Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2010; 6(3):213-224. doi:10.7150/ijbs.6.213 This issue Cite
Research Paper
Analysis of differentially expressed mitochondrial proteins in chromophobe renal cell carcinomas and renal oncocytomas by 2-D gel electrophoresis
1. Laboratory of Molecular Oncology, Medical Faculty, Ruprecht-Karls University, Heidelberg, Germany
2. ZMBH, Core Facility for Mass-Spectrometry and Proteomics, Heidelberg, Germany
Received 2010-1-19; Accepted 2010-4-20; Published 2010-4-23
Abstract
Renal oncocytomas (RO) and chromophobe renal cell carcinomas (RCC) display morphological and functional alterations of the mitochondria. Previous studies showed that accumulation of mitochondria in ROs is associated with somatic mutations of mitochondrial DNA (mtDNA) resulting in decreased activity of the respiratory chain complex I, whereas in chromophobe RCC only heteroplasmic mtDNA mutations were found. To identify proteins associated with these changes, for the first time we have compared the mitochondrial proteomes of mitochondria isolated from ROs and chromophobe RCCs as well as from normal kidney tissues by two-dimensional polyacrylamide gel electrophoresis. The proteome profiles were reproducible within the same group of tissues in subsequent experiments. The expression patterns within each group of samples were compared and 81 in-gel digested spots were subjected to nanoLC-MS/MS-based identification of proteins. Although the list of mitochondrial proteins identified in this study is incomplete, we identified the downregulation of NDUFS3 from complex I of the respiratory chain and upregulation of COX5A, COX5B, and ATP5H from complex IV and V in ROs. In chromophobe RCCs downregulation of ATP5A1, the alpha subunit of complex V, has been observed, but no changes in expression of other complexes of the respiratory chain were detected. To confirm the role of respiratory chain complex alterations in the morphological and/or functional changes in chromophobe RCCs and ROs, further studies will be necessary.
Keywords: chromophobe renal cell carcinoma, mass spectrometry, mitochondria, renal oncocytoma, 2-D PAGE.
1. Introduction
Renal cell carcinoma (RCC) accounts for 2-3% of all malignant tumours. Approximately 75% of the renal cell tumours (RCT) are classified as conventional RCC, around 10% as papillary RCT, 5% each as chromophobe RCC and renal oncocytoma (RO) and the rest belongs to rare types or unclassifiable tumours. Although chromophobe RCC and RO together account only for 10% of the RCTs, their correct diagnosis is important taking into account the different biological nature of the two tumours (1,2). Approximately 90% of patients with chromophobe RCC are alive five years after surgery, but the rest metastasizes and leads to death of patients. In contrary, all ROs have a benign clinical course. The ages of patients with chromophobe RCC and RO varying considerably with a peak incidence in the sixth and seventh decade of life, respectively. Chromophobe RCC occurs equally in males and females, whereas RO occurs in males nearly twice often as in females.
DNA studies identified complex losses of chromosomes 1, 2, 6, 10, 13, 17 and 21 in 70%-97% of the chromophobe RCCs, whereas ROs display loss of chromosomes 1 and 14 and the Y chromosome or translocation between chromosome 11q13 and other chromosomes or show random genetic changes (3-9). The genes located at these chromosomes and involved in the development of chromophobe RCCs are not yet identified. Mutation of the p53 (at chromosome 17p) occurs only 25% of the cases, whereas PTEN (at chromosome 10q) or folliculin (at 11p13) have been excluded to be instrumental in the development of sporadic chromophobe RCCs and ROs (10-12).
Analysis of global gene expression in distinct types of RCTs revealed several hundred genes, which are exclusively expressed in both chromophobe RCCs and ROs (13-17). Only few genes were assigned to be differentially expressed at the mRNA level and none of them was confirmed by protein analysis (14,16,17). A recent study on the expression of total cellular proteins in ROs by applying the 2D-PAGE technique identified two proteins of the metallothionein family, which may be related to the abundancy of mitochondria in this type of tumour (18).
The major cytomorphological characteristic of chromophobe RCCs is the fine reticular cytoplasm. Electron microscopic studies revealed that the characteristic pale “chromophobe” staining of tumour cells is due to the degradation of mitochondria and accumulation of small intracytoplasmic vesicles. In contrast, cells of ROs display a strong eosinophilic staining due to the large number of densely packed mitochondria of variable size. The genetic background of these mitochondrial alterations is not yet cleared. Heteroplasmic mtDNA mutations found in chromophobe RCCs by sequencing the entire mitochondrial genome did not explained the number of mitochondria and vesicles, e.g. the staining characteristics of tumour cells (19). Studies using restriction length polymorphism analysis have provided contradictory data on mtDNA alterations in RO (20-21). However, recently it has been shown that somatic homoplasmic mtDNA mutations lead to the respiratory chain complex I enzyme deficiency (22). The deficiency of complex I activity and protein contents has also been previously demonstrated (23), as well as a lack of its assembly (24). It is very likely that alterations of mitochondrial proteins encoded in the nuclear DNA are responsible for the morphological abnormalities and hyperproliferation of mitochondria in cells of chromophobe RCC and RO, respectively (25).
The intact function and morphology of mitochondria is relying on the coordinated expression of 13 mitochondrial and approximately 1000 nuclear encoded genes (26). Global RNA or protein analysis results in extremely large number of candidate genes. The 2D-PAGE may separate around 1000 proteins and therefore it is an ideal technique to detect protein alterations in mitochondria. In this study, we used a proteomic approach based on two-dimensional gel electrophoresis and mass-spectrometry to compare the mitochondrial proteotypes of RO and chromophobe RCC.
