Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2010; 6(1):89-95. doi:10.7150/ijbs.6.89 This issue Cite
Research Paper
Comparative study of the binding pockets of mammalian proprotein convertases and its implications for the design of specific small molecule inhibitors
Institute of Biomechanics/ School of Bioscience and Bioengineering, South China University of Technology, Guangzhou 510006, China
# Visiting scientist at the Institute of Biomechanics.
Received 2009-11-30; Accepted 2010-1-26; Published 2010-2-3
Abstract
Proprotein convertases are enzymes that proteolytically cleave protein precursors in the secretory pathway to yield functional proteins. Seven mammalian subtilisin/Kex2p-like proprotein convertases have been identified: furin, PC1, PC2, PC4, PACE4, PC5 and PC7. The binding pockets of all seven proprotein convertases are evolutionarily conserved and highly similar. Among the seven proprotein convertases, the furin cleavage site motif has recently been characterized as a 20-residue motif that includes one core region P6-P2´ inside the furin binding pocket. This study extended this information by examining the 3D structural environment of the furin binding pocket surrounding the core region P6-P2´ of furin substrates. The physical properties of mutations in the binding pockets of the other six mammalian proprotein convertases were compared. The results suggest that: 1) mutations at two positions, Glu230 and Glu257, change the overall density of the negative charge of the binding pockets, and govern the substrate specificities of mammalian proprotein convertases; 2) two proprotein convertases (PC1 and PC2) may have reduced sensitivity for positively charged residues at substrate position P5 or P6, whereas the substrate specificities of three proprotein convertases (furin, PACE4, and PC5) are similar to each other. This finding led to a novel design of a short peptide pattern for small molecule inhibitors: [K/R]-X-V-X-K-R. Compared with the widely used small molecule dec-RVKR-cmk that inhibits all seven proprotein convertases, a finely-tuned derivative of the short peptide pattern [K/R]-X-V-X-K-R may have the potential to more effectively inhibit five of the proprotein convertases (furin, PC4, PACE4, PC5 and PC7) compared to the remaining two (PC1 and PC2). The results not only provide insights into the molecular evolution of enzyme function in the proprotein convertase family, but will also aid the study of the functional redundancy of proprotein convertases and the development of therapeutic applications.
Keywords: small molecular inhibitor, mammalian proprotein convertases, substrate specificity, evolution of gene family.
1. Introduction
In the secretory pathway, many secreted proteins or transmembrane proteins are initially synthesized as protein precursors and subsequently cleaved by proprotein convertases before they become fully functional. The cleavage of protein precursors by proprotein convertases is involved in the molecular pathogenesis of a wide range of human diseases including cancer, neurological disorders, autoimmune diseases and various viral infections[1]. So far, seven mammalian subtilisin/Kex2p-like proprotein convertases have been identified: furin, PC1/PC3/SPC3, PC2/SPC2, PC4/SPC5, PACE4/SPC4, PC5/PC6/SPC6 and PC7/PC8/SPC7/LPC. The binding pockets of all seven proprotein convertases are evolutionarily conserved and highly similar[2,3]. Among the seven mammalian proprotein convertases, the cleavage of furin is the best characterized. The 3D crystal structure of dec-RVKR-cmk inhibited mouse furin binding pocket has been solved by Henrich and co-workers[4]. Furthermore, in authors' recent study, instead of a canonical four-residue motif R-X-[K/R]-R↓, the furin cleavage site has been characterized as a 20-residue motif that can be divided into two parts with distinct physical properties: 1) a core region (8 amino acids, position P6-P2´) inside the furin binding pocket that contributes to the binding strength of the substrate and 2) two polar regions (8 amino acids, positions P7-P14; and 4 amino acids, positions P3´-P6´) outside the furin binding pocket that mainly contribute to the solvent accessibility of the substrate[5].
