Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2010; 6(4):333-349. doi:10.7150/ijbs.6.333 This issue Cite
Review
Water Buffalo Genome Science Comes of Age
1. Department of Animal Sciences, Washington State University, Pullman, WA 99164-6351, USA
2. Department of Biologia, UNESP - São Paulo State University, IBILCE, São Jose Rio Preto, SP, Brazil
3. Department of Veterinary Pathobiology, College of Veterinary Medicine, Texas A&M University, College Station, TX 77843-4467, USA
Received 2010-3-29; Accepted 2010-6-14; Published 2010-6-17
Abstract
The water buffalo is vital to the lives of small farmers and to the economy of many countries worldwide. Not only are they draught animals, but they are also a source of meat, horns, skin and particularly the rich and precious milk that may be converted to creams, butter, yogurt and many cheeses. Genome analysis of water buffalo has advanced significantly in recent years. This review focuses on currently available genome resources in water buffalo in terms of cytogenetic characterization, whole genome mapping and next generation sequencing. No doubt, these resources indicate that genome science comes of age in the species and will provide knowledge and technologies to help optimize production potential, reproduction efficiency, product quality, nutritional value and resistance to diseases. As water buffalo and domestic cattle, both members of the Bovidae family, are closely related, the vast amount of cattle genetic/genomic resources might serve as shortcuts for the buffalo community to further advance genome science and biotechnologies in the species.
Keywords: Water buffalo, Genome resources, Cytogenetics, Whole genome mapping, next generation sequencing, Genome biotechnology.
1. Introduction
Since water buffalo were domesticated 3000-6000 years ago, they have had economic importance as dairy, meat, and draught animals, in many highly populated countries [1-3]. The animals are typically found in tropical and subtropical forests, wet grasslands, marshes and swamps. Although they are terrestrial animals, they spend a good portion of time wallowing in mud holes or rivers in order to keep cool. Their habitats typically contain rivers, streams, mud holes, tall grasses, and trees which provide sufficient drinking and wallowing water, food, and coverage [2]. During the Pleistocene epoch, Bubalus was distributed from southern Asia to Europe. As the climate became increasingly dry, the area of distribution shrunk to India, Indonesia, and parts of Southeast Asia. Buffaloes are thought to have been introduced into Italy from central Europe in the sixth century or by the Bay of Tunis in the seventh century at the same time as the Arab conquests. Introduction of water buffalo populations into Australia, Africa, and the Americas has taken place only recently [1, 3].
There are more than 168 million water buffalo in the world with about 161 million in Asia, 3.7 million in Africa, 3.3 million in South America, and the rest are distributed in Europe and Australia [4]. In South America there is a large population of swamp buffalo/river buffalo hybrids, because many of the buffalo were imported from India where river buffalo are predominant, and Australia, which has a high population of swamp buffalo [5]. These 168 million water buffalo comprise only 11.1% of the world's bovid population, but more people depend on the water buffalo than on any other domesticated species in the world [1]. As such, unlike other domesticated bovids, the water buffalo population has increased about 2% per year world-wide in the last 20 years.
Water buffalo provide more than 5% of the world's milk supply, which contains less water and more fat, lactose, protein, and minerals than cow milk [2, 4]. Water buffalo milk is used to make butter, butter oil, high quality cheeses, and other high quality dairy products. They have leaner meat that contains less fat and cholesterol than beef, while having a comparable taste [2]. Their hide can be used to make good quality leather products and they make good beasts of burden, providing 20% to 30% of all farm power, and are superior draught animals in waterlogged conditions such as rice paddies. They can also be used for transportation and can haul heavier loads than cattle [2, 4]. Water buffalo dung is collected, used for heat or fertilizer, and successfully enriches the soil, which reduces or eliminates the need for chemical fertilizers. Water buffalo utilize less digestible feeds than cattle making them easier to maintain using locally available roughages. In addition, water buffalo are used as cash--to be sold when the need arises; thus securing the economic status of many families.
Water buffalo are fairly healthy animals, even though they typically live in hot/humid regions that are favorable to the development of disease. While they are susceptible to most diseases and parasites, that similarly affect cattle, including trypanosomiasis, tuberculosis, brucellosis, rinderpest and piroplasmosis, the effects of disease are often less deleterious [6]. Due to their wallowing behavior, water buffalo are less susceptible to ticks and other ecto-parasites [4]. The resistance to ticks means that tick-borne diseases such as theileriasis, babesiosis, and anaplasmosis have little effect on water buffalo. If inoculated with East Coast fever (a form of theileriasis), water buffalo and cattle are equally susceptible [6]. Wallowing also makes water buffalo resistant to the screwworm fly, which is a major pest of livestock in Central and South America. In areas where cattle are heavily infected with the screwworm fly larvae, water buffalo are not affected. The mud that is caked on water buffalo after wallowing is thought to suffocate the larvae [6]. The adult water buffalo also has a strong resistance to strongyloid nematodes. Water buffalo do not experience the same nutritional deficiencies and the resulting susceptibility to these worms that cattle do because of their ability to utilize low quality roughages [6]. However, despite the resistance to some parasites that wallowing affords, this behavior increases the susceptibility of water buffalo to liver fluke. The buffalo are easily infected with the waterborne stage of liver fluke, although clinical signs of the disease are not usually manifested. Milking water buffalo are less affected than dairy cattle by mastitis although this is likely to change as buffalo milk yield increases [6].
Although there are many advantages to raising water buffalo as described above, these animals remain underutilized. In particular, water buffalo breeders and farmers have been facing many challenges and problems, such as poor reproductive efficiency, sub-optimal production potential, higher than normal incidence of infertility, and lower rates of calf survival.
