Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2009; 5(6):558-569. doi:10.7150/ijbs.5.558 This issue Cite
Research Paper
Comparative characterization of a temperature responsive gene (lactate dehydrogenase-B, ldh-b) in two congeneric tropical fish, Lates calcarifer and Lates niloticus
Molecular Evolution and Ecology Laboratory, School of Marine & Tropical Biology, James Cook University, Townsville QLD 4811, Australia
Received 2009-5-19; Accepted 2009-8-26; Published 2009-9-1
Abstract
The characterization of candidate loci is a critical step in obtaining insight into adaptation and acclimation of organisms. In this study of two non-model tropical (to sub-tropical) congeneric perciformes (Lates calcarifer and Lates niloticus) we characterized both coding and non-coding regions of lactate dehydrogenase-B (ldh-b), a locus which exhibits temperature-adaptive differences among temperate and sub-tropical populations of the North American killifish Fundulus heteroclitus. Ldh-b was 5,004 and 3,527 bp in length in L. calcarifer and L. niloticus, respectively, with coding regions comprising 1,005 bp in both species. A high level of sequence homology existed between species for both coding and non-coding regions of ldh-b (> 97% homology), corresponding to a 98.5% amino acid sequence homology. All six known functional sites within the encoded protein sequence (LDH-B) were conserved between the two Lates species. Ten simple sequence repeat (SSR) motifs (mono-, di-, tri- and tetranucleotide) and thirty putative microRNA elements (miRNAs) were identified within introns 1, 2, 5 and 6 of both Lates species. Five single nucleotide polymorphisms (SNPs) were also identified within miRNA containing intron regions. Such SNPs are implicated in several complex human conditions and/or diseases (as demonstrated by extensive genome-wide association studies). This novel characterization serves as a platform to further examine how non-model species may respond to changes in their native temperatures, which are expected to increase by up to 6°C over the next century.
Keywords: Barramundi, Nile perch, exons, introns, microRNAs, SSRs
Introduction
The lactate dehydrogenase-B enzyme (LDH-B) plays a critical role in maintaining aerobic metabolism by converting lactate, the major by-product of anaerobic glycolysis, to pyruvate via oxidation in the presence of its coenzyme nicotinamide adenine dinucleotide, (NADH) (reviewed by [1, 2]). LDH-B can also convert lactate directly to glucose via gluconeogenesis. This conversion of accumulating lactate from aerobic tissues (e.g. heart, skeletal muscle) occurs in the liver and allows desired aerobic metabolic activity to be sustained for extended lengths of time (reviewed by [1, 2]). In addition to these metabolic functions, the LDH-B enzyme affects the oxygen binding affinity of hemoglobin (Hb) by altering intra-erythrocyte ATP concentrations of Hb in fish [3, 4]. This effect of LDH-B on Hb-O2 binding affinity directly impacts delivery of Hb-bound oxygen to red muscle tissues and may therefore be an alternate mechanism by which LDH-B affects sustainability of aerobic performance such as swimming performance in fish [2, 5, 6].
The dissolved oxygen which is available to aquatic organisms is inversely correlated with water temperature (Henry's Law) and thus leads to the potential for variability in the ability of Hb to uptake and transport oxygen under differing thermal regimes. In natural populations of aquatic organisms the genes involved in aerobic metabolism and oxygen transport, such as ldh-b and hemoglobin may therefore be subjected to strong selective pressure [3, 4, 7-11]. In fact, within thermally distinct populations of the temperate estuarine killifish Fundulus heteroclitus Hb-O2 affinity varies together with intra-erythrocyte ATP concentrations dependent on which LDH-B isozyme (LDH-Ba or LDH-Bb) is fixed [3, 4]. Moreover, a significant difference in transcript abundance of ldh-b was observed in response to both thermal and aerobic stress in discrete North American F. heteroclitus populations, even after acclimation to a common temperature. This transcript response was linked to a one base pair mutation in the glucocorticoid responsive element (GRE) identified within the ldh-b 5' proximal promoter ([7, 10]; see also reviews by [1, 2]). In addition to the extensive characterization and investigation of ldh-b in F. heteroclitus, the translated protein of this candidate gene has also been characterized and investigated in other temperate fishes like rainbow trout, Salmo gairdneri [12, 13] and crested blenny, Anoplarchus purpurescens [14]. However, this gene has not been fully characterized in any tropical perciform to date and there have been no investigations into the role this gene may have in thermal acclimation or the capacity to cope with thermal stress in tropical fishes. Therefore, the ldh-b locus appears to be an ideal candidate gene for the investigation of thermal adaptation and/or acclimation to native thermal regimes [15] in non-model tropical fish..
As a first step in understanding the role ldh-b may have in thermal adaptation or acclimation of tropical fish species, we characterized this gene in two tropical congenerics, the Australian barramundi (Lates calcarifer) and the African nile perch (Lates niloticus). Lates calcarifer is a catadromous, protoandrous hermaphrodite, native to rivers, estuaries and shallow marine environments throughout northern Australia (25°S - 12°S) and the south-east Asian archipelago (13°N - 10°S) [16-18], while Lates niloticus originates from east African rivers and lakes (7°S - 27°N) [19]. These species were targeted in this study for four reasons. Firstly, they occupy a range of different thermal environments; secondly, they are commercially valuable; thirdly, they are well suited for experimental manipulation and, fourthly, they are congeners which allows for comparisons of nucleotide sequences between these congeneric tropical perciformes. In addition to the traditional characterization of coding regions (exons) we also characterize, for the first time in non-model fish, the non-coding regions (introns) of the ldh-b locus in these perciform species to establish if regulatory motifs and/or elements (simple sequence repeats (SSRs) or microRNAs (miRNAs)), which are known to be embedded in or encoded by non-coding regions, are present. Previous studies on a diverse range of taxa, including fish, have demonstrated the presence of such elements within introns of other genes, where they are implicated in regulation or silencing of transcription [20-28].
