Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2009; 5(5):421-427. doi:10.7150/ijbs.5.421 This issue Cite
Research Paper
HCV Antibody Response and Genotype Distribution in Different Areas and Races of China
1. The Institute for Disease Prevention and Control of PLA, Beijing, China.
2. Institute of Basic Medical Sciences, Beijing, China.
3. State Key Laboratory of Biotherapy and Cancer Center, West China Medical School, Sichuan University, Chengdu, China.
4. The 302 nd Hospital of PLA, Beijing, China.
5. Department of Surgery, University of Michigan, Ann Arbor, Michigan, USA
* These authors contributed equally to the work.
Received 2009-4-30; Accepted 2009-6-4; Published 2009-6-13
Abstract
Hepatitis C virus (HCV) heterogeneity accounts for the failure of effective vaccine development and the lack of successful anti-viral therapy in some patients. Little is known about the immune response to HCV peptides and the region or race specific genotypes in China. The objective of this study was to characterize HCV antibody immune response to HCV peptides and HCV genotypes in different regions and races of China. A total of 363 serum samples were collected from HCV carriers in 6 regions in China. The immune response to HCV peptides was evaluated by ELISA. HCV genotypes were examined using nested RT-PCR. We found that the anti-HCV antibody neutralization rates were significantly different among the serum samples from different areas or from different races in the same area. For samples from Tibet and Sinkiang, the rates of neutralization by HCV peptides were only 3.2% and 30.8%, respectively. The genotypes of samples from Tibet and Sinkiang were apparently heterogeneic and included type I, II, III and multiple types (I/II/III, I/II, I/III, II/III). One specific sample with multiple-genotype (I/II/III) HCV infection was found to consist of type I, II, III, II/III and an unclassified genotype. These studies indicate that the anti-HCV antibody immune response to HCV peptides varied across regions and among races. The distribution of HCV genotypes among Tibetans in Tibet and Uighurs in Sinkiang was different from that in the inner areas of China. In addition, a “master” genotype, type II, was found to exist in HCV infection with multiple HCV genotypes.
Keywords: Hepatitis C virus, anti-HCV, ELISA, neutralization test, genotype
Introduction
The incidence of Hepatitis C virus (HCV) infection has been steadily increasing in the last few decades in China and is expected to intensify in the coming years worldwide [1]. Peptide-based vaccines have been generated and tested in pre-clinical and clinical trials [2]. However, the development of effective peptide-based vaccines has been significantly hampered by the high genetic variability of HCV.
Anti-viral humoral immune response plays a fundamental role during HCV infection. Viral clearance is associated with the production of anti-envelope antibodies, and high serum levels of anti-viral envelope antibodies can prevent HCV infection in chimpanzees [3]. Nevertheless, the correlations between antibody production and its potential neutralizing immune response are still largely unknown.
HCV belongs to the hepacivirus genus in the Flaviviridae family [4]. There are 6 different genotypes of HCV and more than 70 sub-types based on the nucleic acid sequences [5]. HCV is therefore characterized by high levels of genetic heterogeneity, which accounts for the difficulties in vaccine development and for the lack of therapeutic efficacies. In China, the prevalence of HCV genotypes I and II has been previously reported [6]. However, little is known about the immune response to HCV peptides, and the region-specific distribution of HCV genotypes in China has not been fully evaluated.
In this report, we conducted serological and molecular studies on a large cohort of HCV carriers in 6 regions of China to determine the HCV antibody immune responses to HCV peptides and the distribution model of HCV genotypes in different areas and races of China.
Materials and Methods
Study population
Human HCV-positive serum samples from 363 HCV-infected patients were collected from hospitals in different areas of China: 35 from Shanghai, 20 from Shaanxi, 19 from Tianjin, 62 Tibetans, 23 Hans from Tibet, 146 Uighurs, 42 Hans from Sinkiang, and 16 from Hebei (Table 1). The selection of the study population was based on the following criteria: presence of HCV RNA in plasma confirmed by nested reverse transcription polymerase chain reaction (nRT-PCR); absence of other concomitant liver diseases; negative HIV test; no prior interferon and/or ribavirine treatment, and neither habitual alcohol nor active intravenous drug users. Control serum samples (n=3) were obtained from subjects who were negative for Hepatitis A, B, C, CMV (cytomegalovirus) and HIV. The peptides CP1, CP2 and NS4 used in this study were prepared in our laboratory:
CP1 (5~46): PKPQRKTKRNTNRRPQDVKFPGGGPIVGGVYLLPRRGPRLGV
CP2(39~80): RRGPRLGVRATRKTAERSQPRGRRQPIQKARRPEGRTWAQPGY
NS4(1694~1735): IIPDREVYREFDEMEECSQHLPYIEQGMMLAEQFKQKALGLL.