Genome research has created a broad basis for promoting and utilizing gene technologies in many fields of livestock production. For example, genome biotechnology will provide a major opportunity to advance sustainable animal production systems of higher productivity through manipulating the variation within and between breeds to realize more rapid and better-targeted gains in breeding value. This type of research will also make it possible to distinguish molecular phenotypes and thus improve the use of genetic resources in domestic animals. Therefore, the present review focuses on the currently available genome resources in water buffalo, thus providing knowledge and technologies that can help optimize production potentials, reproduction efficiency, product quality, nutritional value and resistance to diseases in the species.
2. Genome Anatomies of Water Buffalo
Buffalo belong to the Bovidae family and there are two main species of buffalo: the Asiatic buffalo (Bubalus bubalus) and the African Buffalo (Syncerus caffer) [5]. The Asiatic buffalo originated in India where domestication likely took place in the third millennium BC [7], and China where domestication occurred in the fifth millennium BC [8]. The Asiatic water buffalo can be divided into two subspecies: the river buffalo and the swamp buffalo. The exact phylogenetic relationship between swamp and river buffalo is still in question. Divergence of these two subspecies occurred approximately 10,000 to 1.7 million years ago, long before domestication [1, 9]. Therefore, it is likely that that there were separate domestication events for river buffalo in India and for swamp buffalo in China [10]. As for the African buffalo, there are two subspecies: the cape buffalo (Syncerus caffer caffer) and the forest buffalo (Syncerus caffer nanus) [11].
Cytogenetics of Asiatic Buffalo
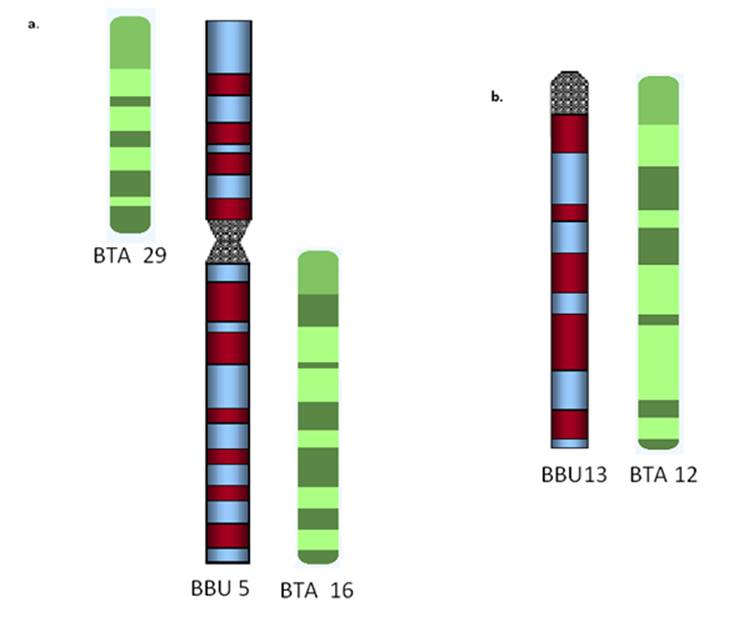
Cytogenetic studies show that river buffalo have 25 chromosome pairs while swamp buffalo have 24 pairs. These subspecies differ by one chromosome; a fusion between river buffalo (BBU) chromosome 4 and 9 is comparable to swamp buffalo chromosome 1 [5], and all chromosomes and chromosome arms are preserved between these two subspecies. Crosses between the two subspecies are fertile but hybrids possess 49 chromosomes, which is thought to lead to lower reproductive values in subsequent matings. River buffalo have 5 biarmed chromosome pairs and all others, including the sex chromosomes are acrocentric. Several studies have shown that river buffalo and domestic cattle, both members of the Bovidae family, are closely related. Indeed, both share chromosome banding and gene order homology, and have been cytogenetically characterized [12]. At the cytogenetic level, cattle and river buffalo chromosomes can be matched arm for arm (see examples in Figure 1). For reference, the cattle genome consists of 29 acrocentric chromosome pairs and a pair of XY sex chromosomes, while the river buffalo genome has 5 biarmed and 19 acrocentric chromosome pairs plus the XY sex chromosomes. The 5 biarmed chromosome pairs correspond to the fusion of two cattle acrocentrics, such as BBU 1 for BTA1 and BTA27, BBU2 for BTA2 and BTA23, BBU 3 for BTA8 and BTA19, BBU4 for BTA5 and BTA28, and BBU5 for BTA16 and BTA29, respectively. Each of the acrocentric river buffalo chromosomes corresponds to one of the remaining cattle chromosomes [12].
At the cytogenetic level, water buffalo chromosomes can be matched to bovine chromosomes arm for arm. Each biarmed water buffalo chromosome is derived from the fusion of two bovine acrocentrics. (a) This shows the similar banding patterns for bovine chromosomes 29 and 16 to water buffalo chromosome 5 [22], (b) This shows similar banding patterns for bovine chromosome 12 and water buffalo chromosome 13 [22].
Cytogenetics of African Buffalo
Cytogenetic studies show that S.c. caffer possess 26 chromosome pairs and S.c. nanus has 27 chromosome pairs. These two species can interbreed although their progeny have 53 chromosomes and have reduced fertility due to unbalanced gametes, which give rise to unbalanced zygotes [5]. The main difference between the two subspecies of African buffalo is the presence of four biarmed chromosomes in S.c. caffer and three biarmed chromosomes in S.c. nanus. The rest of the chromosomes, including the sex chromosomes, are acrocentric in both species. The biarmed pairs in S.c. caffer correspond to the fusion of cattle chromosomes 1, 13; 2, 3; 5, 20; and 11, 29 [13]. In addition, Syncerus and Bubalus share no bi-armed chromosomes pairs, which suggest that there can be no crosses between these two genera because the resulting hybrid would have an unbalanced chromosome set. Therefore, chromosome morphology supports the designation of two separate genera [5].