Materials and methods
Complementary DNA (cDNA) synthesis from hepatic messenger RNA (mRNA)
As no ldh-b sequence information was available for either species, total liver RNA was first required for reverse transcription to obtain a Lates ldh-b cDNA sequence for primer design. This cDNA sequence was necessary to enable the design of ldh-b specific primers targeting the non-coding (intron) sequences of the ldh-b locus that can only be obtained from genomic DNA. Liver RNA was targeted for this initial cDNA sequencing as this tissue exclusively expresses the LDH-B protein as opposed to the alternative isozymes, LDH-A or C, which are expressed in other, non-hepatic tissues [29-31]. Total RNA was extracted from snap-frozen livers from four Darwin Harbour, Northern Territory, L. calcarifer individuals using Trizol as per manufacturer's instructions (Invitrogen Australia Pty, Mount Waverley Victoria). Extracted hepatic RNA was treated with Turbo DNA-free (Ambion, Austin, TX, USA) to remove any remaining trace DNA contamination. To ensure that trace amounts of genomic DNA, if present, do not contribute to the synthesis of cDNA, intron spanning primers were designed (detailed below). RNA purity was verified by NanoDrop Spectrophotometer (Invitrogen Australia Pty, Mount Waverley Victoria) analyses of 260/230 and 260/280nm absorbance. Messenger RNA (mRNA) was reverse transcribed to generate cDNA immediately via IM-Prom II Reverse Transcriptase with Oligo dT20 and random primers (Promega, Madison, WI USA), as per manufacturer's instructions.
Amplification of ldh-b from hepatic cDNA
Amplification and sequencing of ldh-b from the L. calcarifer hepatic cDNA was accomplished with general fish primers designed by aligning ldh-b coding sequences from a taxonomically diverse range of fishes from the National Center for Biotechnology Information (NCBI) sequence database (GenBank) [32]. The following sequences were aligned and primers were designed based on conserved regions - Gadiformes Trachyrincus murrayi [AJ609235], Merlangius merlangus [AJ609234], Gadus morhua [AJ609233] Coryphaenoides armatus [AJ609232]; Cypriniformes Danio rerio and Cyprinus carpio [AY644476]; Cyprinodontiformes, F. heteroclitus [L43525], F. heteroclitus (D. Crawford personal communication); Squaliformes Squalus acanthias [AF059035]. Following the design of several primers the most specific ldh-b product was attained using the forward primer designed from Gadiformes and Cypriniformes (ATGGCCTGTGCCGTCAGC) in conjunction with the reverse primer designed from Cypriniformes (TCTTTCAGGTCTTTCTGGAT), which anneal in exons 2 and 7 respectively. The remaining upstream sequence (exon 1 to 5' end of exon 2) was subsequently obtained using a previously published ldh-b forward primer for F. heteroclitus [33] and the reverse primers L. calcarifer-Intron2-R1 or L. niloticus-Intron2-R1 were designed from intron sequences subsequent to the initial cDNA amplification (Table 1). These intron sequences were obtained from genomic DNA amplification with Lates-specific ldh-b primers designed in exons 2 to 7.
PCR and sequencing primers used to obtain ldh-b sequences in Lates species. Segments 1 to 5 refer to regions depicted in Figure 1. All primers anneal to gDNA (see Methods). Primers used for forward and reverse sequencing reactions only (nested primers) are indicated by (SeqF) and (SeqR), respectively. Final MgCl2 concentrations in brackets refer to L. niloticus amplification requirements, which differed from that required for L. calcarifer.
| Lates spp. ldh-b Primers | 5' to 3' Sequence | MgCl2 | Ta | Amplicon (bp) |
|---|---|---|---|---|
| Segment 1* | ||||
| L. calcarifer-Seg1-R1 | GATTTAGACATGTCGTTCCTCAG | 1.5 - 2.5 | 61°C | 2000 |
| L. calcarifer-Seg1-R2 | ATAATGACACCATCAATGTTCACG | 1.5 - 2.5 | 61°C | 1000 |
| L. calcarifer-Intron1 (SeqR) | ATGGATGAATGTCTCAATCAG | 1.5 - 2.5 | 53°C | 500 |
| L. calcarifer-Intron2-R1 | TCGGATAACAGAAGCACTCAC | 1.5 - 2.5 | 55°C | 1500 |
| L. niloticus-Intron2-R1 | TAATCACTCATGGCCTCGG | 1.5 - 2.5 | 53°C | 1300 |
| L. niloticus-Intron1-R1 | AACTGGAAACTAATCTAGGCC | 1.5 - 2.5 | 55°C | 450 |
| L. niloticus-Intron1 (SeqR) | TCAGGTTAGCACTGCTGC | 1.5 - 2.5 | 51°C | 350 |
| Segment 2 | ||||
| L. calcarifer-F1 | TGATGGAGGATCGTCTGAAAGG | 1.5 - 2.5 | 61°C | 800 |
| L. calcarifer-R1 | TCGGCCATCAGGTAACGGAAG | 1.5 - 2.5 | 61°C | 800 |
| Segment 3 | ||||
| L. calcarifer-F2' | GTTGATGTGCTGACCTACGTC | 1.5 - 2.5 [3.5 - 4.5] | 59°C | 1500 |
| L. calcarifer-R2' | AGCCCTTCAGCTTGATCACC | 1.5 - 2.5 [3.5 - 4.5] | 57°C | 1500 |
| Segment 4 | ||||
| L. calcarifer-F3i | ACAGAGCTTCCACTGTATCAC | 1.5 - 2.5 | 57°C | 850 |
| L. calcarifer-R3i | GCAAAAGGTTCCTAGGCATGTA | 1.5 - 2.5 | 59°C | 850 |
| L. calcarifer-F3 | AGAAGCTGAACCCTGAGATCG | 1.5 - 2.5 [3.5 - 4.5] | 59°C | 800 |
| L. calcarifer-R3 | TTCTGGATGCCCCACAGTGTG | 1.5 - 2.5 [3.5 - 4.5] | 61°C | 800 |
| Segment 5** | ||||
| L. calcarifer-F3i' | TGGTTGCTAGGATAAAGAATGTG | 1.5 - 2.5 | 59°C | 700 |
| L. calcarifer-F3i (SeqF) | AGTTGTAAATAATTCAGGCATC | 1.5 - 2.5 | 53°C | 500 |
| L. niloticus-Intron6-F1 | ATGTGGATAGCCTAGCTTAGC | 1.5 - 2.5 | 55°C | 400 |
* and **: Published forward and reverse F. heteroclitus ldh-b primers [33] used in conjunction with designed primers to amplify terminal (5' and 3') segments in both Lates species, respectively.