The 3D structure of the furin binding pocket[4] and the physical properties of the core region P6-P2´ of the furin cleavage site motif[5] provide a template with which to compare the local structural-functional relationship of all mammalian proprotein convertases. In this report, a multiple sequence alignment of mammalian proprotein convertases was generated, and the physical properties of mutations in the binding pockets were examined. The functional consequences of mutations were interpreted using the 3D structure. The results indicated two residues that play a key role in regulation of the substrate specificity of mammalian proprotein convertases. This finding provides insights into the molecular evolution of the enzyme function of the proprotein convertase family. The comparison of 3D structures also suggested a novel design for specific small molecular inhibitors that will aid the study of the functional redundancy of mammalian proprotein convertases and the development of therapeutic applications.
2. Results and Discussion
2.1. 3D Structural environment of binding pocket of furin
The eight amino acids of the core region P6-P2´ of a furin substrate are located inside the furin binding pocket and are the main factor determining the binding strength of a substrate[5]. The residues of the mouse furin binding pocket surrounding the core region P6-P2´ of furin substrates was analyzed in the 3D crystal structure of the dec-RVKR-cmk inhibited mouse furin binding pocket[4] and also in the authors' recent study on the furin cleavage site motif[5]. Table 1 lists the structural environment surrounding the substrate positions P6-P2´.
List of substrate positions and their interacting residues in the mouse furin binding pocket which explain the substrate specificity of furin and the favoured physical properties at substrate position P6-P2'. The residues were retrieved from the 3D structure of the dec-RVKR-cmk inhibited mouse furin binding pocket (Protein Data Bank ID: IP8J)[4].
| Substrate position | Main interaction partners: residues and positions in the mouse furin binding pocket |
|---|---|
| P1 | His194 Ser253 Trp254 Gly255 Pro256 Glu257 Asp258 Ala292 Ser293 Gly294 Asn295 Asp301 Asp306 Glu331 Ser368 |
| P2 | Asp153 Asp154 Asp191 Asn192 Arg193 His194 |
| P3 | Leu227 Ser253 Trp254 Gly255 Pro256 Glu257 Asp258 |
| P4 | Leu227 Asp228 Gly229 Glu230 Val231 Thr232 Asp233 Glu236 Trp254 Gly255 Pro256 Glu257 Asp258 Asp264 Gly265 Pro266 Ala267 |
| P5, P6 | Leu227 Asp228 Gly229 Glu230 Val231 Thr232 Asp233 Glu236 Trp254 Gly255 Pro256 Glu257 Asp264 Tyr308 |
| P1' | His194 Ser253 Gly294 Asn295 His364 Thr365 Gly366 Thr367 Ser368 Ala369 |
| P2' | His194 Cys198 Ser253 Asn295 Ser319 Try329 Ser342 Ser343 Ile351 His364 Thr365 Gly366 Thr367 Ser368 Ala369 Ser370 Ala371 Pro372 |
2.2. Comparison of the 3D structural environment of binding pockets of seven mammalian proprotein convertases
To study the substrate specificity of the other six proprotein convertases (PC1, PC2, PC4, PACE4, PC5 and PC7), the 3D structural environments provided by these six different mammalian proprotein convertases were compared against furin. A multiple sequence alignment of the binding pockets of seven mammalian proprotein convertases was generated using Clustalx[6] and are publicly available on the associated website www.nuolan.org. The residue positions (listed in Table 1) of the binding pockets surrounding the core region P6-P2´ were examined in the multiple sequence alignment and mutations were identified (Figure 1) (Table 2).
Mutations in the key residues in the binding pockets of mammalian proprotein convertases Black indicates conserved residues. Blue indicates that a mutation changes the amino acid type but does not significantly affect the physical properties of the side chain of an amino acid. Red indicates that a mutation affects the physical properties of the side chain of an amino acid. The multiple sequence alignment was generated with Clustalx[6].