2. Materials and methods
2.1. Tumour samples
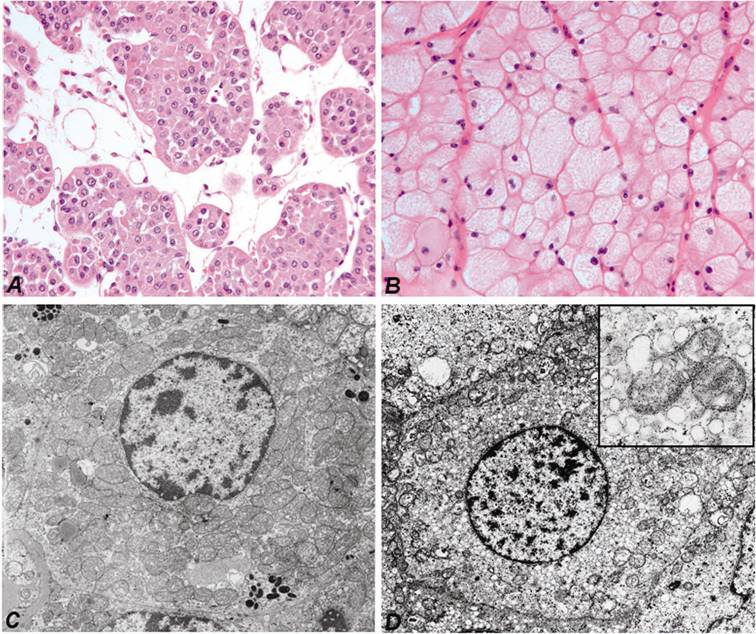
Fresh tumour and corresponding normal parenchymal tissues were obtained by nephrectomy at the Departments of Urology, Hannover Medical School and Ruprecht-Karls-University Heidelberg, Germany. One part of the tumour and normal kidney tissue was immediately snap-frozen in liquid nitrogen and stored at -80°C, whereas the remaining tumour with the nephrectomy specimen was fixed in 4 % buffered formalin and processed for histological examination. The histological diagnosis according to the Heidelberg Classification of Renal Cell Tumours was established by one of the authors (27). For this study we have selected typical cases of ROs with the characteristic morphology (Figure 1A) and electron microscopic features (Figure 1C) and also chromophobe RCCs showing pale “chromophobe” cytoplasmic staining (Figure 1B) and large number of intracytoplasmic microvesicles (Figure 1D). The genetic diagnosis of each tumour sample has been confirmed by the detection of specific chromosomal losses using high-density SNP-arrays (8). The collection and use of tissue samples for this study was approved by the Ethics Committee of the University of Heidelberg.
Characteristic cytoplasmic staining of RO (A) and chromophobe RCC (B). Hematoxilin and eosin staining, the same magnifications. Numerous large mitochondria of variable size are seen in the cytoplasm of cells of RO by electron microscopy (C). The few mitochondria in chromophobe cell are localized near to the cell membrane, whereas the cytoplasma is filled with small vesicles (D). The latter may be originated from the degradation of mitochondria (insert).
2.2. Isolation of mitochondrial protein fractions
All procedures were carried out at 4oC to minimize protease activity. Briefly, the cells of normal and tumour frozen tissues were disrupted by using the Dounce homogenizer (#432-1271, Wheaton, VWR International GmbH, Darmstadt, Germany) with loose and tight pestles. Up and down movements were carried out until getting a homogeneous “milk-like” suspension. The cell disruption was monitored under an inverted microscope. Homogenate was centrifuged at low (500 x g, Megafuge 1.0R, Heraeus, DJB Labcare Ltd, Newport Pagnell, UK) and higher speed (10,000 x g, Centrifuge 5415R, Eppendorf Vertrieb Deutschland GmbH, Wesseling-Berzdorf, Germany) to sediment “nuclei, cell debris and unbroken cells”, and to collect the crude microsomal fraction containing multiple kinds of microsomes (mitochondria, lysosome, peroxisome, Golgi complex, etc.). To obtain a purified fraction of mitochondria, sample of crude mitochondrial pellet was resuspended in homogenization buffer and loaded on the discontinuous sucrose density gradients followed by high-speed centrifugation (50,000 x g, Beckman Optima L70K Ultracentrifuge, SW 28). Mitochondria were concentrated in the lowermost brown band (Supplementry Figure 1), carefully aspirated, washed and precipitated. The purified mitochondrial pellet was stored at -80°C until used.
Mitochondrial integrity was tested in the isolated mitochondria versus the supernatant fraction using anti-mitochondrial antibody (clone MTC02, #MS-1372-PO, Thermo scientific, Fremont, CA, USA) and ß-actin (#A5441, Clone AC-15, Sigma-Aldrich GmbH, Steinhem, Germany), respectively. For this purpose, mitochondrial pellet was dissolved in Lysis buffer (62,5 mM Tris, 2% SDS, 10% Sucrose, pH 6,8) containing protease inhibitors (Roche Diagnostics GmbH, Mannheim, Germany), and protein concentrations were measured by BCATM Protein Assay kit (# 23225, Pirce, Rockford, USA), using BSA as a standard according to the manufacturer's instructions. 20 µg of proteins were size-fractioned by 10% SDS-PAGE, transferred electrophoretically to a nitrocellulose membrane (Protran, Whatman GmbH, Dassel, Germany) and blotted with corresponding antibody. An enhanced chemiluminescence (Western LightningTM Plus-ECL, PerkinElmer LAS GmbH, Rodgau, Germany) system was used for the signal detection.
