Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2010; 6(3):252-267. doi:10.7150/ijbs.6.252 This issue Cite
Research Paper
NIP/DuoxA is essential for Drosophila embryonic development and regulates oxidative stress response
1. Department of Biochemistry and the Siebens-Drake Medical Research Institute, Schulich School of Medicine and Dentistry, The University of Western Ontario, London, Ontario N6A 5C1, Canada;
2. Department of Biology, Schulich School of Medicine and Dentistry, The University of Western Ontario, London, Ontario N6A 5C1, Canada;
3. Centre for Research in Neuroscience, Montreal General Hospital, 1650 Cedar Avenue, Montreal, Quebec H3G 1A4, Canada
# These authors contributed equally to this work.
Received 2009-8-27; Accepted 2009-9-15; Published 2010-5-11
Abstract
NIP/DuoxA, originally cloned as a protein capable of binding to the cell fate determinant Numb in Drosophila, was recently identified as a modulator of reactive oxygen species (ROS) production in mammalian systems. Despite biochemical and cellular studies that link NIP/DuoxA to the generation of ROS through the dual oxidase (Duox) enzyme, the in vivo function of NIP/DuoxA has not been characterized to date. Here we report a genetic and functional characterization of nip in Drosophila melanogaster. We show that nip is essential for Drosophila development as nip null mutants die at the 1st larval instar. Expression of UAS-nip, but not UAS-Duox, rescued the lethality. To understand the function of nip beyond the early larval stage, we generated GAL4 inducible UAS-RNAi transgenes. daG32-GAL4 driven, ubiquitous RNAi-mediated silencing of nip led to profound abnormality in pre-adult development, crinkled wing and markedly reduced lifespan at 29°C. Compared to wild type flies, da-GAL4 induced nip-RNAi transgenic flies exhibited significantly reduced ability to survive under oxidative stress and displayed impaired mitochondrial aconitase function. Our work provides in vivo evidence for a critical role for nip in the development and oxidative stress response in Drosophila.
Keywords: Numb Interacting protein, dual oxidase maturation factor, embryonic development, oxidative stress.
Introduction
NIP was initially cloned as a binding protein to Numb, a cell fate determinant that plays essential roles during the development of the peripheral and central nervous systems in Drosophila and mammals [1-3]. Numb contains an N-terminal phosphotyrosine-binding (PTB) domain[4, 5] and a C-terminal proline-rich region (PRR)[6]. Sequence analysis suggests that NIP is a membrane protein, a prediction that was subsequently confirmed in transfected HEK293 cells and the Drosophila S2 cells [7]. In addition, Drosophila NIP (dNIP) and the mammalian NIP1 and NIP2 homologues all contain a conserved NxxF motif recognized by the Numb PTB domain, with dNIP harbouring two copies of the motif [7]. It was demonstrated that the NxxF motif mediated binding of NIP to Numb PTB domain [7]. These characteristics led to the initial assumption that NIP might regulate the subcellular localization of Numb during asymmetric cell division. Notwithstanding this initial prediction, however, recent work on the mammalian homologues NIP2 and NIP1 (recloned and renamed as DuoxA2 and DuoxA1 for dual oxidase maturation factor A2 and A1, respectively) suggested that it plays an important role in the generation of reactive oxygen species (ROS) by controlling the membrane translocation of the dual oxidases (Duox1 and Duox2) [8, 9]. The human nip2/douxa2 gene is arranged head-to-head with the duox2 gene in the genome; and similarly the nip1/duoxa1 gene is positioned in the same manner with regard to the duox1 gene [9]. This pattern of genomic arrangement suggests that the functions of nip1/nip2 may be coupled to that of duox1/duox2. Indeed, in a reconstituted in vitro ROS generation system DuoxA2 was shown to facilitate the function of Duox2 by allowing rapid ER exit of folded Duox2 or enhanced degradation of misfolded Duox2 [9].
Reactive oxygen species (ROS) such as superoxide (O2.-) and hydroxyl radical (.OH) and hydrogen peroxide (H2O2) have been shown to regulate a number of physiological processes including growth, differentiation and neurotransmission [10]. The generation of ROS in a cell is mediated by the NOX family of enzymes. Dual Oxidase1 and 2 (DUOX 1 and 2) make up a more recently identified subgroup of NOX enzymes that are expressed mainly in the thyroid and epithelial cells of the airways and in the gastrointestinal tract [11, 12]. The Duox enzymes have been shown to generate H2O2 during thyroid hormone biosynthesis [12]. The identification of NIP/DuoxA as a maturation factor for Duox suggests a novel function for NIP that is apparently unrelated to its capacity to bind Numb. In order to characterize the physiological function of NIP, we obtained a piggyBac line (PBac{RB}mole02670, abbreviated as pBac herein) that contained a transposon element insertion in an intron region (at 14977701) for the nip gene which effectively created a functional nip-null mutant. We found that the pBac/pBac embryos were developmentally arrested soon after hatching, exhibited gross developmental defects, and died at the 1st instar larval stage. We further showed that maternal nip was not required for oogenesis and embryo hatch to the 1st instar larvae, because maternal nip-null embryos have the same lethal phenotype as the nip-null embryos in which maternal nip transcripts were carried from the pBac heterozygous females. The nip-null larval lethality could be rescued by expressing a UAS-nip, but not UAS-Duox. In order to decipher the function of nip beyond the 1st larval instar, we generated a GAL4 inducible nip-RNAi transgenic germline. The daG32-GAL4 induced ubiquitous nip-RNAi flies are viable with deformed crinkle wings and markedly increased pre-adult mortality at 29 oC. These flies were also sensitive to oxidative stress and reduced mitochondrial activity.
Materials and Methods
Drosophila Stocks
The nip mutant pBac{RB}mole02670 (Flybase ID: FBst0018073) was obtained from the Bloomington Drosophila stock center. Wild-type and the double Asn to Ala (N1N2/AA) mutant nip cDNA were digested from the Flag-NIP and Flag-NIP-N1N2/AA vectors [7] by restriction enzymes and subcloned into a pP[UAST] vector. The resulting plasmid was injected into the w1118 recipient by a standard P element-mediated transformation. Green balancer and GAL4 drivers were also obtained from the Bloomington fly center:
w[1118]; In(2LR)Gla, wg[Gla-1]/CyO, P{w[+mC]=GAL4-twi.G}2.2, y[1] w[*]; P{w[+mC]=Act5C-GAL4}17bFO1/TM6B, Tb[1], y[1] w[*]; P{w[+mC]=tubP-GAL4}LL7/TM3, Sb[1], w[*]; P{w[+mC]=matalpha4-GAL-VP16}V2H, P{ry[+t7.2]=hsFLP}12, y[1] w[*]; noc[Sco]/CyO.