Characters of 363 HCV-infected patients in different areas of China
| Sample Number | ||||||||
|---|---|---|---|---|---|---|---|---|
| Province | Hebei | Shanghai | Shannxi | Tianjin | Tibet (Tibetans) | Tibet (Hans) | Sinkiang (Uighurs) | Sinkiang (Hans) |
| Total number | 16 | 35 | 20 | 19 | 62 | 23 | 146 | 42 |
| Gender: | ||||||||
| Male | 9 | 14 | 11 | 12 | 30 | 12 | 83 | 22 |
| Female | 7 | 21 | 9 | 7 | 32 | 11 | 63 | 20 |
| Age: (years old) | ||||||||
| 10∼20 | 1 | 3 | 0 | 1 | 2 | 1 | 16 | 3 |
| 21∼30 | 4 | 10 | 6 | 6 | 13 | 5 | 39 | 21 |
| 31∼40 | 5 | 7 | 7 | 6 | 24 | 7 | 52 | 15 |
| 41∼50 | 2 | 5 | 4 | 5 | 11 | 4 | 31 | 2 |
| 51∼60 | 4 | 6 | 3 | 1 | 6 | 5 | 7 | 1 |
| 61∼70 | 0 | 4 | 0 | 0 | 6 | 3 | 1 | 0 |
Anti-HCV ELISA
All 363 serum samples were screened for anti-HCV antibodies using ELISA (Bio-Rad, Marne-La-Coquette, France). The results of the assay were expressed quantitatively as the ratio of the optical density of the test sample to the calculated cut-off absorbance as recommended by the manufacturer. Serum samples with OD values ≥ 0.30 were considered to be positive, while those with OD values < 0.30 were considered negative. Positive and indeterminate serum samples were re-tested with another ELISA assay, Innotest HCV Ab IV (Innogenetics NV HCV, Gand, Belgium). Samples were confirmed as anti-HCV antibody positive when they were tested positive using both ELISA sets.
Neutralization of anti-HCV antibodies
For the neutralization experiments using ELISA, 10µl complex peptides (CP1, CP2, and NS4, 200µg/ml) were added to 90µl serum samples (1:10 dilution in PBS) and incubated 1h at 37°C. Ten microliters of each serum sample and the same sample incubated with the peptides were tested with the HCV ELISA as described previously [7-8]. The cutoff for the assay was established for each test from internal controls. Previously tested anti-HCV negative blood samples and the diluted buffers served as negative controls. An OD value reduced by >50% was used to indicate a positive neutralization. Anti-HCV neutralization ratios in different regions or races = (Anti-HCV neutralization positive sample numbers/Anti-HCV positive sample numbers)×100%.
HCV RNA detection
HCV RNA preparation and cDNA synthesis were performed as described previously[9]. The extracted RNA was used as a template and was amplified by nested RT-PCR with primers specific for the 5'UTR (5' non-coding region) of HCV[10-11]. PCR products were re-amplified using the same cycling program. Negative controls lacking template were included for each pair of primers. If a control was found positive, all of the PCR products in that set were considered to be contaminated and were discarded. Amplified cDNA was electrophoresed by 2.0% agarose gel and stained with ethidium bromide.
Genotyping of HCV
HCV genotypes were determined by amplification of the core region with genotype-specific primers that distinguished between genotypes I, II, III and IV [12-13]. Part of the HCV core gene (272 bp) was amplified from HCV cDNA with universal primers. A portion of the product was then amplified by PCR with universal sense primers and a mixture of five antisense primers deduced from HCV core (HCV-C) gene sequences specific for genotypes I, II, III and IV [12, 14]. The four genotypes were distinguished from one another by the size of the PCR products: 123 bp for genotype I; 211 bp for II; 240 bp for III and 188 bp for IV.