2.3. 2-D PAGE
The pellet of mitochondria was resuspended in 2D Lysis buffer (7M Urea, 2M Thiourea, 4% CHAPS, 40 mM Tris pH 7.4, 1% DTT) and total protein quantitation was performed using PlusOne 2-D Quant kit (#80-6483-56, GE Healthcare Europe GmbH, Freiburg, Germany) according to the manufacturer's instructions. About 200 µg of mitochondrial proteins were precipitated according to the Wessel-Flügge method and resuspended in 200 µl of rehydration buffer (6M Urea, 2M Thiourea, 2% CHAPS, 0.15% DTT, 0.5% Ampholyte). The resuspended proteins were first separated according to pI using 18 cm immobilized non-linear gradient strips (ImmobilineTM DryStrip, #17-1233-01, Amersham Biosciences Europe GmbH, Freiburg, Germany) covering a pH range of 3-10. The strips were rehydrated for 12 h and subsequently subjected to high voltages at 20oC for electric focusing: 200 V for 1 h, 500 V for 1 h, 1000 V for 1 h, 8000 V gradient for 30 min and 8000 V for 6 h. After reduction for 15 min in Equilibration buffer (6M Urea, 30% Glycerin, 2% SDS, 50mM Tris pH 8.8, 1% DTT) and alkylation for an additional 15 min in the same solution containing 5% iodoacetamide, the proteins were separated in the second dimension in 9-16% SDS-PAGE gels. Spots vere visualized by staining wiht Coomassie brilliant blue G-250 (#161-0406, Biorad Labs, Richmond, USA).
To confirm the reproducibility and exclude experimental variations of the 2D-PAGE and also to detect qualitative and quantitative changes of protein spots the experiments we repeated by analyzing three ROs and three chromophobe RCCs and six normal samples.
2.4. Gel analysis and protein identification
The protein patterns in the gel were recorded in digitalized images using a high-resolution scanner (GS-710 Calibrated Imaging Densitometer, Bio-Rad, USA). The scanned images were analyzed using the Image Master 2D Platinum 6.0 software. Differentially expressed proteins were identified in each tumour sample compared to the normal kidney sample from six independent analyses by visual examination.
To facilitate spot picking for protein identification, the gels were restained with silver staining kit (#LC 6070, SilverQuest, Invitrogen). The excised gel pieces were transferred to a 96-well plate and destained (28). Subsequently, the proteins were further reduced with DTT, alkylated with iodacedamide and digested with trypsin, which cleaves at the carboxylic side of arginine and lysine residues, using a Digest Pro MS liquid-handling system (Intavis AG, Cologne, Germany). Following digestion tryptic peptides were extracted from the gel pieces with 50% acetonitrile/0.1% TFA, concentrated nearly to dryness in a SpeedVac vacuum centrifuge and diluted to a total volume of 30 µl with 0.1% TFA. 25 µl of the sample was analysed by a nanoHPLC system (Eksigent, Axel Semrau GmbH, Sprockhövel, Germany) coupled to a ESI LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific Inc., Dreiech, Germany). The sample was loaded on a C18 trapping column (Inertsil, LC Packings-Dionex GmbH, Idstein, Germany) with a flow rate of 10 µl/min 0.1% TFA. Peptides were eluted and separated on an analytical column (75 µm x 150 mm) packed with Inertsil 3 µm C18 material (LC Packings) with a flow rate of 200 µl/min in a gradient of buffer A (0.1% formic acid) and buffer B (0.1% formic acid, acetonitrile): 0-6 min: 3% B; 6-60 min: 3-40% B; 60-65 min: 60-90% B. The column was connected with a nano-ESI emitter (New Objectives Inc., Woburn, USA). 1300 V were applied via liquid junction. One survey scan (res: 30000) was followed by 3 information dependent product ion scans in the Orbitrap (res: 15000). 2+, 3+ and 4+ charged ions were selected for fragmentation.
The uninterpreted MS/MS spectra were searched against the NCBInr-human database (221364 sequences, downloaded on 29.01.2009) using the Mascot software (http://www.matrixscience.com). The algorithm was set to use trypsin as enzyme, allowing at maximum for one missed cleavage site and assuming carbamidomethyl as a fixed modification of cysteine, and oxidized methionine and deamidation of asparagines and glutamine as variable modifications. Mass tolerance was set to 4 ppm and 0.2 Da for MS and MS/MS, respectively.
2.5. Gene and protein expression analysis of HSPB1
Microarray-based gene expression study of four chromophobe RCCs and four ROs was carried out using the HG-U133 Pus2.0 GeneChip platform (Affymetrix, Santa Clara, CA) as previously described (17). The expression data are available at the Gene Expression Omnibus repository (http://www.ncbi.nih.gov/geo) under accession number GSE11151.
For validation of microarray data, real-time PCR analysis was applied on a panel of cDNAs obtained from eight chromophobe RCCs and eight ROs as well as from four normal kidneys as reported previously (17). The relative quantity was calculated by dividing the HSPB1 expression with the expression of ACTB. The following primers were used: 5'-GCAGGACGAGCATGGCTACATCT-3' and 5'-GGTGACTGGGATGGTGATCTCGT-3' for HSPB1 and 5'-ATGGATGATGATATCGCCGCGC-3' and 5'-TTCTGACCCATGCCCACCATCA-3' for ACTB.
Western blot analysis using rabbit polyclonal antibody to HSPB1 (AP0274PU-S, Acris Antibodies GmbH, Herford, Germany) diluted 1:1000 was performed on the tissue protein lysates as described elsewhere (17). Anti-β-actin antibody (AC-15, SIGMA-Aldrich GmbH, Steinhem, Germany) was used as a loading control. The HSPB1 antibody diluted 1:100 was used for immunohistochemical analysis of original paraffin blocks of fetal and adult kidneys, ROs and chRCCs and tissue microarrays containing fetal and adult kidneys as well as of distinct types of kidney tumors as described previously (17).