The UAS-GalT-GFP and UAS-Lys-GFP were kindly provided by Dr. Jennifer Lippincott-Schwartz of the National Institute of Health, USA. Other fly strains used were obtained from Bloomington fly center.
pP[UAST] Mediated UAS-nip RNAi Transgene Constructs
To generate UAS-nip inverted repeat, nip cDNA was amplified by polymerase chain reaction (PCR) with a forward primer containing an EcoRI restriction site and a reverse primer containing an XbaI restriction site. The spacer sequence was also amplified by PCR from the pEGFP vector with EcoRI and Hind III restriction sites at the ends. The target sequence was first inserted into a pBluescript KS+ vector at the EcoRI and XbaI sites and the resulting plasmid was named pBS1. The pBS1 vector was then digested with NotI and HindIII to generate the target sequence that was subsequently cloned into the pcDNA3.1 vector. At the same time, a pBS2 vector was created by inserting the EGFP spacer sequence into pBS1 via the EcoRI and HindIII sites. The pBS2 vector was subsequently linearized by HindIII and XhoI and ligated to the target sequence, which was released from the pcDNA3.1 vector by digest with XhoI and HindIII. Finally, the cDNA fragment was back-cloned into the pBluescript KS+ vector as inverted repeats containing the EGFP spacer in the middle of the two repeats. The inverted repeats were arranged in a head-to-head orientation such that a stem-loop dsRNA may be formed after transcription in vivo. After verification by restriction digestion and DNA sequencing, the inverted repeat sequence was released by XbaI digestion and inserted into a pP[UAST] vector. The UAS-RNAi lines were generated by microinjecting the pP[UAST]-nip-IR construct into w1118 embryos. Ten UAS-nip-RNAi transgenic lines (named 1-1 through 1-10) were created at BestGene Inc. (Chino Hills, CA, USA).
In Situ Hybridyzation
In situ hybridization on whole mount fly embryos was carried out to detect nip transcript expression. RNA probes were generated by PCR using primers directed against the T7 promoter sequence (TAATACGACTCACTATAGGGAGA; for sense RNA probes, T7 promoter was added to 5' primer; for antisense RNA probea, it was added to the 3' primer). The probes were synthesized by in vitro transcription using MEGAscript T7 kit (Ambion). The digoxigenin (DIG) RNA labeling mix (Roche Applied Science) was employed in the labeling reaction.
Aconitase Extraction and Electrophoresis
Sixteen late 3rd instar larvae were homogenized using a blue disposable plastic pestle in 50 ml ice cold extraction buffer containing 2 mM citric acid, 0.6 mM MnCl2, 50 mM Tris-HCl, pH 8.0. The homogenate was centrifuged twice for 5 minutes each at 4 oC. Cellulose membrane electrophoresis was carried out using the GELMAN Science (VWR) system. In a horizontal GELMAN semi-micro electrophoresis chamber, two reservoirs were filled with 200 ml electrophoresis buffer (20 mM K-PO4 and 3.6 mM citric acid, pH 6.5). Cellulose polyacetate membrane (Sepraphore® III, VWR) was equilibrated in the buffer for 10 minutes by floating in the reservoirs. The briefly dried membrane was set on a bridge and draped into the buffer. The samples were loaded onto the membrane near the cathode (-) with 4-applicator (GELMAN Model-51119, Sepratek) and the loaded sample line was moved close to the middle of the bridge. The cytoplasmic and mitochondrial aconitases migrated to the anode (+) and cathode respectively with electrophoresis for 50 minutes at 130 V.
During electrophoresis, staining solution was freshly prepared that contained 100 mM potassium phosphate, pH 6.5, 1 mM p-nicotinamide adenine dinucleotide phosphate (NADPH), 2 mM cis-aconic acid, 1.2 mM 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), 0.3 mM phenazine methosulfate, 25 mM MgCl2, and 5 Units/ml isocitrate dehydrogenase. The membrane (protein side down) was laid onto the solution and incubated in the dark for 6 minutes, then washed in water 3 times for 15 minutes each.
Results
Expression of nip at embryonic and larval stages
Previous cDNA microarray data obtained for the complete life cycle of Drosophila showed that the nip transcript level spikes only in very narrow windows during development and two of such spikes occur immediately after egg laying (AEL) and the other corresponds to early larval stages [13]. To further define the pattern of nip expression during embryonic and larval development, we collected wild type fly embryos at different time points AEL and larvae from the first three instar stages, and determined the corresponding nip mRNA levels in a Northern blot. Since NIP was initially identified as a Numb-interacting protein, the blot was also probed for numb and rp49, the latter of which served as a control. As shown in Fig. 1A, when the nip-5' coding sequence (corresponding to residues 1-111 of nip) was used as a probe, only one specific band corresponding to the nip mRNA was detected in the embryos. The nip mRNA was abundant in the first three hours AEL, which represented the maternal transcript. In contrast, zygotic nip expression was kept at very low levels at all embryonic stages except for a small window (ie. 17-21 hour AEL) at the late embryonic stage just before hatching. The nip transcript was silenced again for the entire larval stages (Fig. 1A). The smear on the nip panel down the heavy bands in 0-3 hours embryos is probably the degradation product during RNA preparation. In situ hybridization was also employed to locate nip mRNA during embryonic development (data not shown). The nip mRNA was ubiquitously distributed in the whole egg in the first few hours AEL. However, during cellularization, nip transcript was excluded from the epidermal cells and only existed inside the eggs. After cellularization, nip mRNA was degraded quickly and became undetectable in the embryos.
Two bands were detected for the numb mRNA in the embryos. The lower band found at E0-2h corresponds to the 3.1 kb maternal transcript and the upper band detected after E3h corresponds to the 3.4 kb zygotic transcript. In the first two hours AEL, only the shorter maternal transcript was detected in the embryos. The larger zygotic transcript and a smaller maternal transcript co-existed in the embryos 2-3 hours AEL. After three hours AEL, maternal numb transcript was completely degraded and zygotic numb expression was increased significantly. Zygotic numb transcript reached the highest level at 7-11 hours AEL and decreased in later embryonic and larval stages. The disparity in expression pattern between nip and numb transcripts suggests that these two genes are genetically uncoupled during embryonic and early larval development.
Expression of nip transcript and localization of NIP protein in Drosophila embryos. (A) Northern blot analysis of nip and numb transcripts in Drosoplila embryos and larvae. Embryos were collected at different time points after egg laying (AEL). Larvae at stages L1, L2 and L3 were collected. The blot was probed with P32-labeled nip, numb and rp49 cDNA fragments, respectively. The position of 18S rRNA (1.98 kb) is indicated with an arrow. Rp49, ribosomal protein 49. (B) Maternal NIP expression and localization during oogenesis. N, nurse cell; O, oocyte. Arrows indicate where NIP immunostain was prominent. (C) localization of NIP protein in embyos of stages 2 to 4. NIP is in red; nuclei (stained with DAPI) in blue.