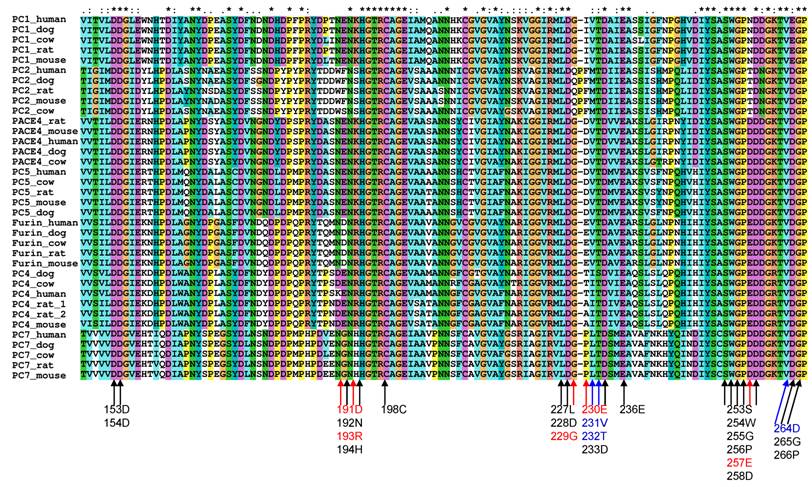
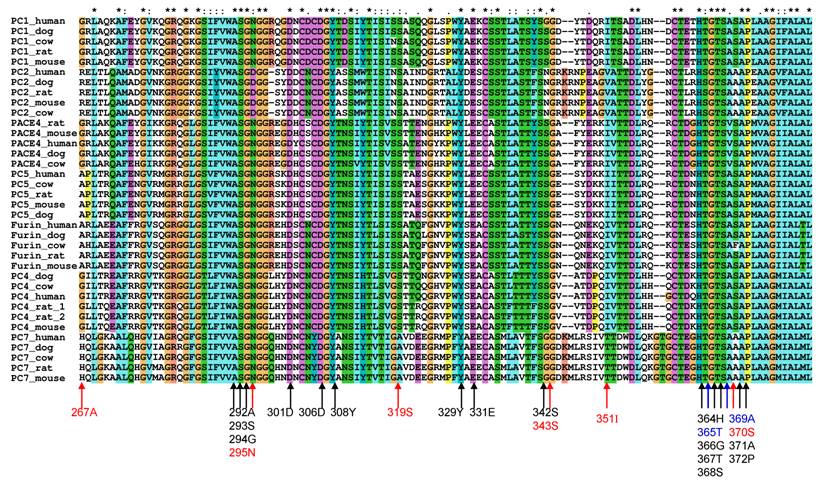
Generally, changes in amino acids at specific positions in the binding pockets can be classified into three types:
- Conserved. Marked in black in Figure 1, e.g. Asp153.
- Amino acid is mutated in one or more proprotein convertases, but the physical properties of the mutated amino acids can be considered as conserved and the favoured interaction between a substrate and a furin binding pocket at this position can still be maintained by the mutated amino acid in the 3D structure. These are marked in blue in Figure 1, e.g. Asp264->Glu264 in PC1.
- Amino acid is mutated in one or more proprotein convertases and the physical properties of the mutated amino acids are changed. These are marked in red in Figure 1, e.g. Glu230->Pro230 in PC7.
Comparison of binding pockets of mammalian proprotein convertases Mutations in the key residues in the binding pocket of mammalian proprotein convertases are listed. Mutations in negatively charged Glu230 and Glu257 are repeatedly observed and highlighted in the table. Both of the negative charges at positions 230 and 257 are lost in the PC1 and PC2 binding pocket. * Mutation is not observed in all species.