3. Results
3.1. 2-D PAGE analysis of mitochondrial proteins
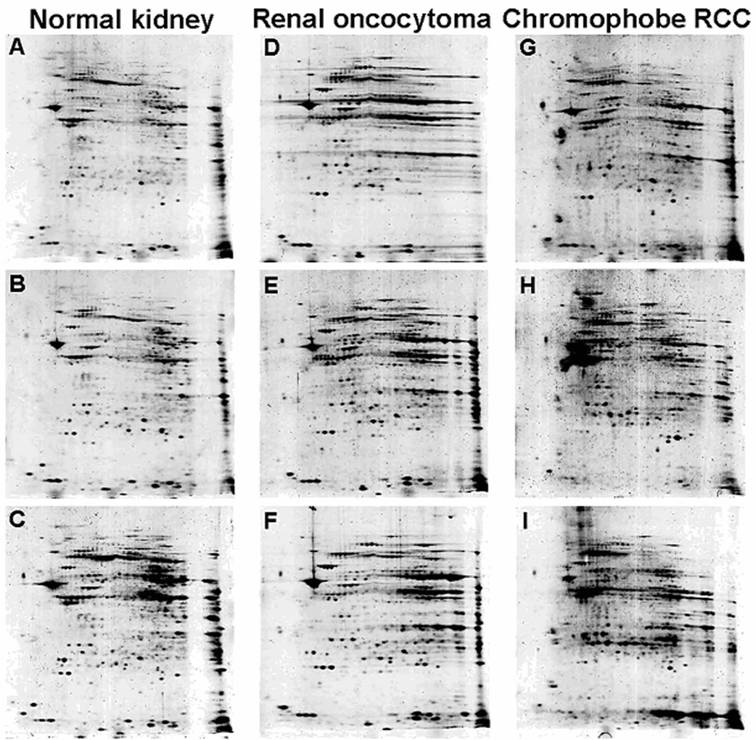
The initial SDS-PAGE blotting of separated mitochondria confirmed the integrity of mitochondrial fraction from both ROs and chromophobe RCCs (Supplementary Figure 1B). Individual 2D mitochondrial protein profiles obtained from three samples of RO and chromophobe RCC each, as well as from six normal kidneys in repeated experiments revealed a near-identical images within a given group (Figure 2). Although the contrast level of the gels of chromophobe RCCs was slightly different, the majority of analogous protein spots showed only minimal differences in relative location and shape. Both tumour types were associated with specific mitochondrial proteotype profile, although intensities of some of the spots were decreased or increased in individual tumour samples as compared to those of normal kidneys.
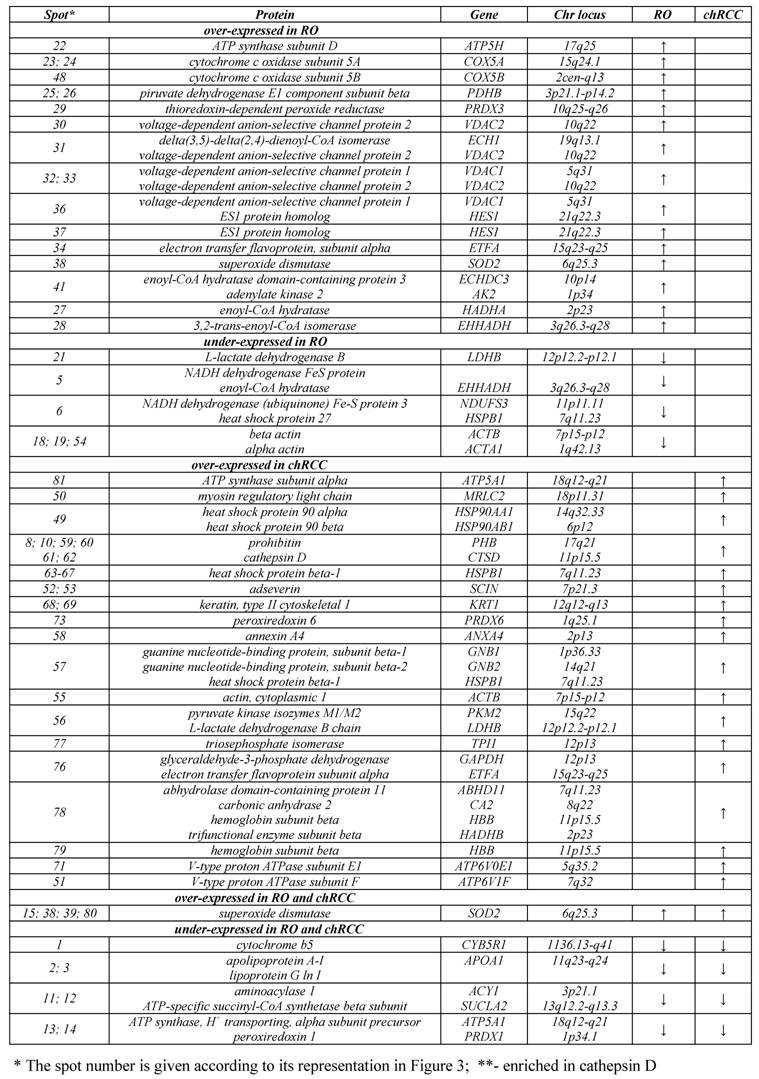
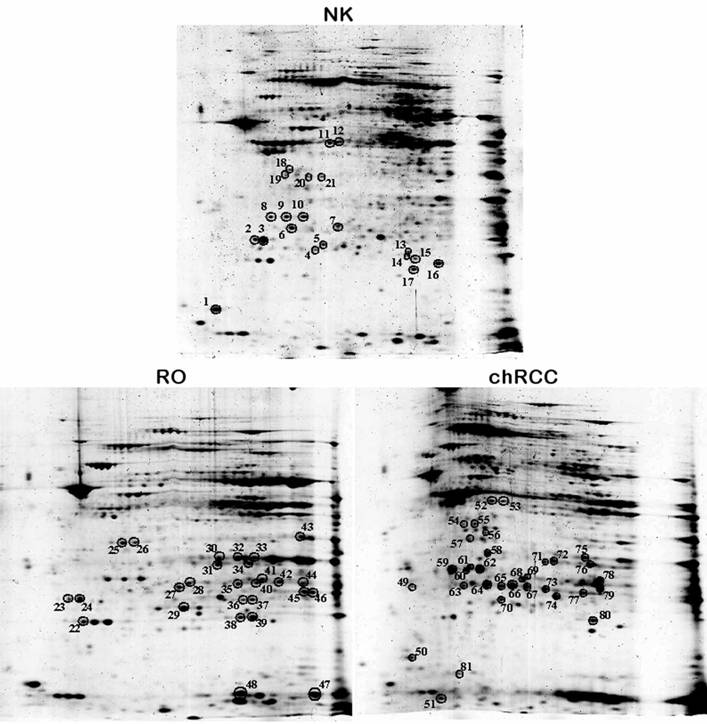
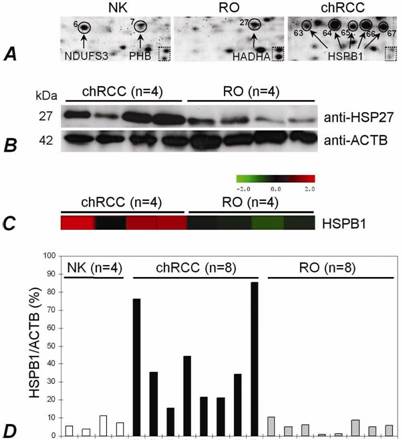
Altogether 81 spots of interest showing constantly decreased or increased signal intensity in all three chromophobe RCCs and ROs were cut from the 2D gel, in-gel digested with trypsin and subjected to nanoLC-MS/MS-based identification of proteins (Figure 3). The peptides were matched to all known human proteins in the NCBI protein database. The theoretical molecular weights and isoelectric points of the proteins, their corresponding accession numbers are listed in Supplementary Table 1, along with the protein sequence coverage in percent. Several horizontal rows were seen with similar molecular size but slightly different isoelectric points. For example, the heat shock protein beta 1 was identified within five spots located along a line (Figure 4A, spots 63-67). Pertinent data of proteins showing increased or decreased expression in ROs and/or chromophobe RCCs are presented in Table 1.
Although numerous protocols for isolation of mitochondria are available, we used an ultracentrifugation with a sucrose gradient density gradient resulting in high enrichment of functional mitochondria compared to other methods as has been recently shown (29). The priority of this method is confirmed by the fact that the vast majority of identified proteins in this study are indeed annotated as mitochondrial proteins. Beside of the proteins with unknown function, some of identified proteins are assigned to cytoskeleton, which is known to be attached to the outer mitochondrial membrane and involved in the movement of the organelle within the cell.
3.2. Proteins differentially expressed in ROs