Cloning, sequencing and analysis of partial HCV-C gene from a multi-genotype (I/II/III) serum sample of a Uighur
The PCR products from a serum sample of a Uighur with multi-genotype (I/II/III) HCV infection were cloned into T-vector which was then transferred into E.coli XL1-Blue and cultured in LB plate at 37°C overnight. Recombinant clones were selected randomly and positive clones were detected and genotyped by PCR as described above. Partial HCV-C genes from the positive clones were amplified by nest-PCR with external primers 5'-ATGAGCACGAACATTCCTAAAACC-3' and 5'-AGCGGAAGCTGGGAGTGGT-3' and internal primers: 5' -CACTCTCGAGCACCCTATCAGGCAGT-3' and 5'-TTCACGCAGAAAGCGTCTAG-3'. Positive and negative controls were included at the extraction step and in both rounds of amplification. PCR products were sequenced using the dideoxy-mediated chain-termination method with a 373A Automatic DNA Sequence Analysis Machine (Applied Biosystems, Weiterstadt, Germany). Individual sequences were analyzed with MegAlign software (DNAStar Inc., Madison, WI).
Statistics
Data are expressed as means ± SD. Statistical analysis was conducted using StatView. Significant differences between groups were determined by ANOVA, and p<0.05 was considered significant. In order to detect the differences among categorical variables of neutralization rate (%), chi-square (χ2) test was applied. In order to detect differences among categorical variables of neutralization OD±SD, t test (t) was applied.
Results
Anti-HCV neutralization
The results in Table 2 show that anti-HCV neutralization rate was significantly different among the human serum samples collected from different areas of China. The neutralization rate in samples collected in Hebei was the highest (87.5%). The rates in Shanghai, Shaanxi, Tianjin were intermediate, and the rates in samples from Tibet and Sinkiang were low. Only 2 of the 62 (3.2%) serum samples from Tibet were neutralized by the HCV complex antigens.
The data in Table 2 also show. that pre-neutralization, the OD value of samples from Tianjin was the highest (1.54±0.64), while that of samples from Tibet was the lowest (0.66±0.26). Post-neutralization, the average OD values of samples from Shanghai, Shaanxi, Tianjin and Hebei appreciably declined (by 50% or nearly 50%), indicating that those samples were significantly neutralized. The OD value of samples from Sinkiang (Uighurs), however, declined slightly (by about 35%), and the OD value of samples from Tibet (Tibetans) barely declined (3.3%).
Table 3 shows that the neutralization rate and average OD values pre- and post-neutralization were significantly different among the serum samples collected from different races in Tibet and Sinkiang. The neutralization rates of Hans in the same area were higher than that of other races, and the OD values of Hans declined more significantly than that of the other races in the same area.
HCV genotypes
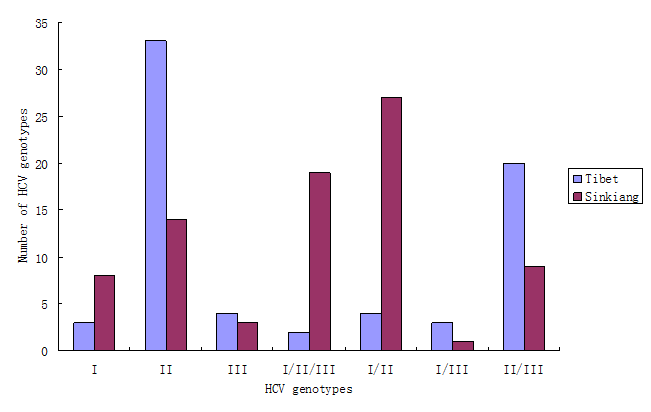
The results in Fig.1 show that the HCV genotype among Tibetans was mainly type II (33/69, 47.8%), followed by type II/III (20/69, 29.1%). Other genotypes were rare. The HCV genotypes among Uighurs in Sinkiang, however, were mainly type I/II (27/81, 33.3%), followed by type I/II/III (19/81, 23.5%), and type II (14/81, 17.3%).