| Proprotein convertase | Mutations in comparison with furin binding pocket |
|---|---|
| PC1/PC3/SPC3 | D191->E, R193->K, E230->I, E257->N, D264->E, A267->G, S343->G* |
| PC2/SPC2 | D191->F, R193->S, G229E230V231->QPFM, E257->T, A267->R, N295->D, S343->N, I351->V, T365->S, S370->A |
| PC4/SPC5 | D191->E, E230V231->A/T I, T232->S*, A267->G |
| PACE4/SPC4 | D191->E , R193 ->K, E230 ->D, E257->D, A267->G, A369->V |
| PC5/PC6/SPC6 | D191->E, R193->K, E230->D, E257->D |
| PC7/PC8/SPC7/LPC | D191->G, R193->H, E230V231->PL, E257->D, A267->H, S319->A, S343->G, I351->T, S370->A |
It is striking that mutations in Glu230 or Glu257 were repeatedly observed, these being key residues interacting with substrate positions P4-P6 (Figure 2). Two of the most notable features and interactions at substrate positions P4-P6 have been observed in previous experiments[2,3,7-9] and can be explained by the 3D structure in this region:
- Sensitivity to positive charge: a positively charged residue such as arginine and lysine at substrate position P5 and P6 could increase the furin cleavage efficiency because it may interact with the negatively charged Glu230 or Glu257 of the furin binding pocket.
- Compensatory effect for positive charge: a positively charged residue such as arginine and lysine is favoured at substrate position P4 because it may interact with the negatively charged Glu236 and Asp264 of the furin binding pocket. Loss of a positively charged residue at position P4 results in loss of favourable interactions between the positive charge and negative charge at substrate position P4. However, this loss can be compensated by the presence of a positively charged residue at substrate position P5 or P6. Known furin cleavage sites with this compensatory effect for positive charges include integrin alpha 4 precursor (HVISKR597↓, GI_124945)[10], pro-parathyroid hormone (KSVKKR31↓, GI_4506267)[11], vascular endothelial growth factor C precursor (HSIIRR227↓, GI_1718154)[12], albumin precursor (RGVFRR24↓, GI_4502027)[13], furin auto-cleavage site (RGVTKR75↓, GI_120611)[14], vitamin K-dependent protein C precursor (LRIRKR42↓, GI_131067)[15] and integral membrane protein 2B (KGIQKR243↓, GI_12643343)[16].
It should be emphasized that what compensates for the loss of the positive charge at substrate position P4 is not the observed presence of a positive charge at substrate position P5 or P6, but the interaction force between the positive charge and negative charge formed at substrate position P5 or P6. Therefore, in order to maintain the sensitivity and compensatory effect of the positive charge at the substrate positions P4-P6, negatively charged residues are required in the binding pocket. Mutations in Glu230 or Glu257 change the overall negatively charged density of the binding pockets of the other six mammalian proprotein convertases, with profound functional consequences:
- For mammalian PC1 and PC2, mutations in Glu230 or Glu257 result in the loss of the negative charge at these two positions. As a consequence, a positively charged residue at the substrate position P5 or P6 cannot interact with the negatively charged residues at position 230 and 257 of the binding pockets of PC1 and PC2, and therefore the presence of a positively charged residue at substrate position P5 and P6 may not effectively contribute to the cleavage efficiency of PC1 or PC2. Moreover, the observed compensatory effect of the positive charge over substrate positions P4-P6 on furin cleavage may be reduced for the cleavage of PC1 and PC2.
- For mammalian PC4 and PC7, mutation in Glu230 results in the loss of one negative charge. However, mutation in Glu257 was not observed in the PC4 binding pocket, and Glu257 is mutated to Asp257, which preserves the negative charge in the PC7 binding pocket. The consequence of the loss of only one negative charge at position 230 is not very clear.
- The mutations in the binding pocket of mammalian PACE4 and PC5 lead to residues with similar physical properties such as charge, hydrophobicity and volume. This suggests that of all the mammalian proprotein convertases, furin, PACE4 and PC5 may have the closest substrate specificity.
The sensitivity and the compensatory effect of the positive charge at substrate positions P4P6 require negatively charged residues in the binding pocket of mammalian proprotein convertases. At position P4, the positively charged residue arginine interacts with Glu236, Asp264, and Tyr308 of the furin binding pocket. Hydrogen bonds of molecular interactions at position P4 are calculated and shown as green lines. If a positively charged residue is absent at position P4 and favourable interactions with negatively charged Glu236 and Asp264 of the furin binding pocket are lost, this loss can be compensated for by the gain of an interaction between the negatively charged Glu230 and Glu257 of the binding pocket and a positively charged residue at substrate position P5 or P6. The distance between Glu230 and Glu257 is only 16.4Å and facilitates flexible interactions in this region. The density of the negative charge in the binding pocket is important for maintaining the sensitivity and the compensatory effect of the positive charge at substrate positions P4-P6. This structure was modeled based on the published 3D structure of the furin catalytic domain in complex with an inhibitor (Protein Data Bank ID: IP8J)[4]. The figure was generated with Swiss-Pdb Viewer[17] and was modified from the authors' previous publication[5].