Sex Chromosomes
Several studies have revealed high degrees of homology among autosomal chromosomes of bovids with similar banding patterns and gene order among the chromosome arms of cattle, river buffalo, sheep, and goats [14, 15]. Bovid sex chromosomes, unlike the highly similar autosomal chromosomes, share a slightly more complex rearrangement of sequences [5]. Chromosome banding comparisons show that while large portions of these chromosomes are conserved, BBU-X has large blocks of constitutive heterochromatin that BTA-X lacks. Cytogenetic studies representing loci order on these sex chromosomes show complex rearrangements that may have occurred during the karyotype evolution of river buffalo and cattle. BBU-X and BTA-X share the same gene order but a different centromere position, indicating a centromere translocation event with the loss of constitutive heterochromatin in BTA-X, which differentiates it from BBU-X [5]. Comparative FISH mapping shows the existence of a similar situation in river buffalo and cattle Y-chromosomes. BTA-Y and BBU-Y differ in an inversion including the centromere and breakage points in both arms (pericentric inversion) where BBU-Y is larger than BTA-Y and gains heterochromatin [5].
Nucleolar Organizing Regions (NORs)
Studies have shown that nucleolar organizing regions (NORs) are present in the telomeres of five cattle, sheep, and goat chromosomes and six water buffalo chromosomes [5]. The presence of these NORs and the highly conserved nature of bovid chromosomes indicated that the same nucleolus organizer chromosomes (NOCs) would also be present in bovids. Research by Gallagher et al. [16] found that some NORs, and not NOCs, were conserved to homologous chromosomes or chromosome arms. Moreover, goats, sheep, cattle, and river buffalo share one NOC; and two of the NOCs are common to river buffalo and cattle showing a close evolutionary proximity to each other [5].
Phylogenetics of Water Buffalo
Barker et al. [17] studied 21 microsatellite loci in 8 swamp and 3 river buffalo populations, and found that there were considerable intra- and interpopulation differences in allelic variability between and within each buffalo type. Ritz et al. [18] studied the relationship between bovid species, including river buffalo. This study compared the analysis of 20 bovine microsatellites and showed that B. bubalis and S. caffer were the most divergent species in the Bos clade. Finally, Kumar et al. [19] used 27 microsatellite loci to show genetic variation among 8 Indian breeds. More recently, amplified fragment length polymorphism (AFLP) fingerprinting analysis was used in the phylogenetic analysis of bovid species that clustered African and water buffalo. This study showed several tree constructions including African buffalo with water buffalo [5]. Analysis of the mitochondrial D-loop DNA sequence of 19 swamp buffaloes and 61 river buffaloes of different breeds was done by Kierstein et al. [1]. The results of this study suggested that there was only one domestication event of water buffalo, which occurred on the Indian subcontinent 5,000 years ago. The authors hypothesized that these water buffalo interbred with wild buffalo or domestic buffalo from China resulting in the buffalo found on the South-East Asian mainland. Alternatively, Kumar et al. [10] suggested an independent domestication for river and swamp buffalo which is supported by the comparison of the mitochondrial D-loop regions of seven Chinese swamp buffalo and other swamp and river buffalo from Australia, India, Brazil, Italy, and Southeast Asia done by Lei et al [20]. These results showed that river buffalo and swamp buffalo diverged before domestication indicating that the domestication of swamp buffalo in China was independent from the domestication of river buffalo on the Indian subcontinent.
Analysis of water buffalo interleukin-12 (IL12) sequences and expression revealed significant sequence identity to bovine IL12 and functional cross-reactivity with bovine immune cells [5]. Furthermore, Indian water buffalo interleukin-18 (IL18) cDNA showed similar amino acid sequence (99%) to cattle.
3. Whole Genome Mapping of Water Buffalo
Several methods have been developed and used in mapping of genomes, such as linkage, radiation hybrid (RH), and in situ hybridization mapping. The linkage map was developed soon after the re-discovery of Mendel's work at the beginning of the 20th century. Linkage maps are generated by counting the number of offspring that receive either parental or recombinant allele combinations from a parent that carries two different alleles at two or more loci. Analyses of this type of data determine whether loci are "linked" to each other and, their relative order and the distance that separate them. Therefore, linkage mapping requires polymorphic markers and reference populations. The starting materials for RH mapping are cell lines that are constructed by fusing irradiated donor cells from the species of interest with a rodent cell line (usually hamster). The irradiation shatters each chromosome into multiple fragments at random locations. The resolution of this technique is as high as linkage analysis but it does not depend upon breeding and polymorphic markers. In situ hybridization uses a labeled probe to detect and localize specific RNA or DNA sequences on a chromosome. The modified in situ protocol that utilizes fluorescent tags is referred to as FISH (for fluorescent in situ hybridization). In general, for the method to be effective, the probe should be at least 500 bp in length. However, the estimated resolution of in situ hybridization is limited to ~10,000 kb. Current research only reports the use of the latter two methods in mapping of the water buffalo genome.
Fluorescent In Situ Hybridization (FISH)
The first cytogenetic map for water buffalo with only 68 loci, mostly assigned using FISH, was reported by Di Meo in 2008 [21]. This map contained at least one bovine molecular marker assigned to each river buffalo chromosome or chromosome arm. Subsequent maps were reported that contain 171 known genes and 122 microsatellites. Of these, 293 were assigned genes and 247 were assigned using FISH [5]. The total number of mapped loci for river buffalo is now 309, which cover all chromosomes and chromosome regions of the river buffalo genome (reviewed in Iannuzzi and Di Meo 2009 [5]).