The results in Table 4 indicate that most type I samples (7/11, >60%) were anti-HCV negative. The type I anti-HCV positive samples could not been neutralized by the complex antigens used in this study. Furthermore, the average OD values for type I was lower than others. Most type II samples (42/47, >80%) were anti-HCV positive and some of these samples could be neutralized. The type II samples had high OD values that clearly declined post-neutralization. In addition, the neutralization rate was different for the same genotype in samples collected from different areas. For example, the neutralization rate of type II from Tibet (Table 5: 6.9%) was much lower than that from Sinkiang (Table 6: 61.5%).
Anti-HCV antibody neutralization in serum samples collected from different areas of China and the average OD values pre- and post-neutralization
| Source of samples | Number of anti-HCV positive samples | Number of samples neutralized with HCV peptides | Neutralization rate(%) | Pre- neutralization OD±SD | Post- neutralization OD±SD |
|---|---|---|---|---|---|
| Shanghai | 35 | 23 | 65.7 | 0.83±0.63 | 0.36±0.34 |
| Shaanxi | 20 | 10 | 50.0 | 0.77±0.57 | 0.33±0.26 |
| Tianjin | 19 | 12 | 63.2 | 1.54±0.64 | 0.79±0.72 |
| Tibet (Tibetans) | 62 | 2 | 3.2 | 0.66±0.26 | 0.64±0.28 |
| Sinkiang (Uighurs) | 146 | 45 | 30.8 | 0.98±0.54 | 0.64±0.36 |
| Hebei | 16 | 14 | 87.5 | 0.84±0.74 | 0.18±0.16 |
χ2=70.57, P<0.001; t=2.978, P<0.05
(χ2 stands for chi-square test; t stands for t test)
Anti-HCV antibody neutralization in serum samples collected from different races in Tibet and Sinkiang
| Source of serum | Number of anti-HCV positive samples | Number of samples neutralized with HCV peptides | Neutralization rate(%) | Pre - neutralization OD±SD | Post- neutralization OD±SD | |
|---|---|---|---|---|---|---|
| Area | Race | |||||
| Tibet | Tibetans | 62 | 2 | 3.2* | 0.66±0.26 | 0.64±0.28Δ |
| Tibet | Hans | 23 | 8 | 34.7* | 0.82±0.34 | 0.61±0.32Δ |
| Sinkiang | Uighurs | 146 | 45 | 30.8** | 0.98±0.54 | 0.64±0.36ΔΔ |
| Sinkiang | Hans | 42 | 25 | 59.5** | 0.87±0.41 | 0.48±0.38ΔΔ |
*χ2=13.20, P<0.001; ** χ2=11.50, P<0.001; Δt=1.41, P>0.05; ΔΔt=3.75, P<0.05
(χ2 stands for chi-square test; t stands for t test)
Relations of genotypes, average OD values and neutralization rates
| Genotype (number) | Anti- HCV (-) | Anti- HCV (+) | Number of samples neutralized with HCV peptides | Neutralization rate(%) | Pre- neutralization OD±SD | Post- neutralization OD±SD |
|---|---|---|---|---|---|---|
| I (11) | 7 | 4 | 0 | 0 | 0.38±0.32 | 0.37±0.28 |
| II (47) | 5 | 42 | 10 | 23.8 | 0.94±0.40 | 0.67±0.42 |
| III (7) | 1 | 6 | 1 | 16.7 | 0.69±0.38 | 0.51±0.43 |
| I/II/III (21) | 4 | 17 | 9 | 52.9 | 0.67±0.52 | 0.52±0.48 |
| I/II (31) | 7 | 24 | 11 | 45.8 | 0.74±0.44 | 0.61±0.34 |
| I/III (4) | 1 | 3 | 1 | 33.3 | 0.74±0.27 | 0.60±0.32 |
| II/III (29) | 6 | 23 | 5 | 21.7 | 0.89±0.37 | 0.62±0.36 |
Relations of genotypes, average OD values and neutralization rates in Tibet
| Genotype (number) | Anti- HCV (-) | Anti- HCV (+) | Number of samples neutralized with HCV peptides | Neutralization rate(%) | Pre- neutralization OD±SD | Post- neutralization OD±SD |
|---|---|---|---|---|---|---|
| I(3) | 2 | 1 | 0 | 0 | 0.32 | 0.31 |
| II(33) | 4 | 29 | 2 | 6.9 | 0.68±0.25 | 0.66±0.27 |
| III(4) | 1 | 3 | 0 | 0 | 0.44±0.34 | 0.41±0.35 |
| I/II/III(2) | 1 | 1 | 0 | 0 | 0.36 | 0.31 |
| I/II(4) | 1 | 3 | 0 | 0 | 0.57±0.32 | 0.54±0.26 |
| I/III(3) | 1 | 2 | 0 | 0 | 0.62±0.27 | 0.57±0.32 |
| II/III(20) | 5 | 15 | 0 | 0 | 0.65±0.28 | 0.60±0.27 |
