stingl J, Poli V, Rich T, Salomoni P, Watson CJ. PML depletion disrupts normal mammary gland development and skews the composition of the mammary luminal cell progenitor pool. Proc Natl Acad Sci U S A. 2009;106:4725-4730
25. de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW. PML is a direct p53 target that modulates p53 effector functions. Mol Cell. 2004;13:523-535
26. Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 2000;14:2015-2027
27. Flenghi L, Fagioli M, Tomassoni L, Pileri S, Gambacorta M, Pacini R, Grignani F, Casini T, Ferrucci PF, Martelli MF. et al. Characterization of a new monoclonal antibody (PG-M3) directed against the aminoterminal portion of the PML gene product: immunocytochemical evidence for high expression of PML proteins on activated macrophages, endothelial cells, and epithelia. Blood. 1995;85:1871-1880
28. Cho Y, Lee I, Maul GG, Yu E. A novel nuclear substructure, ND10: distribution in normal and neoplastic human tissues. Int J Mol Med. 1998;1:717-724
29. Aoto T, Saitoh N, Ichimura T, Niwa H, Nakao M. Nuclear and chromatin reorganization in the MHC-Oct3/4 locus at developmental phases of embryonic stem cell differentiation. Dev Biol. 2006;298:354-367
30. Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223-7233
31. Borden KL. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol Cell Biol. 2002;22:5259-5269
32. Condemine W, Takahashi Y, Zhu J, Puvion-Dutilleul F, Guegan S, Janin A, de The H. Characterization of endogenous human promyelocytic leukemia isoforms. Cancer Res. 2006;66:6192-6198
33. Yoshida H, Ichikawa H, Tagata Y, Katsumoto T, Ohnishi K, Akao Y, Naoe T, Pandolfi PP, Kitabayashi I. PML-retinoic acid receptor alpha inhibits PML IV enhancement of PU.1-induced C/EBPepsilon expression in myeloid differentiation. Mol Cell Biol. 2007;27:5819-5834
34. Bellodi C, Kindle K, Bernassola F, Cossarizza A, Dinsdale D, Melino G, Heery D, Salomoni P. A cytoplasmic PML mutant inhibits p53 function. Cell Cycle. 2006;5:2688-2692
35. Bellodi C, Kindle K, Bernassola F, Dinsdale D, Cossarizza A, Melino G, Heery D, Salomoni P. Cytoplasmic function of mutant promyelocytic leukemia (PML) and PML-retinoic acid receptor-alpha. J Biol Chem. 2006;281:14465-14473
36. Lin HK, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004;431:205-211
37. Seo SR, Ferrand N, Faresse N, Prunier C, Abecassis L, Pessah M, Bourgeade MF, Atfi A. Nuclear retention of the tumor suppressor cPML by the homeodomain protein TGIF restricts TGF-beta signaling. Mol Cell. 2006;23:547-559
38. McNally BA, Trgovcich J, Maul GG, Liu Y, Zheng P. A role for cytoplasmic PML in cellular resistance to viral infection. PLoS ONE. 2008;3:e2277
39. Kamitani T, Nguyen HP, Kito K, Fukuda-Kamitani T, Yeh ET. Covalent modification of PML by the sentrin family of ubiquitin-like proteins. J Biol Chem. 1998;273:3117-3120
40. Fu J, Wang L, Wang W, Chen Z. [Expression of the Wilms' Tumor Gene WT1 and Detection of Minimal Residual Disease in Acute Leukemia]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2000;8:211-215
41. Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell. 2004;15:5208-5218
42. Fu C, Ahmed K, Ding H, Ding X, Lan J, Yang Z, Miao Y, Zhu Y, Shi Y, Zhu J, Huang H, Yao X. Stabilization of PML nuclear localization by conjugation and oligomerization of SUMO-3. Oncogene. 2005;24:5401-5413
43. Mukhopadhyay D, Ayaydin F, Kolli N, Tan SH, Anan T, Kametaka A, Azuma Y, Wilkinson KD, Dasso M. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J Cell Biol. 2006;174:939-949
44. Duprez E, Saurin AJ, Desterro JM, Lallemand-Breitenbach V, Howe K, Boddy MN, Solomon E, de The H, Hay RT, Freemont PS. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J Cell Sci. 1999;112( Pt 3):381-393
45. Tatham MH, Kim S, Jaffray E, Song J, Chen Y, Hay RT. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat Struct Mol Biol. 2005;12:67-74