Radiation Hybrid Mapping
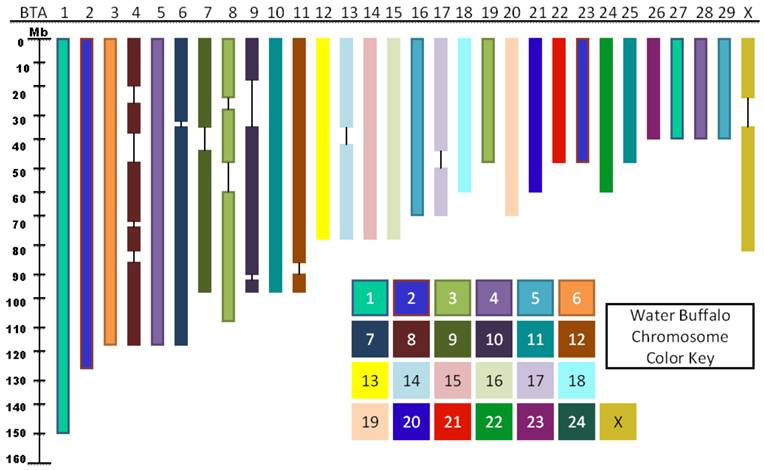
Radiation hybrid mapping is used to generate medium to high resolution maps, and are available for several mammalian species including the cow. The RH panel for river buffalo is a recent development that has been used to construct preliminary RH maps for several of the water buffalo chromosomes [22-26]. The preliminary RH maps for BBU1, BBU3, BBU6, BBU7, BBU10, and BBUX were based on markers derived from cattle, and showed that the bovine genome is a useful source of markers for buffalo genome mapping. This one element is significant and demonstrates that rapid and efficient transfer of genetic information may occur from cattle to water buffalo. Indeed Amaral et al. [12] used the BBURH5000 panel to construct a first generation genome RH map of the river buffalo with 2621 cattle-derived loci covering all chromosomes. This map demonstrates improved coverage with considerable increases in the number of mapped markers, when compared to preliminary maps (previously constructed). After completion of the first generation whole genome RH map for river buffalo, the marker order was compared to the current bovine genome sequence assembly Btau_4.0. This comparison showed that the marker order within the linkage groups for the buffalo chromosomes was consistent with the bovine genome assembly [12]. As such, Stafuzza et al. [27] generated RH maps for BBUY which contain a total of 28 markers distributed within one linkage group. Figure 2 illustrates the bovine genome regions that have been linked to the water buffalo genomes based on the current maps of the species.
4. Whole Genome Sequencing of Water Buffalo
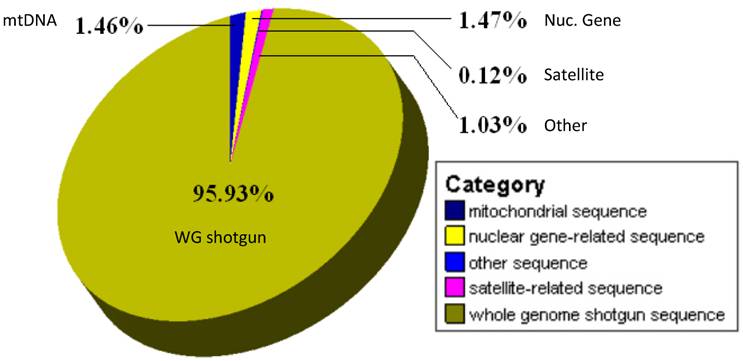
Genome sequencing in farm and other animals has advanced significantly in recent years. For example, sequence data are available in the public domain for many livestock species. As of March 1, 2010, there are 2,509,850 cattle, 3,237,358 pigs, 2,195,532 chicken, 6,259,791 sheep, 470,489 horse, 2,886,083 cat and 2,599,789 dog sequences available in the GenBank nucleotide databases (http://www.ncbi.nlm.nih.gov). Whole genome sequencing has been completed in cattle, horse, chicken and dog and sequencing of the porcine genome is almost completed. A total of 66,935 nucleotide sequences for the water buffalo have been deposited in the GenBank database and are mainly 64,212 whole genome shotgun sequences, while the rest includes 974 mitochondrial genomic sequences and 1,748 nuclear gene/genomic DNA sequences. The latter may be further classified into nuclear gene-related sequences (981), satellite-related sequences (689) and others (78) (Figure 3). The 689 satellite sequences involve satellite (17), microsatellite (311) and minisatellite (361) sequences, respectively, and the 78 other sequences mainly consist of cis-acting regulator, CpG island, repeat sequence, gene-like sequence or some other genomic sequences.
Genome regions that have markers linked between river buffalo and cattle (Data source: [12]).
Categories of 66935 water buffalo sequences deposited in the GenBank Database.
Mitochondrial Genome Sequencing of Water Buffalo
The mitochondrial genome is present as a circular DNA molecule. Unlike nuclear chromosomes that are paired in mammals (except X and Y sex chromosomes), there are many copies of the mitochondrial molecule in every cell. However, the copy number can be extremely variable in different cells. For example, an egg contains 100,000 to 1,000,000 mitochondrial DNA (mtDNA) molecules, while a sperm contains only 100 to 1000. The complete water buffalo mitochondrial genome was sequenced by three groups of scientists at Hainan Medical College, China (see Genbank accession number: AY702168), Centre for Cellular and Molecular Biology, Hyderabad, India (see GenBank accession number: AF547270) and Istituto Spallanzani, Italy (see GenBank accession number: AY488491). Similar to other mammals, each mtDNA molecule in water buffalo harbors genetic material that encodes 37 genes: 13 for proteins (polypeptides), 22 for transfer RNA (tRNA) and one each for the small and large subunits of ribosomal RNA (rRNA) (Figure 4).
Water buffalo mitochondrial genome structure and annotation based on AY488491.