We revealed the deficiency of protein 3 of complex I (NDUFS3, spots 5, 6) as well as upregulation of two subunits of complex IV (COX5A, spots 23, 24; COX5B, spot 48), which are the terminal enzymes of the mitochondrial respiratory chain, and a subunit of complex V (ATP5H, spot 22), which utilizes an electrochemical gradient of protons across the inner membrane during oxidative phosphorylation (OXPHOS). The proteins VDAC1 (spot 36) and VDAC2 (spot 30), which play a role in the control of cellular redox status, were up-regulated in ROs (Figure 3). The VDAC proteins can also regulate the activity of some enzymes involved in the control of redox status. For example, we found the increased level of superoxide dismutase (spots 38, 39) and thioredoxin-dependent peroxide reductase (spot 29) in RO as compared with normal kidney. These enzymes are located in the mitochondrial matrix and convert superoxide radical ions produced by complex I and III of the electron transport chain into H2O2 which can subsequently be converted to water. We found an increased amount of a subunit of the electron transfer flavoprotein (spot 34) and adenylate kinase 2 (spot 41), which is an ADP-generating enzyme localized in the mitochondrial intermembrane space and required for the maintenance of OXPHOS. These proteins might also be involved in the regulation of apoptosis by releasing from the mitochondria together with cytochrome c. The enoyl-CoA hydratase/isomerase superfamily (spots 27, 28, 31, 41) playing important roles in mitochondrial fatty acid β-oxidation and linking glycolysis and the Szent-Györgyi-Krebs cycle, and also a subunit of pyruvate dehydrogenase (spot 25) were also upregulated in ROs.
The low amount of cytochrome b5 (spot 1), a member of the electron transport system associated with the outer membrane, was easily discernable by visual inspection. A group of genes downregulated in RO includes enzymes which are involved in the amino acid (aminoacylase 1, spots 11, 12; lactate dehydrogenase B, spot 21) and lipid (apolipoprotein, spots 2, 3) metabolism. Apolipoprotein is a key element of the reverse cholesterol transport pathway. Decreased expression of members of actin family (spots 18, 19) and of a protein similar to crystallin alpha B (spot 17), which participates in the intracellular architechture, was also observed in RO. These proteins may be bound to the outer membrane of mitochondria, rather than corresponding to contaminating proteins within the mitochondrial fractions.
Two-dimensional PAGE images of a normal kidney (A, B, C), RO (D, E, F) and chromophobe RCC (G, H, I). After isoelectric focusing at pH 3-10 and a 9-16% gradient SDS-PAGE gel electrophoresis independently purified samples of the same histological phenotype showed highly consistent protein profiles, while chromophobe RCC group has partial diversity (see also Supplementary Figure 1).
Proteins with altered expression in renal oncocytoma and chromophobe RCC.
Two-dimensional gel analysis of mitochondrial proteins from normal kidney tissue, RO and chromophobe RCC. Spots with the corresponding numbers represent differentially regulated proteins, which were subsequently identified by mass spectrometry.