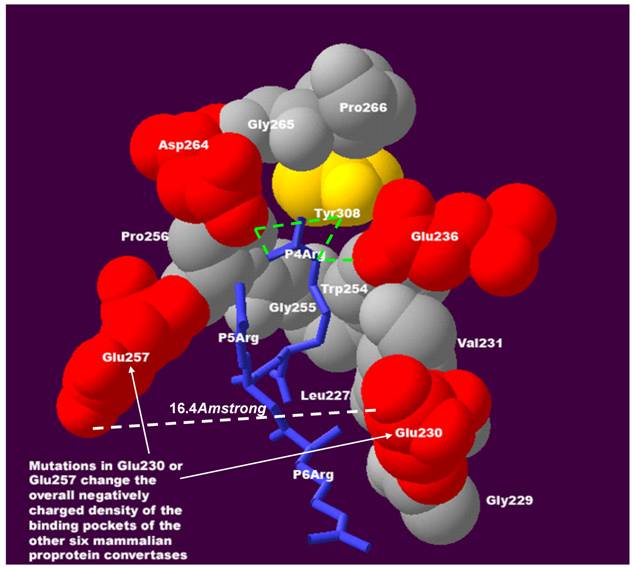
Although these conclusions were entirely derived from the theoretical 3D structure study, they are supported by existing experimental data. The cleavage site of integrin alpha 4 precursor (HVISKR597↓, GI_124945) lacks a positively charged residue at position P4, and compensates for this loss with a positively charged histidine at position P6 that is protonated in the acidic secretory compartments. By co-infecting furin-deficient LOVO cells with individual proprotein convertases and measuring immunoprecipitated products of cleavage, Bergeron and co-workers showed that processing of integrin alpha 4 precursor is performed efficiently by furin, PACE4 and PC5 (similar specificity), and much less efficiently by PC1 (reduced sensitivity for positive charge at position P6), and not by PC7[10]. In another study, Hendy and co-workers co-expressed pro-parathyroid hormone and proprotein convertases in BSC-40 and GH4C1 cells. The mass spectrometric analysis indicated that the cleavage site of pro-parathyroid hormone (KSVKKR31↓, GI_4506267) can be 2-4-fold more efficiently cleaved by furin than PC1 and PC2[11]. Again, the pattern KSVKKR↓ is characterized by the efficient compensation for the absence of a positive charge at position P4 by the presence of a positive charge at P6, which is preserved for furin, but reduced for PC1 and PC2.
To summarize, a negatively charged density at position 230 and 257 is a key factor that regulates the substrate specificity of mammalian proprotein convertases. In the course of molecular evolution, the enzyme function of the mammalian proprotein convertase family seems to have been fine-tuned by adjusting the density of the negative charge of the binding pockets. While this mechanism is supported by both the theoretical approach and experimental data, it is not the only structural mechanism. Which amino acid is present at other substrate positions such as P1' and P2' and the solvent accessibility of the substrates also affect the cleavage efficiency. In addition, physical interaction between the substrates and binding pockets of proprotein convertases is not the only way in which cleavage is regulated; the function of proprotein convertases is also regulated at other levels such as the level of expression in different tissues, their sub-cellular localizations, and the acidity of the different secretory compartments.
2.3. Towards the design of specific small molecule inhibitors targeting furin, PACE4, PC5, PC4 and PC7
Cleavage of proprotein convertases can be efficiently inhibited by a small molecule dec-RVKR-cmk [18,19]. The structure of dec-RVKR-cmk is characterized by positively charged residues at substrate positions P1, P2 and P4. Consequently, this small molecule dec-RVKR-cmk efficiently binds to all seven mammalian proprotein convertases, and only has limited use for studying the functional redundancy of mammalian proprotein convertases.