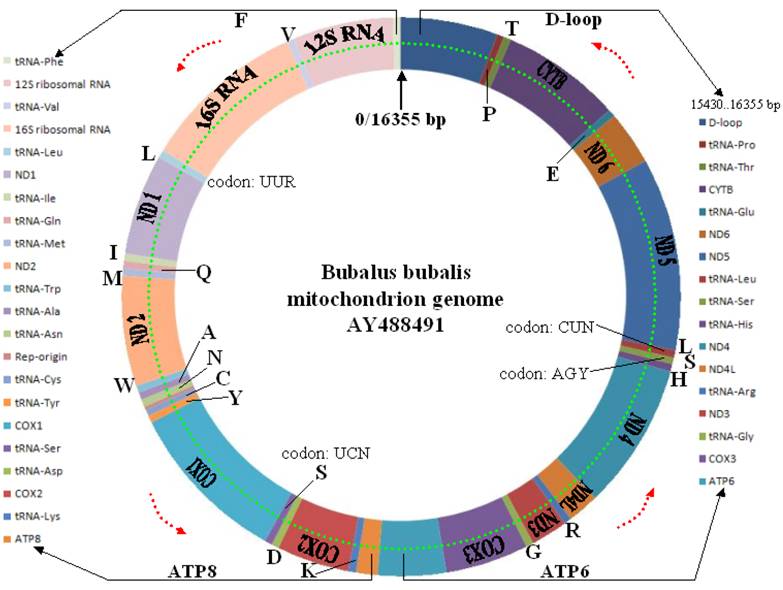
Among 13 coding mitochondrial genes in water buffalo, the length of the coding sequences of four genes - COX1, ND4L, ND4 and ND6 are identical to those in Bison bison (American bison, NC_012346), Bos grunniens (domestic yak, NC_006380), Bos indicus (zebu, AF492350), Bos taurus (domestic cattle, NC_006853), Capra hircus (goat, NC_005044), Equus asinus (donkey, NC_001788), Equus caballus (horse, NC_001640), Lama glama (llama, NC_012102), Oryctolagus cuniculus (rabbit, NC_001913), Ovis aries (sheep, NC_001941) and Sus scrofa (swine, NC_000845). However, the D-loop region (926 bp) is relatively short in water buffalo, whereas it ranges from 888 bp in B. bison to 1800 bp in O. cuniculus. Interestingly, sixteen of these 37 genes overlap in the water buffalo mitochondrial genome, including two three-gene-overlaps (ND1/tRNA-Ile/tRNA-Gln and ATP8/ATP6/COX3), and five two-gene-overlaps (ND2/tRNA-Trp, tRNA-Tyr/COX1, ND4L/ND4, ND5/ND6 and tRNA-Thr/tRNA-Pro) (see GeneBank accession number: AY488491). The overlapping size varies from 1 bp to 40 bp. Certainly some genes are also distanced from each other, but the distance gap ranges only from 1 bp to 4 bp in length. In addition to these complete mitochondrial genome sequences of water buffalo, the D-loop and CYTB regions have also been investigated. Currently, there are 784 entries for the former region and 162 entries for the latter region in the GenBank databases.
Nuclear Gene/Genomic Sequencing of Water Buffalo
As shown in Table 1, 971 known gene sequences have been contributed to the GenBank database for the species. Our annotation (Table 1) revealed that the sequences represent 277 functional genes based on the BLAST searches against orthologous genes in mammals. In particular, we observed that 29 genes/clusters have been heavily investigated, and they may account for 54% (527/971) of the known gene entries. Among them, 243 entries are sequences for genes related to growth and milk production, such as oxidized low density lipoprotein (lectin-like) receptor 1 (OLR1, 53 entries), leptin (LEP, 33 entries), growth hormone receptor (GHR, 29 entries), lactalbumin, alpha (LALBA, 20 entries), casein beta (CSN2, 18 entries), growth hormone 1 (GH1, 15 entries), insulin-like growth factor 1 (IGF1, 14 entries), casein kappa (CSN3, 13 entries), casein alpha s1 (CSN1S1, 13 entries), stearoyl-CoA desaturase (SCD, 10 entries), myostatin (MSTN, 9 entries), diacylglycerol O-acyltransferase homolog 1 (DGAT1, 9 entries) and butyrophilin, subfamily 1, member A1 (BTN1A1, 7 entries). Major histocompatibility complex (BULA@, 86 entries), solute carrier family 11, member 1 (SLC11A1, 24 entries), lactoferrin (LTF, 23 entries), integrin, beta 2 (ITGB2, 11 entries), CD14 molecule (CD14, 11 entries), toll-like receptor 4 (TLR4, 10 entries), lysozyme (LYZ, 10 entries), cathelicidin (CATHL@, 8 entries) and interleukin 2 (IL2, 7 entries) were the targets to study disease resistance in water buffalo. Variables of reproduction is another focus of water buffalo genome research, including studies on follicle stimulating hormone receptor (FSHR, 17 entries), sex determining region Y (SRY, 15 entries), progestagen-associated endometrial protein (PAEP, 14 entries), luteinizing hormone beta polypeptide (LHB, 10 entries), cytochrome P450, family 19, subfamily A, polypeptide 1 (CYP19A1, 9 entries), follicle stimulating hormone, beta polypeptide (FSHB, 8 entries) and estrogen receptor 1 (ESR1, 6 entries).