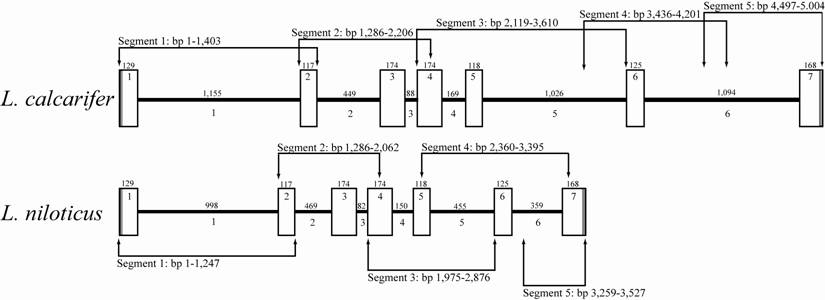
Comparative ldh-b gene map of congeneric Lates species. Exon (white boxes) and intron (black bars) sizes in number of base pairs (bp) are given above their respective graphic representation. Sequential numbering within white boxes and below black bars is for exons and introns, respectively. Arrowhead lines indicate region amplified by specific primer pairs (as per Table 1) along with the size of each segment in number of base pairs.
All PCR reactions were conducted in the following manner: Amplification reactions (20 µL) contained the following final concentrations: 1X Buffer [2.5 mM Tris pH 8.7, 5 mM KCl, 5 mM (NH4)2SO4, containing 1.5 mM MgCl2] (Qiagen, Doncaster, Victoria Australia) or 1x Buffer [2.5 mM Tris pH 8.7, 5 mM KCl, 5 mM (NH4)2SO4 not containing 1.5 mM MgCl2] (Bioline Pty Ltd., Alexandria, New South Wales Australia) (unless more was required as per Table 1)], 250 µM each dNTP, 250 nM each primer (Table 1), 10 ng gDNA template and 0.75 to 1.5 units of Taq Polymerase (Qiagen and Bioline Pty Ltd.). Thermal cycling was conducted on a MJR DNA Engine thermal cycler (Bio-Rad Laboratories Pty., Ltd., Gladesville, New South Wales) as follows: initial denaturation at 94°C for 3 min followed by 35 cycles of 94°C denaturation for 30 sec, annealing at primer specific Ta for 30 sec (Table 1), 72°C extension for 30 to 90 sec depending on target fragment size with larger fragments (> 1,000 bp) requiring longer (> 60 sec) extension times (Table 1) and a final 72°C extension for 10 min. Melting temperature (Tm) was calculated via (A/T x 2 + G/C x 4) method, with the annealing temperature (Ta) set at 5°C less than Tm, for all primer-pairs (Table 1).
Verification of ldh-b sequences amplified from hepatic cDNA
Ldh-b amplification from hepatic RNA derived cDNA with Lates-specific primers (see above) generated a strong single-band product of approximately 800 bp for all L. calcarifer examined (n = 4). Subsequent amplification of genomic DNA with the published forward (F. heteroclitus-F, [33]) and designed reverse (L. calcarifer-Intron2-R1 and L. niloticus-Intron2-R1) primers resulted in a strong single-band product of approximately 1,500 bp and 1,300 bp (L. calcarifer and L. niloticus, respectively) containing the missing exon 1 and 5' end of exon 2 fragment. These products were precipitated with 120 µL isopropanol (70%) for 15 minutes followed by a 500 µL wash with 70% isopropanol prior to drying, re-suspension in water (10 µL) and subsequent sequencing (Macrogen, Inc., South Korea). Sequences were edited in BioEdit [34] and a contig made to give the full length coding sequence (1,005 bp) and produce a consensus cDNA sequence. To check that the correct ldh gene homologue had been obtained the sequence was used in a BLAST search of GenBank and also directly aligned using ClustalW in Mega 3.1 [35] to those ldh-b sequences previously utilized for general fish primer design (see above). The obtained sequence shared 92% homology with F. heteroclitus LDH-B amino acid sequence, a level of homology that is well above that reported for F. heteroclitus LDH-C v. LDH-B and LDH-C v. LDH-A (78% and 70% homology, respectively) [30]). As a further check that the correct ldh gene homologue was obtained all nucleotide (and deduced amino acids) sequences were aligned with those of Danio rerio ldh-b [AF067202] and ldh-a [NM_131246] genes. These two gene homologues differ by 600 bp in length and 34.1% of the nucleotide sequences, respectively. The L. calcarifer ldh-b sequence obtained from hepatic cDNA most closely matched that of the D. rerio ldh-b gene homologue. Once the initial cDNA sequence was obtained from L. calcarifer the full characterization of the ldh-b locus from both Lates species (L. calcarifer and L. niloticus) was accomplished via primer walking along genomic DNA from representative individuals of each species.
Study species and genomic DNA (gDNA) extraction
Genomic DNA was extracted from L. calcarifer samples collected from four locations within tropical Australia. Samples from Gladstone, Queensland (23°S, 151°E) and Darwin, Northern Territory (12°S, 130°E) were obtained directly from fish farms while Archer River (Cape York, Queensland: 13°S, 142°E) and Tully River (Tully, Queensland: 17°S, 145°E) samples originated from wild caught fish. L. niloticus was purchased as two imported frozen fillets at a local supermarket in Townsville, Queensland and therefore the exact geographical origin of the L. niloticus samples examined is unknown but assumed to be from one of the African Rift Valley lakes (4°N - 14°S) where a large export fishery of this species exists. L. niloticus fillets may or may not have been snap-frozen; regardless, extracted genomic DNA appeared to be of equal quality to the DNA from L. calcarifer samples. Fin-clips/muscle tissue were taken from all fish and preserved in ethanol (90%). DNA extractions were performed via proteinase-K digestion (20 mg/mL) in CTAB buffer at 60°C for 1 hr and DNA was subsequently cleaned with a salt and chloroform:isoamyl alcohol (24:1) procedure [36]. All extractions resulted in high molecular weight gDNA, as visualized on a 0.8% agarose gel, with quantities ranging from 20 - 100 ng/µL.