Relations of genotypes, average OD values and neutralization rates in Sinkiang
| Genotype (number) | Anti- HCV (-) | Anti- HCV (+) | Number of samples neutralized with HCV peptides | Neutralization rate(%) | Pre- neutralization OD±SD | Post- neutralization OD±SD |
|---|---|---|---|---|---|---|
| I(8) | 5 | 3 | 0 | 0 | 0.43±0.32 | 0.42±0.28 |
| II(14) | 1 | 13 | 8 | 61.5 | 1.20±0.54 | 0.68±0.56 |
| III(3) | 0 | 3 | 1 | 33.3 | 0.94±0.41 | 0.60±0.51 |
| I/II/III(19) | 3 | 16 | 9 | 56.2 | 0.98±0.52 | 0.72±0.48 |
| I/II(27) | 6 | 21 | 11 | 52.4 | 0.92±0.55 | 0.68±0.42 |
| I/III(1) | 0 | 1 | 1 | 100.0 | 0.85 | 0.62 |
| II/III(9) | 1 | 8 | 5 | 62.5 | 1.12±0.46 | 0.64±0.44 |
The HCV genotype of serum samples collected in Tibet and Sinkiang. The total number of sera collected in Tibet and in Sinkiang was 69 and 81 respectively. The results show that the HCV genotype among Tibetans was mainly type II (33/69, 47.8%), followed by type II/III (20/69, 29.1%). Other genotypes were rare. The HCV genotypes among Uighurs in Sinkiang, however, were mainly type I/II (27/81, 33.3%), followed by type I/II/III (19/81, 23.5%), and type II (14/81, 17.3%).
Cloning, sequencing and analysis of a partial HCV-C gene from a multi-genotype (I/II/III) serum sample of a Uighur
Partial HCV-C gene from a multi-genotype (I/II/III) infected Uighur was cloned and the recombinant clones were selected randomly. One hundred and forty two positive clones were confirmed and genotyped. They consisted of 1 type I (0.70%), 85 type II (59.86%), 40 type III (28.17%), 14 type II/III (9.86%) and 2 unclassified genotypes (1.41%). Genotype II was the most common. The sequences of the five clones that represented each of the five genotypes showed that the method used in this study for HCV genotyping could obviously differentiate among genotypes I, II and III. The genotypes II and III were not clearly distinguishable because the sequence of the type II/III clones was more closely related to genotype II. The sequence of the unclassified HCV genotype clone was also similar to that of type II. The DNA and amino acid sequences (data not shown) of these 5 clones were different from each other, and the DNA sequence homology of these 5 clones were 85%~98%.
Discussion
Genotyping of HCV strains is important in epidemiological studies and clinical practice. In particular, examination of sequence diversity can help understand the different patterns of serological reactivity, virulence and response to treatment [5, 15]. While it has been known that effective T cell response is essential in HCV infection [16-17], neutralizing antibodies also play a critical role [18]. It was observed that immunoglobulin prophylaxis could protect against HCV infection [19-20]. Studies of acutely infected cohorts also show that rapid induction of neutralizing antibodies may provide protection [21-22]. Moreover, antibodies present during the acute phase of infection can neutralize an infectious dose of HCV administered to chimpanzees [23]. Results from a different study also suggested that chronically infected patients develop low levels of or no neutralization antibodies [24-26]. Therefore, sequence differences among viruses may determine their susceptibility to neutralizations by antibodies. Investigating the nature of antigen-antibody interaction in HCV infections may lead to a better understanding of the lack or poor neutralizing efficacy of antibodies, and help developing novel therapeutic strategies using specific anti-HCV immunoglobulin.