3.3. Differentially expressed proteins in chromophobe RCCs
We observed the increased amount of superoxide dismutase (spot 80), which is a mitochondrial matrix enzyme detoxifying superoxides, and peroxiredoxin 6 (spot 73), which is a member of antioxidant protein family. Two proteins involved in organelle trafficking, namely Ca2+-dependent adseverin (spot 52, 53) and annexin A4 (spot 58), were also up-regulated in chromophobe RCCs. The degradation of mitochondria seen in chromophobe RCC might also be associated with an impaired function of the OXPHOS and defective mitophagy. In the present study, we found the over-expression of alpha subunit of OXPHOS complex V (ATP5A1, spot 81) and two subunits of the V-type proton ATPase (spots 71, 51), an enzyme that mediates acidification of intracellular organelles. The expression of prohibitin (spot 60), whose ring complexes in the mitochondrial inner membrane regulate dynamics and function of mitochondria by controlling cristae morphogenesis, was also increased in chromophobe RCC.
A high level of isoforms of the heat shock protein beta-1 (HSPB1, spots 63-67), which plays a role as a molecular chaperon, was also observed in chromophobe RCC (Figure 4A). Moreover, the increased expression of piruvate kinase isozymes M1/M2 (spot 56) determing the glycolytic activity of cells and cathepsin (spots 59, 61, 62), which is the major lysosomal aspartic protease, was observed in chromophobe RCC. Of interest, decreased expression of only three genes, the cytochrome b5 (spot1), pro- and apolipoprotein (spots 2, 3) and aminoacilase (spots 11, 12) was seen in chromophobe RCC.
We extended our study by analysing HSPB1 with several approaches. Using total tissue extract, the HSPB1 was over-expressed in chromophobe RCCs at protein and RNA levels as compared with ROs (Figure 4B,C,D). Immunohistochemically, its expression was seen in the cytoplasm and membrane of the tubular structures of the normal kidney, but not in the glomeruli. The cells of chromophobe RCCs showed strong/moderate and weak diffuse positivity, whereas ROs were weakly positive or negative (not shown).
The HSPB1 expression in normal kidney, RO and chromophobe RCC. (A) The presence of HSPB1 isoforms within five spots situated along a line in chromophobe RCC. The same zoomed areas of 2D-PAGE images shows the position of two spots in the dotted square. (B) Immunodetection of the protein with anti-HSP27 (HSPB1) antibody. Western blot analysis confirmed an increased expression of the HSPB1 protein in chromophobe RCCs in comparison to ROs. ACTB was used as a control. (C) A significantly increased mRNA level in chromophobe RCC as detected by microarray-based gene expression analysis (probe set 201841_s_at). (D) Specific overexpression of the HSPB1 gene in chromophobe RCCs in comparison to ROs has been confirmed by real-time PCR as well.
4. Discussion
The protein compositions and concentrations in cells and organisms are tightly regulated by a complex networks. Advanced protein analysis is a prerequisite to explain the difference between normal and altered function and to differentiate between various stages of development of normal organs or tumours (30). Proteomics has been proven as efficient tools to analyse protein networks even in complex systems (31). However, the complexity of a cellular system often overwhelms even the best instrumentation. Studies on RCTs analysing whole tissue lysates by 2-D electrophoresis revealed a huge number of gene products not related to mitochondria (18). Earlier global RNA analyses searching for mitochondrial changes resulted in an extremely large number of candidate genes (13-17). Therefore, we reduced the complexity of the material by isolating subcellular compartments, e.g. mitochondria in the first step and used a proteomic approach based on 2-D gel electrophoresis and mass-spectrometry to compare exclusively the mitochondrial proteotypes of ROs and chromophobe RCCs to those of normal kidneys.
We have detected several proteins expressed differentially in tumour and normal kidney tissues. Although the list of mitochondrial proteins identified in this study is not complete, we have identified NDUFS3 from complex I of the respiratory chain as downregulated and COX5A, COX5B, and ATP5H from complex IV and V as upregulated in ROs. It is well known that decreased synthesis of ATP by OXPHOS is compensated by glycolysis, e.g. tumor cells shift their energy consumption from oxidative phosphorylation towards glucolysis, (the Warburg effect) (32). The growth of RO is strictly dependent on glucose provision and shows an increased activity of citrate synthase, an enzyme of the Szent-Györgyi-Krebs cycle (33, 22). Therefore, the question arises, whether the mitochondria in ROs are fully functional or their increased number is the result of an attempt to compensate their defective respiratory function. An increased enzymatic activity of cytochrome c oxidase (complex IV) (33, 24) and high amount of ATPase/ATP-synthase (complex V) were found in ROs when compared to those of normal kidneys (33). In contrast, a deficiency in its activity and low protein amount of NADH dehydrogenase (complex I), which is the main gateway to the electron transport chain, was observed in a majority of ROs (23, 22, 24). Enzymatic activities of complex II and III were slightly increased in ROs or they were close to the normal level (33). The decreased activity of complex I may be caused by somatic mutations of genes encoding this subunit rather than by regulation through its mRNA level or by nuclear DNA rearrangement (33, 22, 24). It has been hypothesized that increasing of free radicals or reactive oxygen species (ROS) production due to the high complex III activity could also inactivate complex I and decrease its amount (22). As human complex I contains at least 43 constitutive subunits and at least three other subunits are involved in its assembly, mutations can not be excluded in any one of these subunits.
The decreased protein content of members of complex I chain in RO might be compensated by the increased mitochondrial biogenesis or decreased mitochondria specific autophagy (mitophagy), and a drop in any respiratory chain complex might activate mitochondrial proliferation (34). Indeed, complex I deficiency results in decreased production of ROS, which alter the conformation of proteins in the external membrane of mitochondria and therefore reduce the mitophagy (22). It can not be excluded that low ROS level decreases the membrane signal for mitochondria elimination and programmed cell death (apoptosis), which may shift the balance between growth and apoptosis towards a slow proliferation tumour cells (35).