Amplification of the ldh-b locus from genomic DNA
Full length ldh-b gene sequences (including intron sequences) were obtained from genomic DNA extracts using the primers and primer specific PCR conditions outlined in Table 1 and Figure 1. In some cases different primers were required for amplification of L. niloticus and L. calcarifer introns (e.g. intron 5 & 6 in Figure 1). See Figure 1 for primer binding locations within the ldh-b locus. Thermocycling parameters were the same as that used for cDNA amplification with the exception of the annealing temperature and MgCl2 concentration which varied as listed in Table 1. All PCR products obtained from both L. calcarifer and L. niloticus genomic DNA were verified as ldh-b in two ways: 1) searching the NCBI database (GenBank) via nucleotide (blastn) and protein (blastp) basic local alignment and search tool (b.l.a.s.t.) with resulting matches specific to LDH-B exclusively and 2) direct alignment with F. heteroclitus ldh-b nucleotide and LDH-B protein sequence. After confirmation of product identity full length gene sequences for each individual (L. calcarifer: n = 4 and L. niloticus: n = 2) were assembled in BioEdit and the consensus nucleotide and deduced amino acid sequences from each species were aligned using ClustalW in Mega 3.1. The high level of conservation across individuals from both Lates species allowed for the compilation of L. calcarifer and L. niloticus consensus sequences. Consensus gene sequences for each species were submitted to GenBank under accession numbers [FJ439509] and [FJ439510] for L. calcarifer and L. niloticus respectively. Additionally, L. calcarifer ldh-b coding sequence isolated from hepatic mRNA was submitted to GenBank under accession number [FJ439507]. Deduced LDH-B amino acid sequences were manually assessed for the presence of variation within the NH2-terminal arm (residues 1-20), coenzyme binding domain (residues 21-95 and 118-163), substrate binding domain (residues 164-333) and loop helix ά D region (residues 96-117) [37].
Assessment of non-coding (intron) sequences for micro RNA (miRNA) and simple sequence repeat (SSR) motifs
Several recent studies have demonstrated intron sequences may contain simple sequence repeat (SSR) motifs of functional importance as they potentially bind regulatory machinery (e.g. promoters and/or enhancers) and may affect gene expression levels [25, 38]. Intron sequences from ldh-b of both Lates species were manually assessed for the presence of simple sequence reseat (SSR) motifs. In addition, short microRNA (miRNA) elements (21-23 bp) may be encoded by introns and these are believed to be spliced out of pre-messenger RNA (mRNA) subsequently targeting regions within the 3'UTR of actively expressed mRNAs regulating translation from mRNA transcripts to functional proteins [20-23, 39, 40]. Intron sequences from Lates spp. were therefore assessed for putative miRNA elements with the software package miRanda [40]. All presented miRNA elements are located within conserved intron regions of Lates spp. and matched known miRNA motifs from Danio rerio (zebrafish), Takifugu rubripes (Japanese pufferfish) and Tetraodon nigroviridis (spotted green pufferfish) (miRBase: [41-43]). Several of the identified elements also matched known Xenopus tropicalis (African frog) miRNA motifs (miRBase: [41-43]). A nucleotide match score (score), affinity to bind measure (energy [kcal mol-1]), statistical assessment of match quality (z-score) and homology of motif to query intron sequence (percentage) were all calculated by miRanda for each matching miRNA element [40]. Threshold values were set to 100, -19 kcal mol-1 and 5.0 for score, energy and z-score, respectively, to assess ldh-b for miRNAs under strict parameters so as to avoid false-positive identifications (≤ 5%) [40, 44, 45]. However, overly-stringent thresholds were avoided to maximize the likelihood of identifying putative miRNA as such elements have recently been documented in high abundance within eukaryotic genomes at an average of 100 binding sites per miRNA element, genome-wide [45].
Results and Discussion
Descriptive characterization of ldh-b coding nucleotide (exon) and deduced amino acid sequences
Variation in the primary sequence and level of gene expression of the ldh-b locus has been linked to differences in aerobic performance (reviewed by [1, 2]) and natural selection [7, 46] in the temperate (to sub-tropical) fish F. heteroclitus. The present study is the first to characterize the full length gene sequence of this important metabolic gene in two tropical perciform fish, namely L. calcarifer and L. niloticus. This locus consisted of 5,004 and 3,527 bp, of which 1,005 bp was coding, in these two species, respectively (Figure 1). Consistent with other fish species the ldh-b locus of both L. calcarifer and L. niloticus contained seven exons and six introns, with all seven ldh-b exons being conserved in size between the characterized Lates species (Figure 1). Additional coding nucleotide sequence comparison revealed 29 base differences between L. calcarifer and L. niloticus (2.9% divergence); however, the majority of these occurred in the third codon positions which lead to silent (i.e. synonymous) amino acid substitutions. This high level of conservation was expected between Lates species being that this locus is known to be under functional constraint due to it role in maintaining aerobic metabolism ([7, 46]; see also review by [2]). Moreover, the encoded enzyme (LDH-B) has also been shown to impact on hemoglobin-oxygen binding affinity in fish, with allozyme variants altering this critical interaction [3, 4].
Each residue of the post-translation modified protein sequences has a role in the specific functioning of the molecule, from its internal stability to its external functional interactions. Five amino acid differences (1.5%) are present between the LDH-B of both Lates spp. These variable amino-acids involve the substitution of threonine (T) for methionine (M), leucine (L) for alanine (A), valine (V) for isoleucine (I), lysine (K) for asparagine (N) and valine (V) for isoleucine (I) at residues 34, 35, 126, 127 and 147, respectively (Figure 2). All of these variable amino acid residues reside within the coenzyme (NADH) binding domain (residues 21-95 and 118-163), of LDH-B, which is otherwise conserved between Lates spp. (Figure 2). This is unanticipated in such a conserved functional domain [30, 37]. Noteworthy are the two shifts occurring at residues 34 and 127 as these invoke changes in polarity (polar - non-polar) and acidity (neutral polar - basic polar) between Lates species, respectively (Figure 2). The effect of these amino acid shifts on the catalytic efficiency (kcat) of LDH-B between L. calcarifer and L. niloticus is unknown and warrants further investigation. Conversely, the NH2-terminal arm (residues 1-20), substrate binding domain (residues 164-333) and loop helix ά D region (residues 96-117) [37], as well as the LDH-B substrate binding (residues 100, 107, 139, 170, 249) and proton acceptor (residue 194) sites found within [47], are conserved between both Lates species (Figure 2), as expected [30, 37].