Genomic sequencing in water buffalo also involved satellite, minisatellite and microsatellite sequencing. Satellite sequences were mainly contributed by two groups of researchers, one at the Nagoya University, Japan [28] and one at National Institute of Immunology, India [29]. With digestion of two restriction endonucleases, BamHI and StuI, Tanaka and colleagues [28] identified two types of satellites in water buffalo by sequence analysis: one with ~ 1,400 bp tandem repeat unit and another with ~700 bp tandem repeat unit. The former shows 79% similarity to the bovine satellite I DNA, while the latter is 81% identical to the bovine satellite II DNA. The authors found that both satellite DNAs are localized to the centromeric regions of all chromosomes in either river or swamp type of buffaloes. Furthermore, the hybridization signals with the satellite I DNA on the acrocentric autosomes and X chromosome were much stronger than those on the biarmed autosomes and Y chromosome. However, the hybridization signals with buffalo satellite II DNA was almost the same over all the chromosomes, including the Y chromosome. Pathak and coworkers [29] further confirmed these two types of satellite sequences in the buffalo genome: the 1378- and 673-bp repeat fragments. Using real-time PCR analysis, the authors uncovered 1234 and 3420 copies of 1378- and 673-bp fragments per haploid genome, corresponding to 30 and 68 copies per chromosome, respectively. In addition, both 1378- and 673-bp repeat fragments are abundantly expressed in the spleen and liver.
There are a total of 361 minisatellite sequences deposited in the GenBank database for buffalo, which were mainly contributed by the National Institute of Immunology, India. Srivastava and colleagues [30] performed the minisatellite-associated sequence amplification with an oligo (5' CACCTCTCCACCTGCC 3'), that was designed based on consensus of 33.15 repeat loci using cDNA from the testis, ovary, spleen, kidney, heart, liver, and lung of water buffalo. The authors defined six different sizes of minisatellites with 1,263, 846/847, 602, 576, 487, and 324 bp, respectively. BLAST searches revealed that the 846/847-bp fragment has homology with the adenylate kinase gene, while the 1,263, 324, and 487-bp fragments show homology with the secreted modular calcium binding protein (SMOC-1), leucine-rich repeat neuronal 6A (LRRN6A) mRNA, and human TTTY5 mRNA, respectively. As for the microsatellites, there are a total of 311 submissions in the current GenBank database. Most of them were submitted by the Centre for Cellular and Molecular Biology, India (see AY775830 - AY775944, AY779565 - AY779623, AY787147 - AY787166, AY805331 - AY805389 and AY912133 - AY912182). These microsatellites have yet to be further mapped.
Next Generation Sequencing of Water Buffalo Genome
In the past, two general strategies have been widely used for whole genome sequencing: BAC by BAC sequencing and shotgun sequencing. Both strategies employ the Sanger method, which is relatively costly, time consuming, and labor intensive [31]. Therefore, the high demand for low-cost sequencing has led to the development of high-throughput sequencing technologies, called next-generation sequencing. As recently reviewed by Jiang et al. [32], three such next-generation sequencing technologies have been commercialized, such as Roche/454 life science (http://www.454.com), Illumina/Solexa (http://www.Illumina.com) and Applied Biosystem/SOLiD (http://solid.appliedbiosystems.com). These new generation sequencing methods no longer use the Sanger method for sequencing. Instead, the 454 technology is based on pyrosequencing and emulsion PCR; the Solexa technology utilizes a sequencing-by-synthesis approach for sequencing single DNA molecules attached to microspheres and the SOLiD (supported oligonucleotide ligation and detection) technology is a short-read sequencing method based on ligation. Nevertheless, these next-generation sequencing methods can produce a large amount of sequences in a relatively short time: for example, 500 Mb within 10 hours for Roche 454 GS FLX system, 1.5 Gb within 2.5 days for Illumina Genome Analyzer and 4 Gb within 6 days for Applied Biosystems SOLiD system [33].
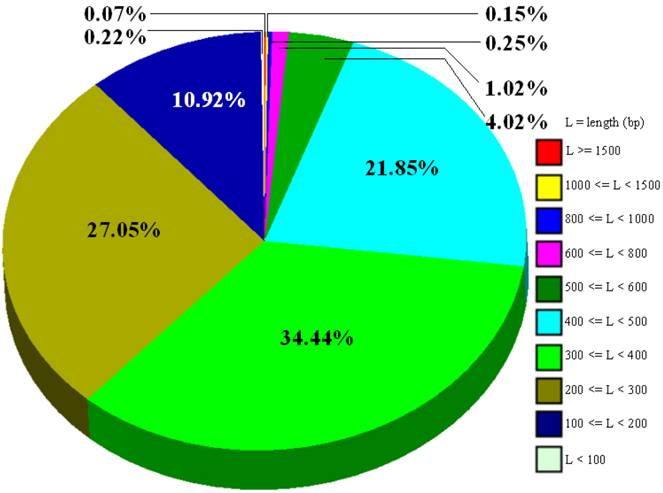
Using Roche 454 GS-FLX Titanium technology, a group of researchers at the Anand Agricultural University, India completed <1x genome sequencing of water buffalo in 2009 with a total of 64,212 sequences submitted to the GenBank database (ACZF01000001-ACZF01064212). The submission information from NCBI shows that a mature buffalo bull of Jaffrabadi breed was used to provide DNA materials for the project. The bull was tested free for bovine tuberculosis, brucellosis and Johne's disease. The submitted sequences range from 92 bp to 4,726 bp in size, but entries with 201 bp - 500 bp account for 83.3% (Figure 5). Overall, the 454 GS-FLX sequencing technologyhas contributed 21,675,247 bp in total to the species.
Also in 2009, Jayaraman [34] reported India's ambitious goal to sequence the complete water buffalo genome (http://news.boloji.com/2009/05/29982.htm), because the species represents the mainstay of the country's dairy industry, producing up to 55 percent of milk in addition to meat, hides and draught power. At that announcement, the Indian Council of Agricultural Research awarded the National Bureau of Animal Genetic Resources in Karnal, Haryana a substantial grant for their research. The Buffalo Genome Resources website at the National Center for Biotechnology Information recently confirmed that collaborations were established among the National Bureau of Animal Genetic Resources in Karnal, the Central Institute for Research on Buffaloes in Hisar, and the Animal Science Division of the Indian Council of Agricultural Research to sequence the buffalo genome. It is anticipated that >95% coverage of the buffalo genome will be sequenced by the end of 2010. In collaboration with the University of Florida, Washington State University will use the Illumina GAIIx technology to produce approximately 40 Gb of sequences for water buffalo during the summer of 2010.