The increased expression of pyruvate kinase isozymes M1/M2 may indicate an increased glycolytic activity of chromophobe cells supporting the Warburg theory, e.g. the shift of energy consumption of tumour cells from OXPHOS towards glucolysis. The upregulation of alpha subunit of OXPHOS complex V (ATP5A1) in chromophobe RCCs suggests that accumulation of ROS may play a role in the morphological changes of mitochondria. The over-expression of HSPB1 protein possibly decreases ROS generation, reduces cytochrome c release from the intermembrane space and does not allow the progression of apoptotic programme (36). In accordance with recently published data (37), we found numerous isoforms of the HSPB1 protein due to post-translational modifications in chromophobe RCCs. Additionally, we showed a high expression of HSPB1 in chromophobe RCC both at the mRNA and protein level when compared with RO. The increased expression of cathepsin, which is the major lysosomal aspartic protease, was also observed in chromophobe RCC. This protein is relocalized to the cytoplasm in response to increased cytosolic levels of ROS and stimulates the translocation of apoptotic factor Bax onto mitochondrial membrane with the subsequent increasing of the mitochondrial VDAC, which in turn leads to the loss of membrane potential and release of cytochrome c involved in cellular apoptosis. Thus, a fine tuning between expression of these genes might be instrumental in the morphological changes of mitochondria in chromophobe RCCs. The intensive changes of mitochondrial membranes and producing a high number of small vesicles support this hypothesis.
Mitochondria with variable morphology undergo constant migration in the cytoplasma. How the spatial distribution and size of mitochondria are controlled in the cell is not yet cleared. It has been suggested that a group of genes and cytoskeletal elements are engaged in the trafficking of these organelles (38). Recent findings indicate that superoxide radicals participate in mitochondrial translocation, since the distribution of mitochondria may serve in compartmentalizing energy production and utilization (39-40). The translocation of mitochondria by cytoskeleton microtubules from an evenly dispersed distribution to the submembraneous spaces resulting in perinuclear clearing in chromophobe cells might be triggered by the increased level of ROS as a protective mechanism to eliminate the damage effects on the nuclei.
In summary, we found differences in the protein map of mitochondria isolated from ROs and chromophobe RCCs that might explain the compensatory proliferation and membrane degradation of mitochondria in these tumours, respectively. The mitochondrial localisation and function of several gene products identified in this study, is not yet known. Disclosing the molecular mechanisms related to the morphological and functional changes of mitochondria in chromophobe RCCs and ROs is important to understand the development of these two unique types of tumours. Therefore, these proteins should further be characterized and assigned to the mitochondrial, nuclear and cytosolic compartments of normal kidney cells by Western blotting. Subsequently, proteins ordered to the mitochondria should be localized by immunoelectron microscopic studies of normal and tumour cells to the specific structures of normal and altered mitochondria, especially to microvesicles in tumour cells.
Supplementary Material
Supplementary Figure 1
Supplementary Table 1
Acknowledgements
This work was supported by a grant from the Wilhelm Sander Stiftung. We thank members of the ZMBH Core Facility for Mass-Spectrometry and Proteomics (Heidelberg, Germany) for their technical assistance in all proteomic analysis.
Conflict of Interest
The authors declare that no conflict of interest exists.
References
1. Crotty TB, Farrow GM, Lieber MM. Chromophobe cell renal carcinoma: clinicopathological feature of 50 cases. J Urol. 1995;154:964-967
2. Davis CJ, Sesterhenn IA, Mostofi FK. et al. Renal oncocytoma. Clinicopathological study of 166 patients. J Urogenital Pathol. 1991;1:41-52
3. Kovacs A, Kovacs G. Low chromosome number in chromophobe renal cell carcinomas. Genes Chromosomes Cancer. 1992;4:267-268
4. Speicher MR, Schoell B, du Manoir S. et al. Specific loss of chromosomes 1, 2, 6, 10, 13, 17 and 21 in chromophobe renal cell carcinomas revealed by comparative genomic hybridisation. Am J Pathol. 1994;145:356-364
5. Bugert P, Gaul C, Weber K. et al. Specific genetic changes of diagnostic importance in chromophobe renal cell carcinomas. Lab Invest. 1997;76:203-208
6. Wilhelm M, Veltman JA, Olshen A. et al. Array based CGH for the differential diagnosis of renal cell cancer. Cancer Res. 2002;62:957-960
7. Nagy A, Buzogany I, Kovacs G. Microsatellite allelotyping differentiates chromophobe renal cell carcinomas from renal oncocytomas and identifies new genetic changes. Histopathology. 2004;44:542-546
8. Yusenko MV, Kuiper RP, Boethe T. et al. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinoma and renal oncocytomas. BMC Cancer. 2009;9:152
9. Füzesi L, Gunawan B, Braun S. et al. Renal oncocytoma with a translocation t(9;11)(p23;q13). J Urol. 2004;152:471-472
10. Contractor H, Zariwala M, Bugert P. et al. Mutation of the p53 tumour suppressor gene occurs preferentially in chromophobe type of renal cell tumours. J Pathol. 1997;181:136-139