Comparison between Lates species and F. heteroclitus full length LDH-B amino acid sequences revealed a relatively extensive divergence, with 30 amino acids (9.0% divergence) observed between these phylogenetically distant species (data not shown). The two residues demonstrated to have fixed differences between thermally and geographically distinct populations of F. heteroclitus were serine (S) v. alanine (A) and alanine (A) v. aspartic acid (D) at residues 185 and 311 for cold northern and warm southern populations, respectively (see review by [1, 2]). L. calcarifer and L. niloticus both possess serine (S) and aspartic acid (D) at residues 185 and 311, respectively (Figure 2). The deduced LDH-B amino acid sequence of both Lates species examined are therefore similar to the cold northern F. heteroclitus population at residue 185 and to the warm southern F. heteroclitus population at residue 311. These residues are located on the internal and external surfaces of the folded LDH-B protein, respectively ([48, 49]; see also reviews by [1, 2]). The former change (residue 185) has been hypothesized to be associated with a variation in thermal stability due to this residue being located at a hairpin turn in the center of the folded protein; whereas the latter change (residue 311) has been hypothesized to be associated with a variation in substrate binding affinity due to this residue being located on the external surface of the conformed protein ([48, 49]; see also reviews by [1, 2]). In light of these previous hypothetical explanations, future studies should strive to compare the structure and function (e.g. enzymatic activity, effect on Hb-O2 binding affinity) of LDH-B in the Lates species in parallel to those studies conducted on within and among thermally discrete F. heteroclitus populations [4, 48, 49] and cod from two different temperatures [50].
Coding nucleotide sequence (CDS) for L. calcarifer and L. niloticus are given on lines 1 and 3, respectively. Deduced amino acid sequence (a.a.) for L. calcarifer and L. niloticus are given on lines 2 and 4, respectively. Bracketed numbers at ends of line 1 and 2 refer to nucleotide position and amino acid residue, respectively. NH2-terminal arm (residues 1-20) is dark-gray box outlined. Coenzyme binding domains (residues 21-95 and 118-163) are boldface box outlined. Substrate binding domain (residues 164-333) and loop helix ά D region (residues 96-117) are light-gray box outlined. Other known functional sites (residues 100, 107, 139, 170, 194 and 249) within domains are medium-gray box outlined. Residues (185 and 311) of known fixed difference between F. heteroclitus populations are boldface box outlined (see Results).

Descriptive characterization of ldh-b introns and identification of embedded putative regulatory motifs and/or elements
Nucleotide sequence comparisons of the homologous introns between the two Lates spp. characterized reveals a high level of homology (97.7%), a level surprisingly similar to that observed between exons of the Lates species (97.1% exon homology). The fact that sequence homology between non-coding introns is similar to that of coding exons suggests similar selective constraint (i.e. functionality) may be driving the high levels of homology observed across these historically non-characterized intronic regions [51, 52]. Interestingly, L. niloticus introns are consistently smaller than L. calcarifer introns with the exception of intron 2, which is 20 bp longer in L. niloticus relative to L. calcarifer (Figure 1). Moreover, one insertion-deletion event (indel) occurs between the two Lates species in all introns, with indels ranging in size across introns 1 to 6 (157, 20, 6, 19, 571 and 735 bp, respectively) (Figure 1). Whether these indels are historical insertion or deletion events and whether putative regulatory elements embedded within indel regions have an impact on the transcriptome or functional proteome of either Lates species is unknown. Therefore, future investigations into the impact these indels may or may not have on L. niloticus ldh-b gene expression, as compared to the variation observed among thermally discrete L. calcarifer populations [53], is warranted.
Numerous simple sequence repeat (SSR) sequences were detected within ldh-b introns of both Lates species (Table 2). Intronic SSRs are of interest because they may regulate gene transcription, lead to abnormal splicing and disrupt export of mRNA to the cytoplasm [25, 38, 52, 54]. Four mononucleotide SSRs (T, C and A), ranging from 5 to 11 repeats, are present within intron 1 of both Lates spp. and these were conserved in size between species (Table 2). In addition to these, two mononucleotide repeats (T10 and C7-11) are also present exclusively in L. calcarifer introns 1 and 6, respectively (Table 2). One dinucleotide repeat (AC) is present at the same location within intron 6 of both L. calcarifer and L. niloticus, but it differs in repeat number between the species (AC5 and AC8, respectively) (Table 2). In addition there is one trinucleotide repeat (CAA) present within intron 1 of L. niloticus and L. calcarifer which varies between 3 and 4 repeats, respectively (Table 2). Another trinucleotide repeat (TCC4) is present within a region in intron 2 and is conserved between both Lates species (Table 2). Noteworthy is that a similar (TCC4) SSR was identified in the 5' flanking untranslated region (UTR) of the ldh-b locus in F. heteroclitus and, more importantly, that variation in repeat number of this SSR in the 5' UTR of ldh-b impacted the level of ldh-b transcription (i.e. gene expression) observed in thermally discrete F. heteroclitus populations [8, 10, 11, 29, 55]. Moreover, this region within the 5' proximal promoter of F. heteroclitus, in addition to the 6fp and Sp1 binding sites also identified, were concluded to be under functional constraint by way of a phylogenetic analysis on the nucleotide sequences of these 5' UTR regulatory motifs which clearly differentiated the cold northern from the warm southern population [7, 46]. Lastly, a tetranucleotide repeat (TGTA4) is observed in a region of intron 6 exclusive to L. calcarifer (Table 2). The variation and potential functional role, if any, of these SSRs on ldh-b gene expression itself and/or on the greater transcriptome functionality within and among thermally discrete L. calcarifer and L. niloticus populations is currently unknown; however, further investigation into such potential implications on the transcriptome is warranted.