46. Gao C, Ho C-C, Reineke EL, Lam M, Cheng X, Stanya K, Chakraborty S, Shih M-S, Kao H-Y. Histone deacetylase 7 promotes PML sumoylation and is essential for PML nuclear body (NB) formation. Molecular and Cellular Biology. submitted
47. Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106-4113
48. Everett RD, Lomonte P, Sternsdorf T, van Driel R, Orr A. Cell cycle regulation of PML modification and ND10 composition. J Cell Sci. 1999;112( Pt 24):4581-4588
49. Ohbayashi N, Kawakami S, Muromoto R, Togi S, Ikeda O, Kamitani S, Sekine Y, Honjoh T, Matsuda T. The IL-6 family of cytokines modulates STAT3 activation by desumoylation of PML through SENP1 induction. Biochem Biophys Res Commun. 2008;371:823-828
50. Gong L, Yeh ET. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem. 2006;281:15869-15877
51. Muller S, Miller WHJr, Dejean A. Trivalent antimonials induce degradation of the PML-RAR oncoprotein and reorganization of the promyelocytic leukemia nuclear bodies in acute promyelocytic leukemia NB4 cells. Blood. 1998;92:4308-4316
52. Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de The H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547-555
53. Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538-546
54. Weisshaar SR, Keusekotten K, Krause A, Horst C, Springer HM, Gottsche K, Dohmen RJ, Praefcke GJ. Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett. 2008;582:3174-3178
55. Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6:665-672
56. Yang S, Kuo C, Bisi JE, Kim MK. PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2. Nat Cell Biol. 2002;4:865-870
57. Scaglioni PP, Yung TM, Cai LF, Erdjument-Bromage H, Kaufman AJ, Singh B, Teruya-Feldstein J, Tempst P, Pandolfi PP. A CK2-dependent mechanism for degradation of the PML tumor suppressor. Cell. 2006;126:269-283
58. Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599-609
59. Okumura F, Zou W, Zhang DE. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 2007;21:255-260
60. Jeon YJ, Choi JS, Lee JY, Yu KR, Kim SM, Ka SH, Oh KH, Kim KI, Zhang DE, Bang OS, Chung CH. ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep. 2009;10:374-380
61. Okumura A, Pitha PM, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci U S A. 2008;105:3974-3979
62. Cohen N, Sharma M, Kentsis A, Perez JM, Strudwick S, Borden KL. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. Embo J. 2001;20:4547-4559
63. Shah SJ, Blumen S, Pitha-Rowe I, Kitareewan S, Freemantle SJ, Feng Q, Dmitrovsky E. UBE1L represses PML/RAR{alpha} by targeting the PML domain for ISG15ylation. Mol Cancer Ther. 2008;7:905-914
64. Hayakawa F, Abe A, Kitabayashi I, Pandolfi PP, Naoe T. Acetylation of PML is involved in histone deacetylase inhibitor-mediated apoptosis. J Biol Chem. 2008;283:24420-24425
65. Matsuzawa S, Takayama S, Froesch BA, Zapata JM, Reed JC. p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. Embo J. 1998;17:2736-2747
66. Fanelli M, Fantozzi A, De Luca P, Caprodossi S, Matsuzawa S, Lazar MA, Pelicci PG, Minucci S. The coiled-coil domain is the structural determinant for mammalian homologues of Drosophila Sina-mediated degradation of promyelocytic leukemia protein and other tripartite motif proteins by the proteasome. J Biol Chem. 2004;279:5374-5379
67. Chan JY, Li L, Fan YH, Mu ZM, Zhang WW, Chang KS. Cell-cycle regulation of DNA damage-induced expression of the suppressor gene PML. Biochem Biophys Res Commun. 1997;240:640-646
68. Barr SM, Leung CG, Chang EE, Cimprich KA. ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Curr Biol. 2003;13:1047-1051
69. Varadaraj A, Dovey CL, Laredj L, Ferguson B, Alexander CE, Lubben N, Wyllie AH, Rich T. Evidence for the receipt of DNA damage stimuli by PML nuclear domains. J Pathol. 2007;211:471-480
70. Yang L, Yeh SD, Xie S, Altuwaijri S, Ni J, Hu YC, Chen YT, Bao BY, Su CH, Chang C. Androgen suppresses PML protein expression in prostate cancer CWR22R cells. Biochem Biophys Res Commun. 2004;314:69-75
71. Cao X, Clavijo C, Li X, Lin HH, Chen Y, Shih HM, Ann DK. SUMOylation of HMGA2: selective destabilization of promyelocytic leukemia protein via proteasome. Mol Cancer Ther. 2008;7:923-934