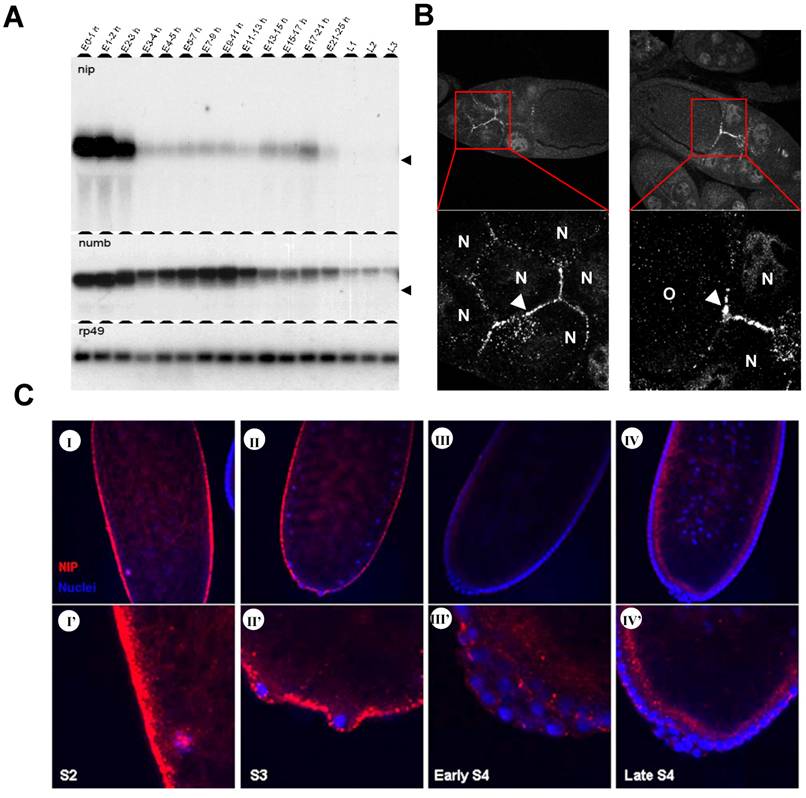
We next carried out whole mount immunostaining of NIP on embryos of stages S2 to S4 before cellularization occurred. NIP was highly expressed in the preblastoderm embryos (Fig. 1C-I & I') where it was found to localize to the cytoplasmic membrane as discrete dots, consistent with our previous prediction of NIP being a membrane protein [7]. NIP immunostain weakened significantly at the pole bud formation stage (Fig. 1C-II & II') and was further reduced at the syncytial blastoderm (Fig. 1C-III & III') and cellularization stages (Fig. 1C-IV & IV').
Nip is an essential gene in Drosophila development
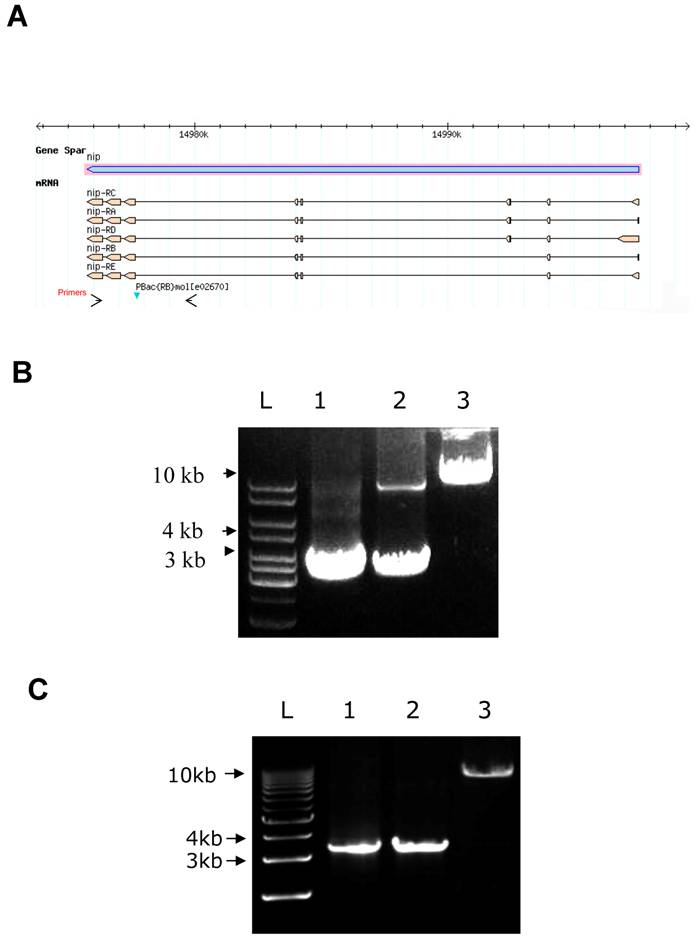
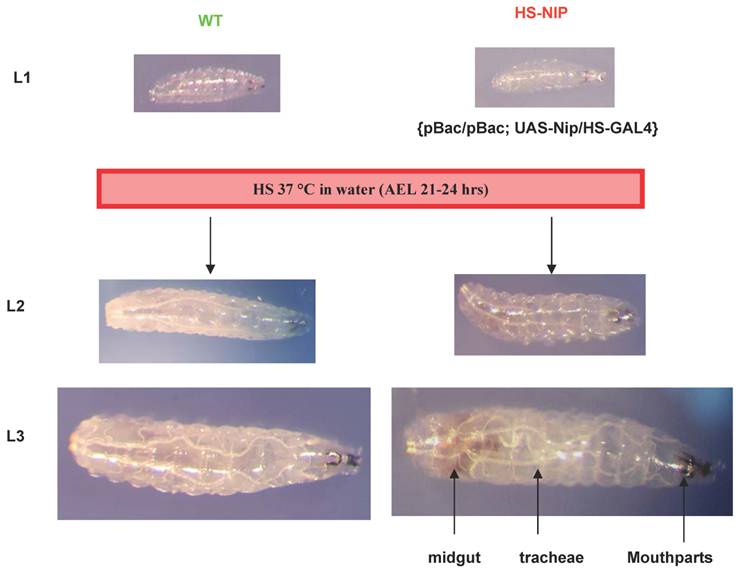
By searching the Drosophila database (Flybase), we found a nip insertion mutant strain named PBac{RB}mole02670 (or pBac for short) with a piggyBac{RB} transposable element inserted in an intron of the nip gene (Fig. S1). This nip allele is recessively lethal.. All nip mutant homozygous flies died at embryonic or the 1st instar larval stages. To address whether the recessive lethality of the pBac allele was caused by the loss of functional nip expression, rescue experiments were carried out using a UAS-nip transgene driven by TubP-GAL4, Act5c-Gal4 or Hs-GAL4. The lethality of pBac homozygous flies could be rescued by UAS-nip driven by ubiquitous and heatshock Gal4 drivers (Figs. S1 & S2), suggesting the phenotype was caused specifically by a loss of nip.
The pBac/pBac; UAS-nip/TubP-Gal4 adult flies were obtained by crossing between pBac/CyO;UAS-nip/TM3, Ser Act-GFP and pBac/CyO;TubP-Gal4(or Act5c-Gal4, Hs-Gal4)/TM3, Ser Act-GFP. These pBac homozygous flies appeared healthy and were fertile. To confirm the pBac insertion on both the second homozygous chromosomes in these flies, genomic DNA was isolated for PCR analysis. PCR primers were chosen to amplify a nip gene fragment including the pBac insertion site. A single DNA fragment of 3.4 kb was obtained from the y w genome, which contained no pBac insertion in the nip gene. Two PCR products of 3.4 kb and 10 kb were obtained from the heterozygous genomic DNA (Fig. S1B). The larger PCR product was generated from the chromosome bearing the pBac insertion. It appeared weaker in comparison to the 3.4 kb band due probably to the fact that longer extension time was needed to amplify a larger cDNA fragment by PCR. For pBac homozygous flies, only the longer DNA product was amplified by PCR, confirming the homozygous pBac insertion in these flies (Fig. S1B). Importantly, pBac/pBac;TubP-Gal4/TM3,Ser,actGFP or pBac/pBac;UAS-nip/TM3,Ser,actGFP flies never appeared in the adult progenies, suggesting the pBac/pBac;+/+ flies were pre-adult lethal. Rescue of pBac homozygote lethality by the UAS-nip transgene suggests that the piggyBac{RB} insertion mutant is a nip loss-of-function allele and that nip is an essential gene in fly development.