Specific short peptide inhibitors for only a specific group of proprotein convertases will aid the study of the functional redundancy of proprotein convertases. The results from a comparative study of the 3D structure immediately suggested a novel pattern for small molecule inhibitors targeting a specific group of mammalian proprotein convertases. The general 3D structure of this small molecule is similar to that of short peptides [K/R]-X-V-X-K-R, which omit a positively charged residue at position P4, but add a positively charged residue at position P6. The loss of the negative charge on Glu230 and Glu257 of the PC1 and PC2 binding pocket likely results in reduced sensitivity to the positive charge at substrate positions P5 and P6, whereas at least one negative charge is conserved at Glu230 or Glu257 in the binding pockets of the other five mammalian proprotein convertases: furin, PACE4, PC5, PC4 and PC7. Therefore, a short peptide [K/R]-X-V-X-K-R, with no positive charge at position P4 and one positive charge at position P6, should have a higher affinity to furin, PACE4, PC5, PC4 and PC7 than to PC1 and PC2.
Interestingly, in the course of molecular evolution, PC1 and PC2 have acquired very different tissue distributions from the other five mammalian proprotein convertases[2]: PC1 and PC2 are highly expressed in neuroendocrine tissue; PC4 is highly expressed in the testicular and ovarian tissues; furin, PACE4, PC5 and PC7 are widely expressed. Given the different expression levels of the different mammalian proprotein convertases in the various tissue types, a small specific molecular inhibitor specifically targeting furin, PACE4, PC5, PC4 and PC7 would be especially interesting for the development of therapeutic applications. Various derivatives of this novel short peptide pattern [K/R]-X-V-X-K-R need to be tested for the optimal binding affinity and specificity, and further work is needed to confirm their potential.
3. Materials and Methods
3.1 Analysis of 3D structural environment of the mouse furin binding pocket
The 3D crystal structure of the dec-RVKR-cmk inhibited mouse furin binding pocket was retrieved from the Protein Data Bank ID:IP8J[4]. To analyse the 3D structure of the active site and calculate hydrogen bonds of molecular interactions, Swiss-PDB Viewer[17] was used and the procedures described by Guex et al. on the Swiss-Pdb Viewer site http://www.expasy.org/spdbv/ were followed.
3.2 Multiple sequence alignment of mammalian proprotein convertases
The mammalian orthologues of seven proprotein convertases (furin, PC1/PC3/SPC3, PC2/SPC2, PC4/SPC5, PACE4/SPC4, PC5/PC6/SPC6 and PC7/PC8/SPC7/LPC) were identified by the BLAST algorithm[20]. Five mammal species with relatively well-assembled genomes were selected: Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus and Canis familiaris. A multiple sequence alignment was produced by Clustalx[6].
4. Conclusions
Our results extended our previous study on the physical properties of a 20-residue furin cleavage motif by determining the functional consequences of mutations at two specific positions (230 and 257) of the binding pockets of the other six mammalian proprotein convertases. We showed that the residues at positions 230 and 257 are frequently mutated in the binding pocket of mammalian proprotein convertases, resulting in a change in the density of the negative charge of the binding pockets. Comparison of the 3D structures suggests that different proprotein convertases have different sensitivities for the positive charge at substrate positions P5 and P6. Specifically, PC1 and PC2 may have reduced sensitivity for positively charged residues at the substrate position P5 or P6, whereas the substrate specificities of furin, PACE4, and PC5 may be very similar to each other. The density of the negative charge of the binding pockets seems to have played a key role in the molecular evolution of the enzyme function of the mammalian proprotein convertase family. This may allow the rational design of novel specific molecular inhibitors targeting specific members of the mammalian proprotein convertase family.