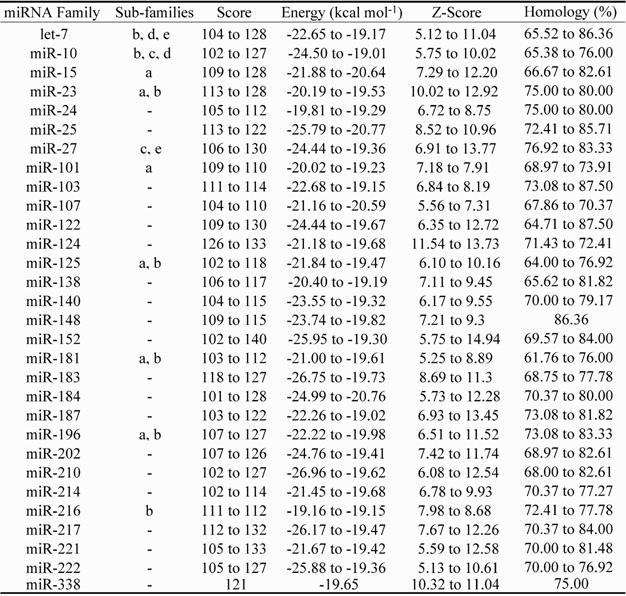
Numerous potential microRNA elements (miRNAs) were also identified within Lates species intron sequences (Table 3). The encoding of thirty putative miRNA elements was identified within conserved intron regions and these had a score, energy (kcal mol-1), z-score and homology ranging from 102 to 140, -26.96 to -19.01, 5.12 to 14.94 and 61.76 to 87.5%, respectively (see Methods). Four of the putative miRNA elements identified (Table 3: miR-let7b, miR-124, miR-181 and miR-223) have previously been associated with specific regulatory functions [21, 39, 56-59]. Of these, miR-let7b and miR-223 knockout in mouse HeLa cells [59] and neutrophils [21] resulted in reduced expression of approx. 2,700 and approx. 3,800 proteins, respectively. Two additional sub-families (d and e) from the highly investigated let-7 miRNA element family were identified within Lates spp. ldh-b introns (see Table 3), which provides additional data consistent with the widespread dispersal and high abundance of targets and/or functions of miRNA elements encoded by introns throughout the genome [21, 22, 45, 59]. Noteworthy is that the dre-let-7 (-b, -c, -d) miRNA elements have also been recently identified within the 5' UTR of the L. calcarifer myostatin gene (mstn) [60].
Five intronic single nucleotide polymorphisms (SNPs) also were identified in intronic regions of the ldh-b locus of individuals representing eight discrete L. calcarifer populations in a pilot population genetics screening [53]. Interestingly, these SNPs are present at sites where multiple miRNA elements overlap [53]. The fact that a single SNP can impact multiple putative miRNA elements concurrently may provide insight in regard to recent findings of genome-wide association studies, which show a relationship between such SNPs and variation in complex human behaviour (e.g. schizophrenia and bipolar disorder) and/or susceptibility to complex diseases (e.g. type 2 diabetes and Crohn's disease) [61-65].
Simple sequence repeat (SSR) motifs present within non-coding intron sequences of Lates species.
| SSR Motifs | Location | # of Repeats: L. calcarifer | # of Repeats: L. niloticus |
|---|---|---|---|
| Mononucleotide | |||
| T^ | Intron 1 | 10 | - |
| C | Intron 1 | 6 | 5 |
| C | Intron 1 | 8 | 5 |
| T | Intron 5 | 8 | 5 |
| A | Intron 6 | 7 | 8 |
| C^ | Intron 6 | 7 - 11 | - |
| Dinucleotide | |||
| AC | Intron 6 | 5 | 8 |
| Trinucleotide | |||
| CAA | Intron 1 | 4 | 3 |
| TCC* | Intron 2 | 4 | 4 |
| Tetranucleotide | |||
| TGTA^ | Intron 6 | 4 | - |
Putative microRNA (miRNA) motifs identified within ldh-b intron sequences of Lates spp. Score: nucleotide match score; Energy: affinity to bind measure; Z-Score: statistical assessment of match quality; Homology: motif homology to query intron sequence.
Evidence for selective constraint on non-coding (intron) sequences
The putative encoding of known functional motifs and/or elements (SSRs and miRNAs) by intronic regions of loci, in addition to the even distribution of such motifs and/or elements throughout longer introns [66], provides evidence that functional constraint is arguably acting on these historically less characterized non-coding regions. Indeed, such functional constraint is more likely to occur for introns of loci whose coding nucleotide or amino acid sequences are known to be under selection [22, 51, 52]. More specifically, longer introns ( > 87 bp) have been found to exhibit less divergence than shorter introns (< 87 bp) because of either an increased likelihood of embedded functional motifs being present in longer introns or the potential impact any mutations may have on the secondary structure of precursor messenger RNA (pre-mRNA) [66]. A recent pairwise and cross-taxa comparison of intron sequences between three mammalian species (human, whale and seal) revealed sequence homologies 14% and 12% higher, respectively, than those expected from a neutral model of evolution based on expected rates of substitution for non-coding DNA, further suggesting functional constraint acting upon non-coding intronic sequences [51]. Moreover, the existence of numerous intron motifs and/or elements is known to be essential for the functioning of complex multi-cellular organisms, as they permit a two-fold regulatory system in eukaryotic organisms: one for the transcriptome [20, 22, 25, 26, 28, 67] and one for the proteome [21, 39, 59]. Regardless, further research is required to determine if these motifs and/or elements (SSRs and/or miRNAs) have similar target sites and/or impacts on the transcriptome (i.e. gene expression) and/or the proteome (i.e. gene silencing) within and among Lates species and/or fish in general.
Conclusions
The ldh-b locus was found to be highly conserved between two tropical perciformes, L. calcarifer and L. niloticus, with just 2.9% divergence of coding regions and five amino acid differences between deduced LDH-B protein sequences. Variation within the coenzyme binding domains (residues 21-95 and 118-163) of the deduced protein sequences may possibly confer a variation in the specific catalytic efficiency of LDH-B in geographically isolated L. calcarifer and L. niloticus and warrants further exploration. Non-coding (intron) sequences of the ldh-b locus in both Lates species were as conserved as the coding regions of this gene, despite comprising 72.5 to 80% of the entire gene sequence. Ten SSR motifs and thirty putative miRNA elements were observed within the introns of the ldh-b locus in both Lates species. These putative regulatory elements and/or motifs warrant further investigation for their potentially functional importance in the regulation and/or expression of ldh-b or other constituent loci contributing to the transcriptome and proteome. The characterization of the ldh-b locus has spawned additional studies, one of which confirmed that ldh-b expression differs significantly between L. calcarifer populations from different temperature environments, suggesting that ldh-b is adaptive in this species [53], as was shown for the temperate (to sub-tropical) killifish Fundulus heteroclitus [7, 46]. These findings, in conjunction with a pilot population genetics screening of the ldh-b locus among and within discrete L. calcarifer populations [53], are suggestive of thermal adaptation occurring within this tropical estuarine species. Moreover, future studies on other non-model tropical species are also permitted by the ldh-b characterization presented herein. Indeed, future studies of this adaptive locus in other non-model tropical species will provide insight into the response of thermally sensitive species (living at the edge of their thermal maxima) to changes in their native temperatures, which are expected to increase by up to 6°C over the next century.