Surprisingly, the lethality caused by pBac insertion could be rescued by a nip mutant transgene whose protein product was deficient in Numb-binding [7]. To test the importance of the two NxxF motifs in NIP function in Drosophila, UAS-nipNN/AAt flies carrying double Asn to Ala mutations were generated for the rescue experiment. Adult pBac/pBac; UAS-nipNN/AA/TubP-Gal4 flies were produced by crosses and could be kept as a stock. The homozygous insertion of pBac was confirmed as well (Fig. S1C). Our observation that the UAS-nipNN/AA rescued the lethality of the homozygous pBac mutant flies as efficiently as UAS-nip suggests that the capacity of NIP to bind Numb is not required for its in vivo function.
Given the important role Numb plays in Drosophila neurogenesis, we repeated the rescue experiment using UAS-nip under the control of elav-Gal4, a specific driver in the nervous system. In contrast to ubiquitous Gal4 drivers, the pBac/pBac, elav-Gal4/UAS-nip flies were not viable to adulthood, suggesting that the restore of NIP in the nervous system is not sufficient for the rescue of the pBac lethality.
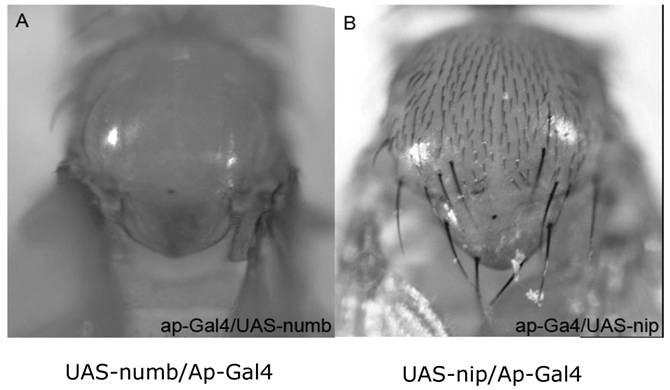
To investigate whether ectopic expression would result in a phenotype, UAS-nip was employed to over-express NIP under one of following GAL4 drivers: Act5c-Gal4, TubP-Gal4 (ubiquitous); ap-Gal4, ptc-Gal4, 1096-Gal4, dpp-Gal4, wg-Gal4, vg-Gal4 (wing disc specific); elev-Gal4, PO163-Gal4, GMR-Gal4 (nervous system specific). Neither a ubiquitous nor tissue specific driver resulted in a visible phenotype. For instance, the ap-Gal4 was used to drive numb or nip expression in the wing disc. Whereas over-expression of Numb inhibited the production of hairs during asymmetric division of the sensory organ precursors, which resulted in bald nota in all progenies, over-expression of NIP did not cause any defect in hair production (Fig. S3). Moreover, when both Numb and NIP were over-expressed in the same fly, the phenotype of bald notum was not enhanced. These results suggest that it is unlikely for numb and nip to interact genetically during fly development. Because NIP/DuoxA was shown to be a regulator of Duox activity in mammalian cells, we expressed a UAS-Duox transgene under the daG32-Gal4 driver in the nip null background (data not shown). However, overexpression of Duox failed to rescue the nip null lethality, suggesting that genetically, Duox may not be directly downstream of nip or that the function of nip is not limited to its ability to regulate Duox activity in the fly.
Nonessential role for maternal NIP in embryogenesis
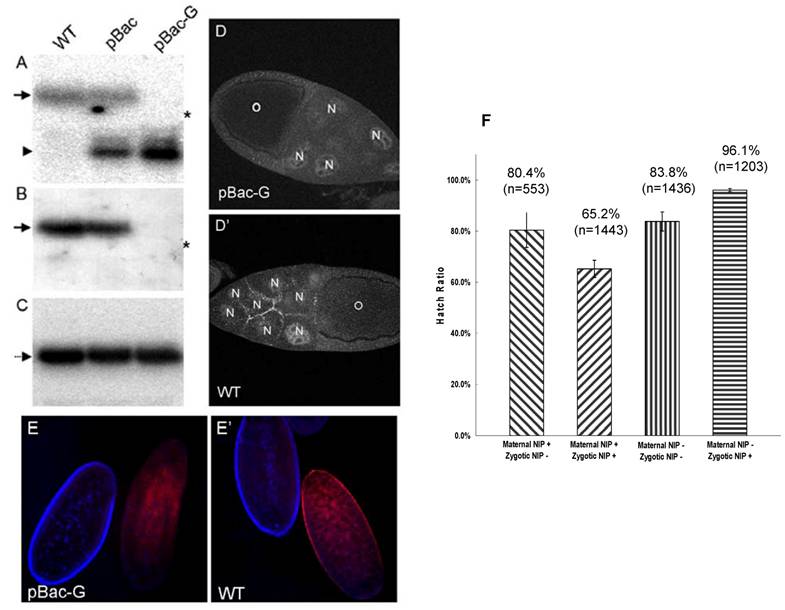
To investigate whether maternal NIP plays a role in fly embryogenesis, we employed the autosomal flipase-dominant female sterile (FLP-DFS) technique to generate a pBac homozygous germline clone (GLC) [14], in which the maternal NIP expression would be blocked by the piggyBac transposon in the nip gene. To ascertain that the maternal NIP expression was indeed lost in the pBac GLC females, mRNA isolated from eggs laid by pBac GLC females were analyzed by Northern blot in comparison to the pBac/CyO or y w (wt) females. As shown in Fig. 2A, a single band representing the full-length nip transcript was detected in the wt embryos. The pBac/CyO eggs produced two bands, with the upper band corresponding to the complete nip mRNA and the lower band representing the truncated nip transcript from the nip allele carrying the pBac insertion. In contrast, only the truncated nip transcript was detected in pBac GLC embryos, suggesting a deficiency in maternal expression of full-length nip mRNA by the pBac GLC clones (Fig. 2A). The lack of full-length mRNA transcription in the pBac GLC flies was confirmed when the blot was probed with a nip exon sequence downstream of the pBac insertion site (Fig. 2B). The numb mRNA level, which was used as a control, remained unchanged from the wt to the mutant flies (Fig. 2C).
The loss of NIP expression in the pBac GLC mutant was further verified by immunostaining of the ovaries and embryos from germline nip mutant females. As shown in Fig. 2D & E, the characteristic cell junctional immunostaining of NIP on the nurse cells seen in the wild type egg chamber was no longer observed in the nip mutant GLC egg; similarly, no specific NIP immunostaining signal on the cell membrane was observed for the nip mutant GLC embryos. These data demonstrated that the nip transcription was disrupted by the pBac transposable element, and no full-length nip mRNA or functional protein was maternally produced in the pBac GLC flies.
Since NIP is abundant in the first two hours AEL, we were interested in finding out whether maternal NIP plays a role in embryogenesis. To this end, pBac GLC females or pBac/CyO, twi-EGFP females were crossed with pBac/CyO, twi-EGFP males and the hatch ratios of the corresponding progeny embryos were scored. For the pBac GLC females, only two types of embryos were produced: non-GFP embryos that lacked both maternal and zygotic NIP and GFP embryos that lacked only the maternal NIP. Surprisingly, more than 80% of either type of embryos was hatched (Fig. 2F). The development of these embryos was similar to the pBac/CyO, twi-EGFP females, but none of the non-GFP pBac homozygous flies passed the 1st instar larval stage. In contrast, the GFP pBac GLC heterozygous flies developed normally to adult. These results suggest that no obvious defect in embryogenesis is associated with maternal NIP depletion and the lethality of homozygous pBac flies stems from deficiency in zygotic NIP expression.