In this report, we demonstrated that the anti-HCV neutralization rate and average OD values of pre- and post-neutralization were significantly different among human serum samples from different areas of China or from different races in the same area. In particular, in serum samples from Tibet and Sinkiang, only 3.2% and 30.8% of anti-HCV positive serum samples were neutralized by the HCV peptides (Table 2). These data suggest that the immune response to HCV peptides derived from a single HCV genotype varies depending on region and race. One reason for this may be that the titer of anti-HCV antibody in serum is different in different regions or among different races. Alternatively, the HCV genotypes may be different.
The response to therapy is dependent on several factors, e.g. viral genotypes and patient characteristics [1]. Therefore, it is important to track different genotypes of HCV virus in clinic [27]. In this study, we showed that the distribution of HCV genotypes among Tibetans and Uighurs was different from that in inner areas of China. In HCV RNA positive samples from Tibetans and Uighurs, there were 3 types, type I, type II, type III and multiple types (Fig.1). In most provinces of inner China, the prevalence of HCV genotypes I and II has been previously reported [6]. Genotype I viral strain is the most prevalent and most difficult to treat in Europe and North America and represents the greatest unmet treatment need. Genotypes II and III appear to be more prevalent in the Far East. Among the other genotypes, genotype IV was first found predominantly in the Middle East [28] and then in central Africa [6, 29-31], while genotypes V and VI are predominant in South Africa and South-East Asia, respectively [27]. This HCV heterogeneity could bring about a series of puzzles in the diagnosis of HCV infection and in the development of HCV vaccines.
In this study, multiple-genotype (I/II/III) HCV infection was detected by nest-PCR (Fig.1, Table 4, 5, 6). In particular, we found that a “master” genotype, type II (59.86%), exists in multiple infection with different HCV genotypes, and this is probably region-specific or race-specific.
The infection of different genotypes with different polypeptide antigens for anti-HCV positive patients in different regions of China is still unknown. Vicente Sentandreu et al. suggested that recombination should be considered as a potentially relevant mechanism of genetic variation in HCV [32]. Further studies focused on recombination among different HCV genotypes/subtypes may lead to more effective prevention and treatment for HCV infection.
Acknowledgements
This work was supported by grants from the Chinese Ministry of Science and Technology 863 project (No.2007AA02Z144) and the Natural Science Foundation (No. 30772001; No. 30671927).
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Manns MP, Foster GR, Rockstroh JK, Zeuzem S, Zoulim F, Houghton M. The way forward in HCV treatment--finding the right path. Nat Rev Drug Discov. 2007;6(12):991-1000
2. Lang K, Weiner DB. Immunotherapy for HCV infection: next steps. Expert Rev Vaccines. 2008;7(7):915-23
3. Missale G, Cariani E, Ferrari C. Role of viral and host factors in HCV persistence: which lesson for therapeutic and preventive strategies? Dig Liver Dis. 2004;36(11):703-11
4. Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39(1):5-19
5. Halfon P, Neumann AU, Bourliere M. et al. Slow viral dynamics of hepatitis C virus genotype 4. J Viral Hepat. 2003;10(5):351-3
6. Simmonds P. Genetic diversity and evolution of hepatitis C virus--15 years on. J Gen Virol. 2004;85(Pt 11):3173-88
7. Eiras A, Franco E, Montoro JA, Planelles D, Villaescusa R. HCV NAT (minipool RT-PCR) and HCV core antigen ELISA. Transfusion. 2003;43(1):118
8. Rodriguez M, Riezu-Boj JL, Ruiz M. et al. Immunogenicity of variable regions of hepatitis C virus proteins: selection and modi?cation of peptide epitopes to assess hepatitis C virus genotypes by ELISA. Journal of General Virology. 1999;80:727-38
9. Hermida M, Ferreiro MC, Barral S, Laredo R, Castro A, Diz DP. Detection of HCV RNA in saliva of patients with hepatitis C virus infection by using a highly sensitive test. J Virol Methods. 2002;101(1-2):29-35