11. Sükösd F, Digon B, Fischer J. et al. Allelic loss at chromosome 10q23.3 but lack of mutation of the PTEN/MMAC1 in chromophobe renal cell carcinoma. Cancer Genet Cytogenet. 2001;128:161-163
12. Nagy A, Zubakov D, Stupar Z. et al. Lack of mutation of the folliculin gene in sporadic chromophobe RCC and renal oncocytoma. Int J Cancer. 2004;109:472-475
13. Boer JM, Huber WK, Sültmann H. et al. Identification and classification of differentially expressed genes in renal cell carcinoma by expression profiling on a global human 31,500-element cDNA array. Genome Res. 2001;11:1861-1870
14. Takahashi M, Yang XY, Sugimura J. et al. Molecular subclassification of kidney tumors and the discovery of new diagnostic markers. Oncogene. 2003;22:6810-6818
15. Young AN, Amin MB, Moreno CS. et al. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. Am J Pathol. 2001;158:1639-1651
16. Higgins JPT, Shinghal R, Gill H. et al. Gene Expression Patterns in Renal Cell Carcinoma Assessed by Complementary DNA Microarray. Am J Pathol. 2003;162:925-932
17. Yusenko MV, Zubakov D, Kovacs G. Gene expression profiling of chromophobe renal cell carcinomas and renal oncocytomas by Affymetrix GeneChip using pooled and individual tumours. Int J Biol Sci. 2009;5:517-527
18. Zhuang Z, Huang S, Kowalak JA. et al. From tissue phenotype to proteotype: Sensitive protein identification in microdissected tumor tissue. Int J Oncol. 2006;28:103-110
19. Nagy A, Wilhelm M, Sükösd F. et al. Somatic mitochondrial DNA mutations in human chromophobe renal cell carcinomas. Genes Chromosomes Cancer. 2002;35:256-260
20. Welter C, Kovacs G, Seitz G. et al. Alteration of mitochondrial DANN in human oncocytomas. Genes Chromosomes Cancer. 1989;1:79-82
21. Brooks JD, Marshall FF, Isaacs WB. et al. Absence of HinfI restriction abnormalities in renal oncocytoma mitochondrial DNA. Mol Urol. 1999;3:1-3
22. Gasparre G, Hervouet E, de Laplanche E. et al. Clonal expansion of mutated mitochondrial DNA is associated with tumor formation and complex I deficiency in the benign renal oncocytoma. Hum Mol Genet. 2008;17:986-995
23. Simonnet H, Demont J, Pfeiffer K. et al. Mitochondrial complex I is deficient in renal oncocytomas. Carcinogenesis. 2003;24:1461-1466
24. Mayr JA, Meierhofer D, Zimmermann F. et al. Loss of Complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin Cancer Res. 2008;14:2270-2275
25. Heddi A, Faure-Vigny H, Wallace DC. et al. Coordinate expression of nuclear and mitochondrial genes involved in energy production in carcinoma and oncocytoma. Biochim Biophys Acta. 1996;1316:203-209
26. McDonald TG, van Eyk JE. Mitochondrial proteomics. Undercover in the lipid bilayer. Basic Res Cardiol. 2003;98:219-227
27. Kovacs G, Akhtar M, Beckwith BJ. et al. The Heidelberg classification of renal cell tumours. J Pathol. 1997;183:131-133
28. Catrein I, Herrmann R, Bosserhoff A. et al. Experimental proof for a signal peptidase I like activity in Mycoplasma pneumoniae, but absence of a gene encoding a conserved bacterial type I SPase. FEBS J. 2005;272:2892-2900
29. Hartwig S, Feckler C, Lehr S. et al. A critical comparison between two classical and a kit-based method for mitochondria isolation. Proteomics. 2009;9:3209-3214
30. Abbott A. How to spot a protein in a crowd. Nature. 1999;402:716-717
31. Lopez MF, Kristal BS, Chernokalskaya E. et al. High-throughput profiling of the mitochondrial proteome using affinity fractionation and automation. Electrophoresis. 2000;21:3427-3440
32. Gudinot C, de Laplanche E, Hervouet E. et al. Actuality of Warburg's views in our understanding of renal cancer metabolism. J Bioenerg Biomembr. 2007;39:235-241
33. Simonnet H, Alazard N, Pfeiffer K. et al. Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis. 2002;23:759-768
34. Hervouet E, Godinot C. Mitochondrial disorders in renal tumors. Mitochondrion. 2006;6:105-117
35. Hervouet E, Simonnet H, Godinot C. Mitochondria and reactive oxygen species in renal cancer. Biochimie. 2007;89:1080-1088
36. Arrigo AP. The cellular "networking" of mammalian Hsp27 and its functions in the control of protein folding, redox state and apoptosis. Adv Exp Med Biol. 2007;594:14-26
37. Sarto C, Valsecchi C, Magni F. et al. Expression of heat shock protein 27 in human renal cell carcinoma. Proteomics. 2004;4:2252-2260
38. Knowles MK, Guenza MG, Capaldi RA. et al. Cytoskeletal-assisted dynamics of the mitochondrial reticulum in living cells. Proc Natl Acad Sci USA. 2002;99:14772-14777
39. Hallmann A, Milczarek R, Lipinski M. et al. Fast perinuclear clustering of mitochondria in oxidatively stressed human choriocarcinoma cells. Folia Morphol (Warsz). 2004;63:407-412
40. Pletjushkina OY, Lyamzaev KG, Popova EN. et al. Effect of oxidative stress on dynamics of mitochondrial reticulum. Biochim Biophys Acta. 2006;1757:518-524
Author contact
![]() Corresponding author: Laboratory of Molecular Oncology, Medical Faculty, Ruprecht-Karls-University, Im Neuenheimer Feld 325, D-69120 Heidelberg, Germany. Phone: 49-6221-566519, Fax: 49-6221-564634, E-mail: gyula.kovacsuni-heidelberg.de
Corresponding author: Laboratory of Molecular Oncology, Medical Faculty, Ruprecht-Karls-University, Im Neuenheimer Feld 325, D-69120 Heidelberg, Germany. Phone: 49-6221-566519, Fax: 49-6221-564634, E-mail: gyula.kovacsuni-heidelberg.de

 Global reach, higher impact
Global reach, higher impact