Growth arrest, developmental defects and lethality for nip mutants
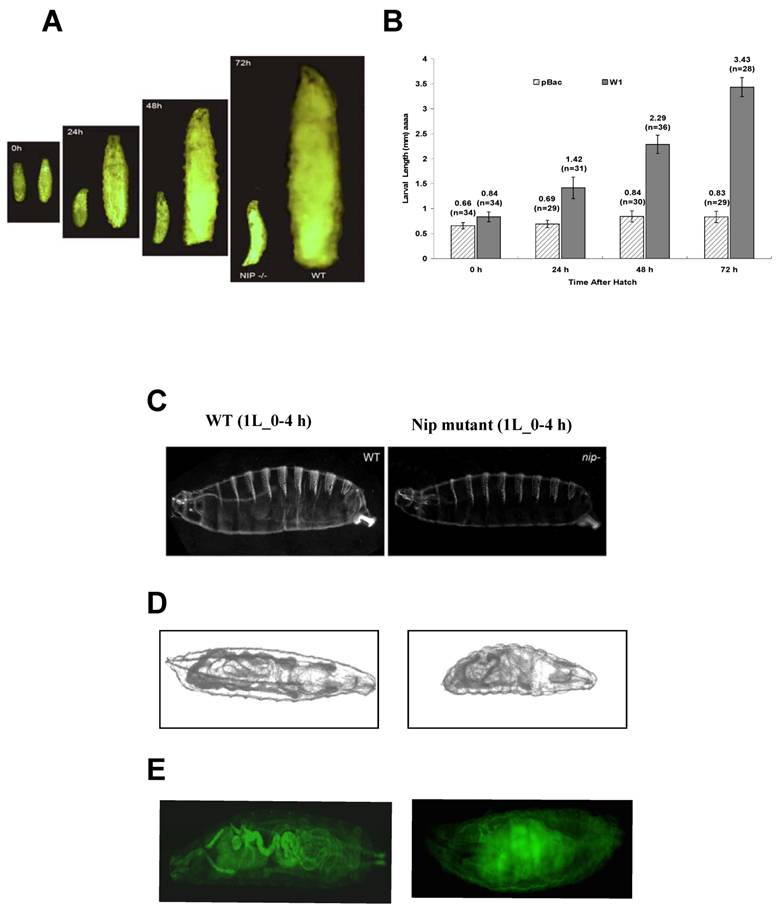
Notwithstanding the above observation that more than 80% of the pBac or the pBac-GLC flies hatched, , none of the mutant larvae lived past the 1st instar. Moreover, pBac homozygotes exhibited extreme retardation in larval growth. Although the size of wt or mutant embryos were comparable when they were hatched, in 24 hours, the wt larvae were approximately twice as long as the mutant (Fig. 3A). In fact, the mutant larvae hardly grew at all during an extended 1st instar larval period during which time a wt larvae reached a body length 4-5 times of a mutant (Fig. 3A & B). Despite the remarkable growth retardation of the mutant embryo, cuticle of the 1st instar larvae (L1) showed no major defect in comparison to the wt embryo. However, the tracheal system of the mutant embryo appeared underdeveloped. Mallory stain of the L1 larvae revealed gross defects in the development of inner organs for the nip mutant (Fig. 3D). To gain a better understanding of the developmental defects associated with loss of nip function, we generated both wt and mutant flies expressing the green fluorescent protein GFP under the control of the daG32-GAL4. This allowed direct visualization of the major inner organs of the embryo under a fluorescent microscope. As shown in Fig. 3E, while the wt larva exhibited a well developed intestinal tract, the mutant larva showed no distinctive features in the gut and annexes, suggesting gross developmental defects in this lineage. These defects were manifested also in the feeding pattern of the mutant larvae since they were lethargic in feeding and had much less food intake than the wt larvae.
Intriguingly, although some of the pBac homozygous larvae were dead by 24 hours after hatching, most of them remained alive for a longer period of time and died after an extended first larval instar. As shown in Fig. 3B, pBac homozygous larvae remained in the first instar stage and had an average length of 0.8 mm three days after hatching when wild type larvae already progressed into the third instar stage and had an average length of 4mm. Some pBac homozygous larvae lived much longer than three days, and a few of them survived longer than ten days. Therefore, nip-null mutants appeared to be developmentally arrested at an extended first instar.
Transgenic nip-RNAi flies show defects in wing development and have a dramatically shortened life span at 29 oC
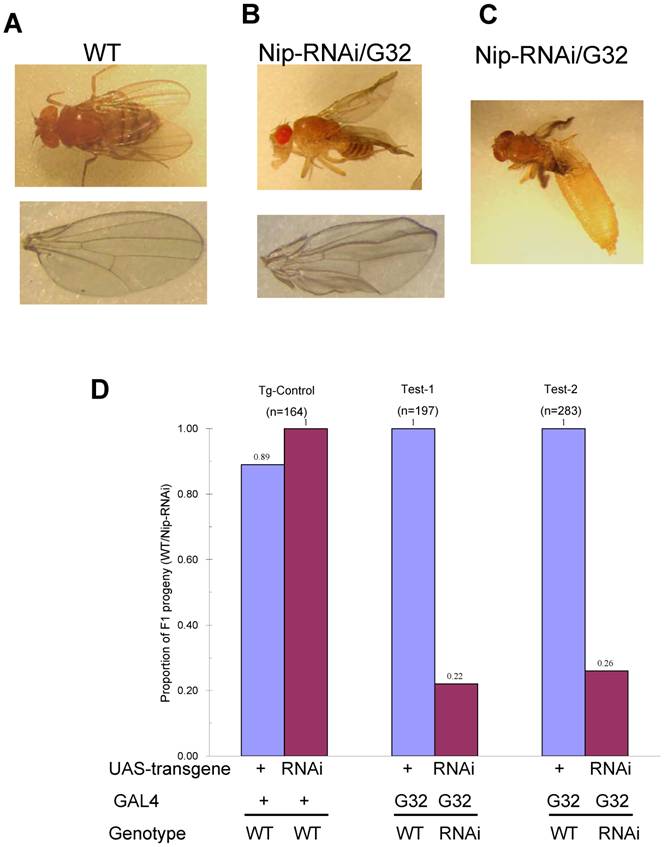
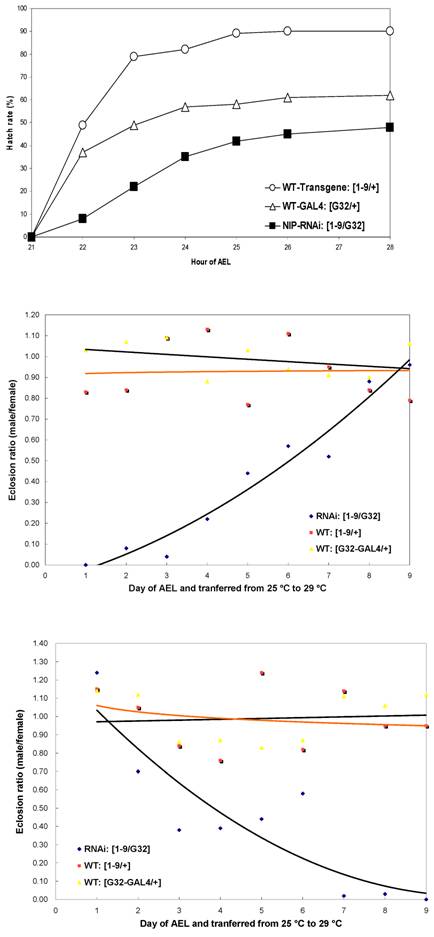
Since nip-null mutant were 1st instar larval lethal, we used RNA interference to knockdown nip expression and created transgenic UAS-nip-RNAi lines driven by TubP-GAL4 or daG32-GAL4. Compared to transgene/+ and the Gal4/+ control flies, the nip-RNAi flies showed a significantly reduced rate of hatching (Fig. S4). The nip-RNAi flies exhibited defects in eclosion when they were transferred from 25 oC to 29 oC (Fig. 4A and Fig. S4). Moreover, all nip-RNAi progenies displayed crinkled wings compared to the smooth morphology for the wt flies (Fig. 4B&C). This phenotype is reminiscent of those observed on the withered and Sod1 mutant flies [15-17]. Interestingly these mutants also displayed hypersensitivity to oxidative stress as observed also for the nip-RNAi flies (see below).