Size distributions of 64,212 water buffalo whole genome shotgun sequences deposited in the GenBank Database.
Annotation of known gene sequences in water buffalo.
| Gene symbol | Description | N | ACCESSION |
|---|---|---|---|
| ABCG2 | ATP-binding cassette, sub-family G, member 2 | 2 | DQ272524 DQ205444 |
| ACTA2 | actin, alpha 2, smooth muscle, aorta | 1 | DQ846868 |
| ACTB | actin, beta | 4 | FJ468013 AF156978 DQ087263 DQ661647* |
| ACTC1 | actin, alpha, cardiac muscle 1 | 1 | DQ904371 |
| AKR1C4 | aldo-keto reductase family 1, member C4 | 1 | DQ884879 |
| AMELX | amelogenin (amelogenesis imperfecta 1, X-linked) | 2 | DQ469598 EF062996 |
| AMELY | amelogenin, Y-linked | 2 | DQ469599 EF062997 |
| ANGPT1 | angiopoietin 1 | 1 | AM237586 |
| ANGPT2 | angiopoietin 2 | 1 | AM237589 |
| ART4 | ADP-ribosyltransferase 4 (Dombrock blood group) | 1 | EF118387 |
| BMP15 | bone morphogenetic protein 15 | 3 | FJ613788 FJ613792 EF375880* |
| BOP1 | block of proliferation 1 | 1 | DQ082844 |
| BPI | bactericidal/permeability-increasing protein | 1 | AB234871 |
| BTN1A1 | butyrophilin, subfamily 1, member A1 | 7 | EU199797-8 AY491471-4 EU194868 |
| BULA-A** | MHC class I antigen-like | 20 | AY894407-10 AY188812-13 AY785758-61 AY925136 AY785759 DQ157373-81 |
| BULA-DIB** | MHC class II DI-beta antigen, Bubu-DIB*0101 allele | 1 | AY155224 |
| BULA-DQA** | MHC class II antigen, BuLA-DQA*01 allele | 7 | DQ822570-3 DQ868979-81 |
| BULA-DQA1** | MHC class II antigen | 3 | DQ116959 AY954685 DQ440647* |
| BULA-DQA2** | MHC class II antigen precursor | 1 | DQ440648* |
| BULA-DQB** | MHC class II antigen | 24 | DQ908903-4 EU025857-66 AY699876-87 |
| BULA-DRA** | MHC class II antigen, Bula-DRA*01 allele | 3 | AF385488-9 DQ016629* |
| BULA-DRB** | MHC class II antigen, Bula-DRB*01 allele | 8 | AF385473-80 |
| BULA-DRB1** | MHC class II antigen precursor | 1 | DQ146947* |
| BULA-DRB3** | MHC class II antigen | 28 | AF261955-6 AF270653-74 DQ057985* DQ187336-7 AY496063 |
| BULA-DYA** | MHC class II antigen | 3 | DQ188097* AY155211-2 |
| BYS1** | Y chromosome repeat sequence | 1 | X93551 |
| CAPN1 | calpain 1, (mu/I) large subunit | 1 | AB362998 |
| CAPN2 | calpain 2, (m/II) large subunit | 1 | AB362999 |
| CAST | calpastatin | 1 | AB363000 |
| CATHL@** | cathelicidin-like cluster | 8 | DQ661043* AJ812216 AY762972 DQ832665* EF050453* DQ832666 GQ396636 GQ354416 |
| CATSPER1 | cation channel, sperm associated 1 | 1 | GU372967 |
| CCL3 | chemokine (C-C motif) ligand 3 | 1 | EF562448* |
| CCNB2 | cyclin B2 | 1 | EF367720 |
| CCT8 | chaperonin containing TCP1, subunit 8 (theta) | 2 | EF121558 EF113594 |
| CD14 | CD14 molecule | 11 | DQ444324 EU370398-404 DQ457089* EU871785 GU368103 |
| CD2 | CD2 molecule | 1 | EF118474 |
| CD247 | CD247 molecule | 1 | DQ057984* |
| CGA | glycoprotein hormones, alpha polypeptide | 1 | AJ302670 |
| CHI3L1 | chitinase 3-like 1 (cartilage glycoprotein-39) | 1 | AY295929 |
| CLEC7A | C-type lectin domain family 7, member A | 1 | EF636900* |
| CP | ceruloplasmin (ferroxidase) | 1 | FN649762 |
| CS | citrate synthase | 1 | FJ468012 |
| CSF2 | colony stimulating factor 2 (granulocyte-macrophage) | 1 | AY553190* |
| CSN1S1 | casein alpha s1 | 13 | AF529305 FJ392261* EF025978-83 AY948385* DQ111783* AY514427 AJ005430 EF133464 |
| CSN1S2** | casein alpha-S2 | 15 | AJ005431 AY514428 DQ133467* DQ173244* EF025984-9 EF066480 FM865618-21 |
| CSN2 | casein beta | 18 | AY352050 DQ191170-2 Y17836-9 FM946182 FN424088 EF115306 AY599833 DQ317447* DQ631829* EF066481 AJ005165 AJ005432 FM986648 |
| CSN3 | casein kappa | 13 | AF164024 AM900443 EF133463 DQ191173-4 AY750857* DQ645429* EF066482 FJ770200 U96662 AJ011387 AJ628346 D14370^ |
| CSRP3 | cysteine and glycine-rich protein 3 | 1 | GQ231523 |
| CTLA4 | cytotoxic T-lymphocyte-associated protein 4 | 1 | FJ827143 |
| CXCR2 | chemokine (C-X-C motif) receptor 2 | 1 | AY864732 |
| CYM** | chymosin | 2 | EU265816* AF177290* |
| CYP11A1 | cytochrome P450, family 11, subfamily A, polypeptide 1 | 1 | AM237596 |