Acknowledgements
Thanks to Paolo Momigliano and Dianne Rowe for help in the Molecular Evolution and Ecology lab, Christian De Santis for L. niloticus gDNA samples, Manue Botte for RNA precipitation protocols, the Kuranda Fish Farm and Darwin Aquaculture Center for L. calcarifer fingerlings and the James Cook University Research Advancement Program for Fin-fish Genetics (RAP) for funding.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Powers D.A, Lauerman T, Crawford D. et al. Genetic Mechanisms for Adapting to a Changing Environment. Annu Rev Genet. 1991;25:629-659
2. Powers D.A, Schulte P.M. Evolutionary Adaptations of Gene Structure and Expression in Natural Populations in Relation to a Changing Environment: A Multidisciplinary Approach to Address the Million-Year Saga of a Small Fish. J Exp Zool Comp Exp Biol. 1998;282:71-94
3. Powers D.A. Molecular Ecology of Teleost Fish Hemoglobins: Strategies for Adapting to Changing Environments. Am Zool. 1980;20:139-163
4. Powers D.A, Greaney G.S, Place A.R. Physiological correlation between lactate dehydrogenase genotype and haemoglobin function in killifish. Nature. 1979;277:240-241
5. DiMichele L, Powers D.A. Physiological Basis for Swimming Endurance Differences between LDH-B Genotypes of Fundulus heteroclitus. Science. 1982;216:1014-1016
6. Fangue N.A, Mandic M, Richards J.G. et al. Swimming Performance and Energetics as a Function of Temperature in Killifish Fundulus heteroclitus. Physiol Biochem Zool. 2008;81:389-401
7. Crawford D.L, Pierce V.A, Segal J.A. Evolutionary Physiology of Closely Related Taxa: Analyses of Enzyme Expression. Am Zool. 1999;39:389-400
8. Crawford D.L, Segal J.A, Barnett J.L. Evolutionary Analysis of TATA-less Proximal Promoter Function. Mol Biol Evol. 1999;16:194-207
9. Rees B.B, Bowman J.A.L, Schulte P.M. Structure and Sequence Conservation of a Putative Hypoxia Response Element in the Lactate Dehydrogenase-B Gene of Fundulus. Biol Bull. 2001;200:247-251
10. Schulte P.M, Glemet H.C, Fiebig A.A. et al. Adaptive variation in lactate dehydrogenase-B gene expression: Role of a stress-responsive regulatory element. Proc Nati Acad Sci USA. 2000;97:6597-6602
11. Segal J.A, Schulte P.M, Powers D.A. et al. Descriptive and Functional Characterization of Variation in the Fundulus heteroclitus Ldh-B Proximal Promoter. J Exp Zool Comp Exp Biol. 1996;275:355-364
12. Kao Y.-H.J, Farley T.M. Purification and properties of allelic lactate dehydrogenase isozymes at the B2 locus in rainbow trout, Salmo gairdneri. Comp Biochem & Physio Pt B. 1978;61:507-512
13. Klar G.T, Stalnaker G.B, Farley T.M. Comparative blood lactate response to low oxygen concentrations in rainbow trout, Salmo gairdneri, LDH-B2 phenotypes. Comp Biochem & Physio Pt A. 1979;63:237-240
14. Johnson M.S. Association of allozymes and temperature in the crested blenny Anoplarchus purpurescens. Mar Biol. 1977;41:147-152
15. Cossins A.R, Crawford D.L. Opinion: Fish as models for environmental genomics. Nat Rev Genet. 2005;6:324-333
16. Chenoweth S.F, Hughes J.M, Keenan C.P. et al. When Oceans Meet: A Teleost Shows Secondary Intergradation at an Indian-Pacific Interface. Proc R Soc Lond B Biol Sci. 1998;265:415-420
17. Doupé R.G, Horwitz P, Lymbery A.J. Mitochondrial genealogy of Western Australian barramundi: applications of inbreeding coefficients and coalescent analysis for separating temporal population processes. J Fish Biol. 1999;54:1197-1209
18. Keenan C.P. Recent Evolution of Population Structure in Australian barramundi, Lates calcarifer (Bloch): An Example of Isolation by Distance in One Dimension. Aust J Mar Freshwat Res. 1994;45:1123-1148
19. Ribbink A.J. African lakes and their fishes: conservation and suggestions. Environ Biol Fish. 1987;19:3-26
20. Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-356
21. Baek D, Villen J, Shin C. et al. The impact of microRNAs on protein output. Nature. 2008;455:64-73
22. Bartel D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281-298
23. Doench J.G, Sharp P.A. Specificity of microRNA target selection in translational repression. Genes & Dev. 2004;18:504-511
24. Lai E.C. microRNAs: Runts of the Genome Assert Themselves. Curr Biol. 2003;13:R925-R936
25. Li Y.-C, Korol A.B, Fahima T. et al. Microsatellites Within Genes: Structure, Function, and Evolution. Mol Biol Evol. 2004;21:991-1008
26. Meloni R, Albanèse V, Ravassard P. et al. A tetranucleotide polymorphic microsatellite, located in the first intron of the tyrosine hydroxylase gene, acts as a transcription regulatory element in vitro. Hum Mol Genet. 1998;7:423-428
27. Reisman D, Greenberg M, Rotter V. Human p53 oncogene contains one promoter upstream of exon 1 and a second, stronger promoter within intron 1. Proc Nati Acad Sci USA. 1988;85:5146-5150
28. Wittekindt N.E, Hortnagel K, Geltinger C. et al. Activation of the c-myc promoter P1 by immunoglobulin  gene enhancers in Burkitt lymphoma: functional characterization of the intron enhancer motifs B, E box 1 and E box 2, and of the 3' enhancer motif PU. Nucleic Acids Res. 2000;28:800-808
29. Crawford D.L, Powers D.A. Evolutionary Adaptation to Different Thermal Environments via Transcriptional Regulation. Mol Biol Evol. 1992;9:806-813
30. Quattro J.M, Woods H.A, Powers D.A. Sequence analysis of teleost retina-specific lactate dehydrogenase C: Evolutionary implications for the vertebrate lactate dehydrogenase gene family. Proc Natl Acad Sci USA. 1993;90:242-246
31. Segal J.A, Crawford D.L. LDH-B enzyme expression: the mechanisms of altered gene expression in acclimation and evolutionary adaptation. Am J Physiol Regulatory Integrative Comp Physiol. 1994;267:1150-1153