Maternal NIP is not essential for embryogenesis. (A-C) Maternal NIP expression is lost in the pBac germline clone flies. Total RNA was isolated from eggs laid by w1118 (WT), pBac/CyO (pBac), pBac GLC (pBac-G) females at one hour AEL. The nip and numb cDNA probes used were: (A) nip-RA n176-472, which is upstream of the pBac insertion site; (B) nip-RA n602-1117, which is downstream of pBac insertion site; (C) numb n286-816. The full-length nip mRNA (arrow) is detected in wt and pBac eggs, while truncated nip mRNA (arrow head) is detected in pBac and pBac-GLC eggs. The asterisk (*) shows the position of Drosophila 18S rRNA (1.98 kb). (D&D') Staining of a pBac-GLC (D) and a wild type (D') egg chamber with an anti-NIP antibody. No NIP protein was detected in the oocyte (O) or nurse cells (N) in the pBac-GLC egg chamber. (E&E') Staining of eggs laid by pBac-GLC females (E) and wt females (E'). The developmental stages are distinguishable by the nucleus staining (blue): the right eggs are at post-cellularization stages and left eggs at the pre-blastoderm stage. No NIP protein is detected in the pBac-GLC embryo at the pre-blastoderm stage. (F) Hatch ratios of embryos from the nip mutant and the germline flies.
Lack of NIP expression leads to growth and developmental arrest. (A) Comparison in body size between wt (right) and nip-/- pBac homozygous (left) larvae at different time points after larval hatching. (B) Graphical representation of body lengths of pBac homozygus and wt larvae at different time points after hatch. (C) Cuticles of wt and nip mutant larvae. Anterior is left and dorsal is down. Larvae are not actual size. (D) Mallory staining pattern of wt and mutant L1 larvae showing defects in inner organ development for the nip mutant. (E) G32-Gal4 induced GFP expression to visualize morphology of wt and nip mutant larvae. Note the gross developmental defects for the mutant fly, in particular in the digestive track.
Developmental defects of nip-RNAi flies. Compared to wt (A), nip-RNAi flies show vestigial wing (B) and eclosion defect (C) at 29oC. Moreover, Nip-RNAi/G32 flies exhibited dramatic preadult lethality at 29oC.
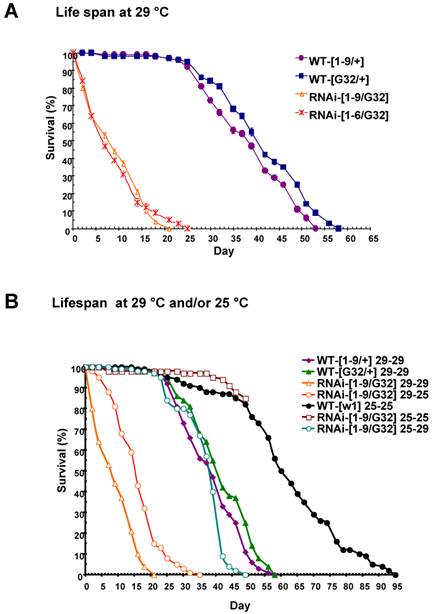
Some progenies of the nip-RNAi strains showed pre-adult lethality but most survived to adulthood and had a normal life span at 25oC. Since an eclosion defect was observed at 29oC, we checked the survival rate of the nip-RNAi flies reared at 29 oC. Both the nip-RNAi [1-9]/ daG32-GAL4] and the nip-RNAi [1-6]/ daG32-GAL4] lines examined had a significantly reduced life span compared to the two control lines. The nip-RNAi flies, on average, lived for 22 days at 29oC compared to 55 days for the wt flies, a reduction of 60% in life span (Fig. 5A). Interestingly, the preadult lethality of nip-RNAi appeared to be temperature-dependent as flies hatched at 25 oC, but switched to 29 oC at L1 lived as long as the wt flies reared at 29 oC throughout. In contrast, nip-RNAi flies hatched at 29 oC that were transferred to 25 oC at L1 did almost as poorly as flies hatched and grown at 29 oC (Fig. 5B). These results suggest the NIP-depleted flies were incapable of coping with the stress caused by an elevated temperature.
NIP knockdown leads to defects in oxidative stress response
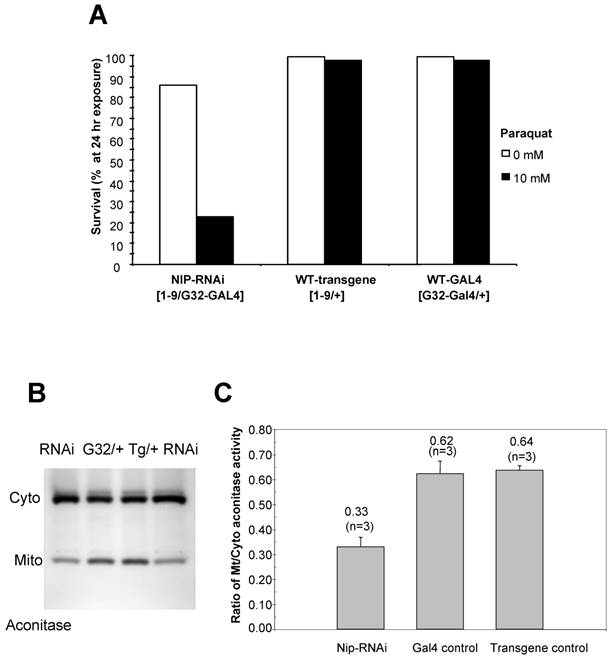
Since the nip-RNAi phenotype at 29 oC mimics that observed for Sod1-null flies [17] in that both displayed crinkled wings and increased level of superoxide, we next examined whether the nip-RNAi flies were as capable as the wt flies in coping with external oxidative stress. We treated the flies with paraquat, a redox-cycling agent [17]. As shown in Fig. 6A, a 24-hour exposure to paraquat led to a 80% lethality for the nip-RNAi flies compared to the controls, suggesting that the former were hypersensitive to external oxidative stress. As aconitase activity is commonly used as a biomarker for oxidative damage as the enzyme's [4Fe-4S]2+ cubane cluster is susceptible to oxidation-induced disassembly [18, 19], we assessed the impact of NIP depletion on mitochondrial aconitase activity, an indicator of altered superoxide flux in mitochondria [16]. Superoxide inactivates Fe-S cluster-containing proteins such aconitase, which in turn serve as sentinels of oxidative stress. An aconitase assay method developed by Huang et al [20] allows for simultaneous analysis of mitochondrial and cytosolic aconitase activity, and this method was adapted for the measurement of aconitase activity associated with wt or nip-RNAi flies, respectively. As shown in Fig. 6B & C, the ratio of mitochondrial to cytosolic aconitase activity in the nip-RNAi flies was reduced to approximately half of that obtained for the control flies, suggesting diminished mitochondrial function caused by NIP depletion. This reduction in mitochondrial function may underlie the compromised ability of the nip-RNAi flies to survive under cellular stress such as exposure to oxidative compounds or mild heat shock [21].