Acknowledgements
This project was started as Sun Tian's coursework for 445.039 Projekt Medizinische Informatik und Neuroinformatik at TUGraz and was fund by the GENAU Bioinformatics Integration Network PhD programme until June 2007 (PhD programme and grant leader: Professor Zlatko Trajanoski at TUGraz) and NFSC grants 10372118 (JW). Sun Tian gratefully acknowledges the support of Professor Zlatko Trajanoski.
Conflict of Interests
Sun Tian has had a full time job since PhD graduation in June 2007. Sun Tian and this project have been self-financed since then and this study was completed in the author's own time. The authors declare that no conflict of interest exists.
References
1. Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753-766
2. Nakayama K. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem J. 1997;327( Pt 3):625-635
3. Seidah NG, Chretien M. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 1999;848:45-62
4. Henrich S, Cameron A, Bourenkov GP, Kiefersauer R, Huber R, Lindberg I. et al. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat Struct Biol. 2003;10:520-526
5. Sun T. A 20 Residues Motif Delineates the Furin Cleavage Site and its Physical Properties May Influence Viral Fusion. Biochemistry Insights. 2009;2:9-20
6. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876-4882
7. Steiner DF. The proprotein convertases. Curr Opin Chem Biol. 1998;2:31-39
8. Lazure C, Gauthier D, Jean F, Boudreault A, Seidah NG, Bennett HP. et al. In vitro cleavage of internally quenched fluorogenic human proparathyroid hormone and proparathyroid-related peptide substrates by furin. Generation of a potent inhibitor. J Biol Chem. 1998;273:8572-8580
9. Rockwell NC, Krysan DJ, Komiyama T, Fuller RS. Precursor processing by kex2/furin proteases. Chem Rev. 2002;102:4525-4548
10. Bergeron E, Basak A, Decroly E, Seidah NG. Processing of alpha4 integrin by the proprotein convertases: histidine at position P6 regulates cleavage. Biochem J. 2003;373:475-484
11. Hendy GN, Bennett HP, Gibbs BF, Lazure C, Day R, Seidah NG. Proparathyroid hormone is preferentially cleaved to parathyroid hormone by the prohormone convertase furin. A mass spectrometric study. J Biol Chem. 1995;270:9517-9525
12. Siegfried G, Basak A, Cromlish JA, Benjannet S, Marcinkiewicz J, Chretien M. et al. The secretory proprotein convertases furin, PC5, and PC7 activate VEGF-C to induce tumorigenesis. J Clin Invest. 2003;111:1723-1732
13. Brennan SO, Nakayama K. Furin has the proalbumin substrate specificity and serpin inhibitory properties of an in situ hepatic convertase. FEBS Lett. 1994;338:147-151
14. Anderson ED, VanSlyke JK, Thulin CD, Jean F, Thomas G. Activation of the furin endoprotease is a multiple-step process: requirements for acidification and internal propeptide cleavage. EMBO J. 1997;16:1508-1518
15. Drews R, Paleyanda RK, Lee TK, Chang RR, Rehemtulla A, Kaufman RJ. et al. Proteolytic maturation of protein C upon engineering the mouse mammary gland to express furin. Proc Natl Acad Sci U S A. 1995;92:10462-10466
16. Kim SH, Wang R, Gordon DJ, Bass J, Steiner DF, Lynn DG. et al. Furin mediates enhanced production of fibrillogenic ABri peptides in familial British dementia. Nat Neurosci. 1999;2:984-988
17. Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714-2723
18. Angliker H, Wikstrom P, Shaw E, Brenner C, Fuller RS. The synthesis of inhibitors for processing proteinases and their action on the Kex2 proteinase of yeast. Biochem J. 1993;293( Pt 1):75-81
19. Stieneke-Grober A, Vey M, Angliker H, Shaw E, Thomas G, Roberts C. et al. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992;11:2407-2414
20. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403-410
Author contact
![]() Corresponding author: E-Mail: sun.tianorg; Tel.: 00862039380608; Fax: 00862039380607
Corresponding author: E-Mail: sun.tianorg; Tel.: 00862039380608; Fax: 00862039380607

 Global reach, higher impact
Global reach, higher impact