32. Zhang J, Madden T.L. PowerBLAST: A new network BLAST application for interactive or automated sequence analysis and annotation. Genome Res. 1997;7:649-656
33. Bernardi G, Sordino P, Powers D.A. Concordant mitochondrial and nuclear DNA phylogenies for populations of the teleost fish Fundulus heteroclitus. Proc Natl Acad Sci USA. 1993;90:9271-9274
34. Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95-98
35. Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150-163
36. Sambrook J, Fritsch E, Maniatus T. Molecular cloning: a laboratory manual. New York, USA: Cold Spring Harbour Laboratory Press. 1989
37. Li S.S, Fitch W.M, Y.- Pan C.P. et al. Evolutionary Relationships of Vertebrate Lactate Dehydrogenase Isozymes A4 (Muscle), B4, (Heart), and C4, (Testis). J Biol Chem. 1983;258:7029-7032
38. Yue G, Li Y, Orban L. Characterization of Microsatellites in the IGF-2 and GH Genes of Asian Seabass (Lates calcarifer). Mar Biotechnol. 2001;3:1-3
39. Chen C.-Z, Li L, Lodish H.F. et al. MicroRNAs Modulate Hematopoietic Lineage Differentiation. Science. 2004;303:83-86
40. Enright A.J, John B, Gaul U. et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1-14
41. Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109- D111
42. Griffiths-Jones S, Grocock R.J, van Dongen S. et al. miRBase: micro sequences RNA, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140-D144
43. Griffiths-Jones S, Sain H.K, van Dongen S. et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2007;36:D154-D158
44. Bonnet E, Wuyts J, Rouzé P. et al. Evidence that micro precursors RNA, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911-2818
45. Brennecke J, Stark A, Russell R.B. et al. Principles of microRNA-target recognition. PLoS Biol. 2005;3:0404-0418
46. Pierce V.A, Crawford D.L. Phylogenetic Analysis of Glycolytic Enzyme Expression. Science. 1997;276:256-259
47. The UniProt C. The Universal Protein Resource (UniProt). Nucl Acids Res. 2008;36:D190-195
48. Place A.R, Powers D.A. Purification and Characterization of the Lactate Dehydrogenase (LDH-B4) Allozymes of Fundulus heteroclitus. J Biol Chem. 1984;259:1299-1308
49. Place A.R, Powers D.A. Kinetic Characterization of the Lactate Dehydrogenase (LDH-B4) Allozymes of Fundulus heteroclitus. J Biol Chem. 1984;259:1309-1318
50. Zakhartsev M, Johansen T, Pörtner H.O. et al. Effects of temperature acclimation on lactate dehydrogenase of cod (Gadus morhua): genetic, kinetic and thermodynamic aspects. J Exp Biol. 2004;207:95-112
51. Hare M.P, Palumbi S.R. High Intron Sequence Conservation Across Three Mammalian Orders Suggests Functional Constraints. Mol Biol Evol. 2003;20:969-978
52. Mattick J.S. Introns: evolution and function. Curr Opin Genet Dev. 1994;4:823-831
53. Edmunds R.C. Evidence for thermal adaptation among geographically, genetically and thermally distinct populations of the Australian barramundi, Lates calcarifer (Bloch 1790): a multi-level approach. PhD Thesis. Townsville: School of Marine and Tropical Biology, James Cook University. 2009
54. Silva M.C, Edwards S.V. Structure and Evolution of a New Avian MHC Class II B Gene in a Sub-Antarctic Seabird, the Thin-Billed Prion (Procellariiformes: Pachyptila belcheri). J Mol Evol. 2009;68:279-291
55. Schulte P.M, Gomez-Chiarri M, Powers D.A. Structural and Functional Differences in the Promoter and 5' Flanking Region of Ldh- Within B and Between Populations of the Teleost Fundulus heteroclitus. Genetics. 1997;145:759-769
56. Lagos-Quintana M, Rauhut R, Yalcin A. et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735-739
57. Lai E.C. MicroRNAs are complementary to 3' UTR motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363-364
58. Lai E.C, Tomancak P, Williams R.W. et al. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:1-20
59. Selbach M, Schwanhausser B, Thierfelder N. et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58-63
60. De Santis C, Evans B, Smith-Keune C. et al. Molecular characterization, tissue expression and sequence variability of the barramundi (Lates calcarifer) myostatin gene. BMC Genomics. 2008;9:82
61. Duerr R.H, Taylor K.D, Brant S.R. et al. A Genome-Wide Association Study Identifies IL23R as an Inflammatory Bowel Disease Gene. Science. 2006;314:1461-1463
62. Hirschhorn J.N, Daly M.J. Genome-wide association studies for common diseases and complex traits. Nature Reviews: Genetics. 2005;6:95-108
63. Scott L.J, Mohlke K.L, Bonnycastle L.L. et al. A Genome-Wide Association Study of Type 2 Diabetes in Finns Detects Multiple Susceptibility Variants. Science. 2007;316:1341-1345
64. Sladek R, Rocheleau G, Rung J. et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881-885
65. Wang W.Y.S, Barratt B.J, Clayton D.G. et al. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109-118
66. Haddrill P.R, Charlesworth B, Halligan D.L. et al. Patterns of intron sequence evolution in Drosophila are dependent upon length and content GC. Genome Biol. 2005;6:R67
67. Nakaya H.I, Amaral P.P, Louro R. et al. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007;8:R43
Author contact
![]() Correspondence to: Richard C. Edmunds, School of Marine & Tropical Biology, James Cook University, Townsville QLD 4811 Australia. Tel: +61 7 4781 5735; Fax: +61 7 4725 1570; E-mail: richard.c.edmundscom
Correspondence to: Richard C. Edmunds, School of Marine & Tropical Biology, James Cook University, Townsville QLD 4811 Australia. Tel: +61 7 4781 5735; Fax: +61 7 4725 1570; E-mail: richard.c.edmundscom

 Global reach, higher impact
Global reach, higher impact