nip-RNAi/G32 flies have a temperature-dependent reduction in lifespan. (A) Drastically reduced lifespan observed for two lines of nip-RNAi/G32 flies compared to the controls. (B) Effect of temperature shift from 25°C to 29°C or from 29°C to 25°C on the lifespan of nip-RNAi/G32 and wt flies. The survival percentage was determined for 20 flies/vial x 5 vials on standard cornmeal food and transferred with a 2-3 day interval.
(A) nip-RNAi flies are hypersensitive to paraquat induced oxidative stress compared to the wild types. Female adults reared at 29°C were scored as 5 vial X 20 flies/vial after 5 hr eclosion. The flies were allowed to recover overnight from CO2 anesthesia and then exposed to 10 mM paraqut in 1 % sucrose. The survival rate was determined at 24 hr after exposure. Data shown are representative of two independent sets of experiments. (B) nip-RNAi flies exhibit a reduced ratio of mitochondrial/cytosolic aconitase activity. Aconitase activity was measured for flies in the 3rd instar and compared to the wild type control. Cyto, cytosolic; mito, mitochondrial. (C) A graphical representation of the mitochondrial/cytosolic aconitase activity ratio. Data shown are averages from three independent experiments.
Discussion
The identification of NIP/DuoxA in both Drosophila and vertebrates suggest an evolutionarily conserved function for this protein [7] (Qin et al). Despite biochemical and cellular studies that link DuoxA to the modulation of dual oxidase activity, a whole animal study is necessary to fully understand the physiological function of NIP/DouxA. By using genetic and biochemical assays, we show here that NIP/DuoxA plays an essential role in the embryonic and larval development of Drosophila. Both zygotic and germinal deletion mutants of nip exhibited 1st instar larval lethality and gross embryonic developmental defects underscored by growth arrest and underdevelopment in the intestinal tract. This latter defect may underlie the severe growth retardation defect observed for the mutant animals as they were unable to intake food. These phenotypes, however, could be fully rescued by ubiquitous expression of a UAS-nip or a UAS-nipNN/AA. Because this mutant NIP was shown in a previous study to be incapable of Numb-binding, the ability of UAS-nipNN/AA to rescue the mutant lethality suggests the function of NIP is unrelated to Numb in Drosophila embryonic and larval development. Intriguingly, despite recent reports demonstrating a regulatory role for NIP/DuoxA on Duox activity and an early study showing that dDuox mutant flies showed larval/pupal lethality[22], expression of a UAS-dDuox transgene under the control of a ubiquitous daG32-GAL4 failed to rescue the nip mutant lethality (data not shown). This result suggests that dnip and dDuox may not be genetically coupled as tightly as their mammalian counterparts [9]. Indeed, although both the dnip/mol and the dDuox genes are found in chromosome 2L, the former is located at the cytogenetic region 23B2-23B3 while the latter at 35B7-35B8.
Although an early larval lethality prevented analysis of nip function in later development, transgenic flies expressing a GAL4-regulated, inverted repeat nip-RNA-interference transgene survived to adulthood at 25 oC. However, NIP ablation resulted in crinkled wings and a marked reduction in the life span of the transgenic flies at 29 oC, suggesting that these animals were incapable of coping with cellular stress associated with mild heat shock. Because oxidative stress is an important contributor to aging, it is likely that the reduced life span observed on the nip-RNAi flies stems from excessive oxidative stress or an inability in coping with oxidative stress. Indeed, we found that, in comparison to wt flies, the RNAi flies showed enhanced sensitivity to applied oxidative stress and incurred early onset lethality in young adults. In agreement with this observation, we found that the RNAi files exhibited reduced aconitase activity in the mitochondria relative to the cytosol. Because mitochondrial respiration is the principal source of reactive oxygen within cells, it is possible that NIP depletion elevated endogenous oxidative stress in the cell or diminished the ability of mitochondia to effectively eliminate reactive oxygen species.
The drastically reduced lifespan observed for the nip-RNAi flies at 29 oC likely originated from a hypersensitivity to oxidative stress as paraquat treatment markedly enhanced the lethality of the RNAi flies. Moreover, the mitochondrial aconitase activity was significantly reduced in the RNAi flies. Decreased mitochondrial efficiency caused by oxidative stress has been considered a major cause of aging [23-27]. Oxidative damage in aging is mediated through specific molecular targets such as mitochondrial DNA and aconitase [28-30]. Conversely, mitochondrial oxidative stress can promote tissue aging through apoptosis [25]. In accordance with this possibility, trypan blue staining of nip-RNAi flies showed enhanced apoptosis compared to wt flies (data not shown). The nip-RNAi phenotype at 29 oC was reminiscent of that obtained for the Sod1 mutant [31]. A potential relationship between Nip and Sod1 was investigated by measuring Sod1 activity in the nip-RNAi flies. However, no change in SOD1 activity was detected (data not shown). Unlike Sod1, which extends Drosophila lifespan when overexpressed in the motorneurons, no significant effect on lifespan was seen when NIP was overexpressed. Therefore, although both NIP and SOD1 or SOD2) are involved in oxidative stress response, the mechanism of their action may be distinct.
Despite our earlier study demonstrating a Numb-NIP interaction at the protein level, a genetic interaction between numb and nip was not observed. The expression patterns of numb and nip are not closely correlated: numb is active in most of embryonic stages when zygotic nip is silent. In the nip mutant embryos, Numb is asymmetrically localized on the plasma membrane during the neuroblast division (data not shown), indicating that NIP is not critical for the Numb localization. Unlike the over-expression of Numb that results in bald notum, over-expression of NIP did not result in an apparent phenotype. Our immunohistological staining data suggest the existence of maternal NIP. The location of NIP during oogenesis and early embryonic stages was therefore determined. However, the embryos that lack both maternal and zygotic NIP did not show more severe developmental defects than the embryos that lack only zygotic NIP. These data suggest that the maternal NIP is not essential for egg formation and fly development.
Taken together, we have identified an essential role for nip in Drosophila embryonic and larval development and in oxidative stress response. The markedly shortened life span and reduced ability to survive under oxidative stress observed for Nip-ablated flies suggest that NIP is involved in defending against ROS damage, likely through modulating the mitochondrial activity of aconitase. Although our rescuing experiments preclude Duox as a direct downstream gene for dNip/dDuoxA, it remains to be seen whether the activity of Duox or an NAGPH oxidase is affected by the loss of NIP. Moreover, recent studies suggest that Duox regulates Drosophila gut immunity by the Gαq-phospholipase Cβ-Ca2+ pathway [32]. Our preliminary data showed that the nip RNAi flies were compromised in their ability to clear bacterial infection (data not shown), suggesting an immune defect associated with loss of NIP function. These and other aspects of NIP function await further investigation.