72. Lee HR, Kim DJ, Lee JM, Choi CY, Ahn BY, Hayward GS, Ahn JH. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J Virol. 2004;78:6527-6542
73. Kang H, Kim ET, Lee HR, Park JJ, Go YY, Choi CY, Ahn JH. Inhibition of SUMO-independent PML oligomerization by the human cytomegalovirus IE1 protein. J Gen Virol. 2006;87:2181-2190
74. Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol. 2006;80:7995-8005
75. Leppard KN, Everett RD. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J Gen Virol. 1999;80( Pt 4):997-1008
76. Hoppe A, Beech SJ, Dimmock J, Leppard KN. Interaction of the adenovirus type 5 E4 Orf3 protein with promyelocytic leukemia protein isoform II is required for ND10 disruption. J Virol. 2006;80:3042-3049
77. Boutell C, Orr A, Everett RD. PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J Virol. 2003;77:8686-8694
78. Everett RD, Sourvinos G, Orr A. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J Virol. 2003;77:3680-3689
79. Ling PD, Tan J, Sewatanon J, Peng R. Murine gammaherpesvirus 68 open reading frame 75c tegument protein induces the degradation of PML and is essential for production of infectious virus. J Virol. 2008;82:8000-8012
80. Herzer K, Weyer S, Krammer PH, Galle PR, Hofmann TG. Hepatitis C virus core protein inhibits tumor suppressor protein promyelocytic leukemia function in human hepatoma cells. Cancer Res. 2005;65:10830-10837
81. Bischof O, Nacerddine K, Dejean A. Human papillomavirus oncoprotein E7 targets the promyelocytic leukemia protein and circumvents cellular senescence via the Rb and p53 tumor suppressor pathways. Mol Cell Biol. 2005;25:1013-1024
82. Nakahara T, Lambert PF. Induction of promyelocytic leukemia (PML) oncogenic domains (PODs) by papillomavirus. Virology. 2007;366:316-329
83. Kofod-Olsen E, Ross-Hansen K, Mikkelsen JG, Hollsberg P. Human herpesvirus 6B U19 protein is a PML-regulated transcriptional activator that localizes to nuclear foci in a PML-independent manner. J Gen Virol. 2008;89:106-116
84. Turelli P, Doucas V, Craig E, Mangeat B, Klages N, Evans R, Kalpana G, Trono D. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol Cell. 2001;7:1245-1254
85. Borden KL, Campbell Dwyer EJ, Salvato MS. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J Virol. 1998;72:758-766
86. Sivachandran N, Sarkari F, Frappier L. Epstein-Barr nuclear antigen 1 contributes to nasopharyngeal carcinoma through disruption of PML nuclear bodies. PLoS Pathog. 2008;4:e1000170
87. Wang ZG, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi PP. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547-1551
88. Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ, Verbel DA, Cordon-Cardo C, Pandolfi PP. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J Natl Cancer Inst. 2004;96:269-279
89. Ito K, Bernardi R, Morotti A, Matsuoka S, Saglio G, Ikeda Y, Rosenblatt J, Avigan DE, Teruya-Feldstein J, Pandolfi PP. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453:1072-1078
90. Zimber A, Nguyen QD, Gespach C. Nuclear bodies and compartments: functional roles and cellular signalling in health and disease. Cell Signal. 2004;16:1085-1104
91. Regad T, Bellodi C, Nicotera P, Salomoni P. The tumor suppressor Pml regulates cell fate in the developing neocortex. Nat Neurosci. 2009;12:132-140
Author contact
![]() Correspondence to: Hung-Ying Kao, Department of Biochemistry, School of Medicine, Case Western Reserve University (CASE) the Comprehensive Cancer Center of CASE. 10900 Euclid Avenue, Cleveland, Ohio 44106, USA. TEL (216)368-1150; Fax (216)368-3419; Email hxk43edu
Correspondence to: Hung-Ying Kao, Department of Biochemistry, School of Medicine, Case Western Reserve University (CASE) the Comprehensive Cancer Center of CASE. 10900 Euclid Avenue, Cleveland, Ohio 44106, USA. TEL (216)368-1150; Fax (216)368-3419; Email hxk43edu

 Global reach, higher impact
Global reach, higher impact