10. Bukh J, Purcell RH, Miller RH. Importance of primer selection for the detection of hepatitis C virus RNA with the polymerase chain reaction assay. Proc Natl Acad Sci U S A. 1992;89(1):187-91
11. Ohno O, Mizokami M, Wu RR. et al. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microbiol. 1997;35(1):201-7
12. Okamoto H, Sugiyama Y, Okada S. et al. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992;73( Pt 3):673-9
13. Song P, Duc DD, Hien B. et al. Markers of hepatitis C and B virus infections among blood donors in Ho Chi Minh City and Hanoi, Vietnam. Clin Diagn Lab Immunol. 1994;1(4):413-8
14. Okamoto H, Tokita H, Sakamoto M. et al. Characterization of the genomic sequence of type V (or 3a) hepatitis C virus isolates and PCR primers for specific detection. J Gen Virol. 1993;74( Pt 11):2385-90
15. Fried MW. Viral factors affecting the outcome of therapy for chronic hepatitis C. Rev Gastroenterol Disord. 2004;4(Suppl 1):S8-S13
16. Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436(7053):946-52
17. Shoukry NH, Cawthon AG, Walker CM. Cell-mediated immunity and the outcome of hepatitis C virus infection. Annu Rev Microbiol. 2004;58:391-424
18. Owsianka AM, Tarr AW, Keck ZY. et al. Broadly neutralizing human monoclonal antibodies to the hepatitis C virus E2 glycoprotein. J Gen Virol. 2008;89(Pt 3):653-9
19. Conrad ME, Lemon SM. Prevention of endemic icteric viral hepatitis by administration of immune serum gamma globulin. J Infect Dis. 1987;156(1):56-63
20. Knodell RG, Conrad ME, Ginsberg AL, Bell CJ. Efficacy of prophylactic gamma-globulin in preventing non-A, non-B post-transfusion hepatitis. Lancet. 1976;1(7959):557-61
21. Lavillette D, Morice Y, Germanidis G. et al. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J Virol. 2005;79(10):6023-34
22. Pestka JM, Zeisel MB, Blaser E. et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci U S A. 2007;104(14):6025-30
23. Farci P, Alter HJ, Wong DC. et al. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci U S A. 1994;91(16):7792-6
24. Ishii K, Rosa D, Watanabe Y. et al. High titers of antibodies inhibiting the binding of envelope to human cells correlate with natural resolution of chronic hepatitis C. Hepatology. 1998;28(4):1117-20
25. Rosa D, Campagnoli S, Moretto C. et al. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc Natl Acad Sci U S A. 1996;93(5):1759-63
26. Meyer K, Ait-Goughoulte M, Keck ZY, Foung S, Ray R. Antibody-dependent enhancement of hepatitis C virus infection. J Virol. 2008;82(5):2140-9
27. Sy T, Jamal MM. Epidemiology of hepatitis C virus (HCV) infection. Int J Med Sci. 2006;3:41-46
28. Ray SC, Arthur RR, Carella A, Bukh J, Thomas DL. Genetic epidemiology of hepatitis C virus throughout egypt. J Infect Dis. 2000;182(3):698-707
29. Fretz C, Jeannel D, Stuyver L. et al. HCV infection in a rural population of the Central African Republic (CAR): evidence for three additional subtypes of genotype 4. J Med Virol. 1995;47(4):435-7
30. Simmonds P. The origin and evolution of hepatitis viruses in humans. J Gen Virol. 2001;82(Pt 4):693-712
31. Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36(5 Suppl 1):S21-9
32. Sentandreu V, Jimenez-Hernandez N, Torres-Puente M. et al. Evidence of recombination in intrapatient populations of hepatitis C virus. PLoS ONE. 2008;3(9):e3239
Author contact
![]() Correspondence to: Hongbin Song or Qiao Li, Center for Disease Control and Prevention, Academy of Military Medical Sciences, Beijing 100071, China. Tel: 86-10-66948475; Fax: 86-10-66948475; Email: hongbinsongnet; or Department of Surgery, University of Michigan, Ann Arbor; MI 48109-0666, USA. Tel: 734-615-1977, Fax: 734-763-4135; Email: qiaoliedu
Correspondence to: Hongbin Song or Qiao Li, Center for Disease Control and Prevention, Academy of Military Medical Sciences, Beijing 100071, China. Tel: 86-10-66948475; Fax: 86-10-66948475; Email: hongbinsongnet; or Department of Surgery, University of Michigan, Ann Arbor; MI 48109-0666, USA. Tel: 734-615-1977, Fax: 734-763-4135; Email: qiaoliedu

 Global reach, higher impact
Global reach, higher impact