Acknowledgements
This work was supported by a grant (to SSCL) from the Canadian Institute of Health Research (CIHR). SSCL holds a Canada Research Chair in Functional Genomics and Cellular proteomics.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58(2):349-360
2. Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377(6550):624-627
3. Knoblich JA. Asymmetric cell division during animal development. Nat Rev Mol Cell Biol. 2001;2(1):11-20
4. Li SC, Zwahlen C, Vincent SJ, McGlade CJ, Kay LE, Pawson T, Forman-Kay JD. Structure of a Numb PTB domain-peptide complex suggests a basis for diverse binding specificity. Nat Struct Biol. 1998;5(12):1075-1083
5. Schlessinger J, Lemmon MA. SH2 and PTB domains in tyrosine kinase signaling. Sci STKE. 2003;2003(191):RE12
6. Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390(Pt 3):641-653
7. Qin H, Percival-Smith A, Li C, Jia CY, Gloor G, Li SS. A novel transmembrane protein recruits numb to the plasma membrane during asymmetric cell division. J Biol Chem. 2004;279(12):11304-11312
8. Grasberger H, De Deken X, Miot F, Pohlenz J, Refetoff S. Missense mutations of dual oxidase 2 (DUOX2) implicated in congenital hypothyroidism have impaired trafficking in cells reconstituted with DUOX2 maturation factor. Mol Endocrinol. 2007;21(6):1408-1421
9. Grasberger H, Refetoff S. Identification of the maturation factor for dual oxidase. Evolution of an eukaryotic operon equivalent. J Biol Chem. 2006;281(27):18269-18272
10. Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87(1):245-313
11. Lambeth JD, Kawahara T, Diebold B. Regulation of Nox and Duox enzymatic activity and expression. Free Radic Biol Med. 2007;43(3):319-331
12. Milenkovic M, De Deken X, Jin L, De Felice M, Di Lauro R, Dumont JE, Corvilain B, Miot F. Duox expression and related H2O2 measurement in mouse thyroid: onset in embryonic development and regulation by TSH in adult. J Endocrinol. 2007;192(3):615-626
13. Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297(5590):2270-2275
14. Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144(4):1673-1679
15. Strub BR, Parkes TL, Mukai ST, Bahadorani S, Coulthard AB, Hall N, Phillips JP, Hilliker AJ. Mutations of the withered (whd) gene in Drosophila melanogaster confer hypersensitivity to oxidative stress and are lesions of the carnitine palmitoyltransferase I (CPT I) gene. Genome. 2008;51(6):409-420
16. Kirby K, Hu J, Hilliker AJ, Phillips JP. RNA interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress. Proc Natl Acad Sci U S A. 2002;99(25):16162-16167
17. Woodruff RC, Phillips JP, Hilliker AJ. Increased spontaneous DNA damage in Cu/Zn superoxide dismutase (SOD1) deficient Drosophila. Genome. 2004;47(6):1029-1035
18. Beinert H, Kiley PJ. Fe-S proteins in sensing and regulatory functions. Curr Opin Chem Biol. 1999;3(2):152-157
19. Vasquez-Vivar J, Kalyanaraman B, Kennedy MC. Mitochondrial aconitase is a source of hydroxyl radical. An electron spin resonance investigation. J Biol Chem. 2000;275(19):14064-14069
20. Huang TT, Raineri I, Eggerding F, Epstein CJ. Transgenic and mutant mice for oxygen free radical studies. Methods Enzymol. 2002;349:191-213
21. Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105(32):11364-11369
22. Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310(5749):847-850
23. Sastre J, Pallardo FV, Vina J. Mitochondrial oxidative stress plays a key role in aging and apoptosis. IUBMB Life. 2000;49(5):427-435
24. Sastre J, Pallardo FV, Garcia de la Asuncion J, Vina J. Mitochondria, oxidative stress and aging. Free Radic Res. 2000;32(3):189-198
25. Sastre J, Borras C, Garcia-Sala D, Lloret A, Pallardo FV, Vina J. Mitochondrial damage in aging and apoptosis. Ann N Y Acad Sci. 2002;959:448-451
26. Vina J, Sastre J, Pallardo F, Borras C. Mitochondrial theory of aging: importance to explain why females live longer than males. Antioxid Redox Signal. 2003;5(5):549-556
27. Sastre J, Pallardo FV, Vina J. The role of mitochondrial oxidative stress in aging. Free Radic Biol Med. 2003;35(1):1-8
28. Genova ML, Pich MM, Bernacchia A, Bianchi C, Biondi A, Bovina C, Falasca AI, Formiggini G, Castelli GP, Lenaz G. The mitochondrial production of reactive oxygen species in relation to aging and pathology. Ann N Y Acad Sci. 2004;1011:86-100
29. Lenaz G, Baracca A, Fato R, Genova ML, Solaini G. New insights into structure and function of mitochondria and their role in aging and disease. Antioxid Redox Signal. 2006;8(3-4):417-437
30. Lenaz G, D'Aurelio M, Merlo Pich M, Genova ML, Ventura B, Bovina C, Formiggini G, Parenti Castelli G. Mitochondrial bioenergetics in aging. Biochim Biophys Acta. 2000;1459(2-3):397-404
31. Parkes TL, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, Boulianne GL. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 1998;19(2):171-174
32. Ha EM, Lee KA, Park SH, Kim SH, Nam HJ, Lee HY, Kang D, Lee WJ. Regulation of DUOX by the Galphaq-phospholipase Cbeta-Ca2+ pathway in Drosophila gut immunity. Dev Cell. 2009;16(3):386-397
Figures
A DNA map of the nip mutant PBac{RB}mole02670 and rescue of pBac homozygous flie (A) A piggyBac{RB} element is inserted in an intron of the nip gene. The arrows show the location of primers used for PCR to identify pBac insertion in B&C. (B&C) PCR products obtained from genomic DNA of y w (lane 1), pBac/CyO (lane 2) and pBac/pBac;UAS-nip/TubP-Gal4 (lane 3 in B) or pBac/pBac;UAS-nipNN/AA/TubP-Gal4 (lane 3 in C) flies. The short band in lane 2 of (B) was too weak to detect in the lane 2 of (C).
Expression of NIP under an HS-GAL4 driver completely rescued the pBac/pBac mutant phenotype.
Phenotypes of flies with ectopic expression of Numb or NIP in the wing disc. The ap-Gal4 was used to induce the ectopic expression of Numb (A) and NIP (B) in the wing disc. Ectopic expression of Numb resulted in a bald notum (A) whereas over-expression of NIP did not result in a significant defect. The absence of a few hairs in the ap-Gal4/UAS-nip flies (B) was most likely due to Gal4 instead of NIP because it was also observed when ap-Gal4 was crossed with wild type flies (data not shown).
Nip-RNAi flies display hatching and eclosion defects at 29°C compared to wt flies.
Author contact
![]() Corresponding author: email: slica
Corresponding author: email: slica

 Global reach, higher impact
Global reach, higher impact