| CYP19A1 | cytochrome P450, family 19, subfamily A, polypeptide 1 | 13 | EF126034-6 EF178281 DQ407274 EU621845 FJ572657-9 AH015824 DQ489713 DQ489714 EU308111 |
| CYP1A1 | cytochrome P450, family 1, subfamily A, polypeptide 1 | 2 | FJ984598-9 |
| CYP21** | cytochrome P450, subfamily XXI | 1 | AF163768 |
| DAZ1 | deleted in azoospermia 1 | 1 | EU106872 |
| DDX3Y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | 5 | AY928814 AY928821 AY936546 AY936552 GQ259328 |
| DEFB@** | beta defensin cluster | 10 | GQ231526-9 EF418029* EF418030* EF418031 AY392452* AY301005* EF489402 |
| DGAT1 | diacylglycerol O-acyltransferase homolog 1 (mouse) | 9 | AY999090^ DQ886485^ DQ182702 FJ014704-6 DQ120929* DQ173243 AJ971291 |
| DHX36 | DEAH (Asp-Glu-Ala-His) box polypeptide 36 | 1 | EF088288 |
| EIF1AD | eukaryotic translation initiation factor 1A domain containing | 1 | FJ415608 |
| ERO1L | ERO1-like (S. cerevisiae) | 1 | EF088290 |
| ESR1 | estrogen receptor 1 | 6 | AM237582 EU662287-91 |
| ESR2 | estrogen receptor 2 (ER beta) | 1 | AM237583 |
| F11 | coagulation factor XI | 2 | DQ232885 DQ233653 |
| FABP3 | fatty acid binding protein 3, muscle and heart | 2 | GQ402490 AY758205* |
| FAM13A1** | family with sequence similarity 13, member A1 | 2 | DQ246517 DQ297675 |
| FBXL6 | F-box and leucine-rich repeat protein 6 | 2 | DQ097508 DQ082845 |
| FCGRT | Fc fragment of IgG, receptor, transporter, alpha | 1 | EU262263 |
| FGF1 | fibroblast growth factor 1 (acidic) | 1 | AM237590 |
| FGF2 | fibroblast growth factor 2 (basic) | 1 | AM237591 |
| FN1 | fibronectin 1 | 1 | AY862395 |
| FOXD3 | forkhead box D3 | 1 | DQ487024 |
| FSHB | follicle stimulating hormone, beta polypeptide | 11 | EU340285* AY449463^ DQ666186* DQ862125-7 EF650039-42 EF710660* |
| FSHR | follicle stimulating hormone receptor | 17 | EU148059 DQ785802* EU016216* EF650043-50 AM237585 DQ845245* EU662283-6 |
| FST | follistatin | 1 | EF585672* |
| G6PD | glucose-6-phosphate dehydrogenase | 3 | AJ56462-8 GU324292 |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase | 2 | GU324291 AY974798 |
| GATA4 | GATA binding protein 4 | 1 | DQ487031 |
| GCNT3 | glucosaminyl (N-acetyl) transferase 3, mucin type | 3 | AY847312-3^ AY847321^ |
| GDF9 | growth differentiation factor 9 | 3 | FJ529501* FJ529502 EF202171* |
| GGT1 | gamma-glutamyltransferase 1 | 1 | AM260200 |
| GH1 | growth hormone 1 | 16 | AJ000549 AF177288 EF417860 EF417870 EU344998-9 FJ754322 GU223914 AJ005116 AJ011533 AY940159 X72947 AJ011513-4 D16297 DQ307367 |
| GHR | growth hormone receptor | 29 | AY608917* AY053546-69 EF207441^ EU178741* AY739705 AY775296 |
| GHRH | growth hormone releasing hormone | 1 | DQ064594 |
| GHRHR | growth hormone releasing hormone receptor | 2 | EF600712^ EF600713* |
| GHRL | ghrelin/obestatin prepropeptide | 5 | GU071074-5 DQ118139 EF583468^ EU604028 |
| GJA1 | gap junction protein, alpha 1, 43kDa | 1 | GQ896281 |
| GNL3L | guanine nucleotide binding protein-like 3 like | 1 | GQ853551 |
| GNLY | granulysin | 1 | EF583467 |
| GNRH1 | gonadotropin-releasing hormone 1 | 1 | EU395713* |
| GNRH2 | gonadotropin-releasing hormone 2 | 1 | EU887534# |
| GNRHR | gonadotropin-releasing hormone receptor | 4 | AJ422122 AJ621497 DQ821403* EU621854* |
| GNRHR2 | gonadotropin-releasing hormone (type 2) receptor 2 | 1 | EU541504 |
| GOT1 | glutamic-oxaloacetic transaminase 1, soluble | 1 | AB437282* |
| GPAA1 | glycosylphosphatidylinositol anchor attachment protein 1 homolog (yeast) | 1 | DQ082846 |
| GPR33 | G protein-coupled receptor 33 (gene/pseudogene) | 2 | AY490667-8 |
| GYG1 | glycogenin 1 | 1 | DQ885595 |
| H2B | H2B histone | 1 | EF526308* |
| HAMP | hepcidin antimicrobial peptide | 1 | EU399814* |
| HAS2 | hyaluronan synthase 2 | 1 | DQ989300 |
| HBA1 | hemoglobin, alpha 1 | 4 | AJ242731-4 |
| HBB | hemoglobin, beta | 5 | AM886147-51 (AM886149#) |
| HERC3 | hect domain and RLD 3 | 3 | AH015380 DQ272525-6 |
| HEXA | hexosaminidase A (alpha polypeptide) | 1 | EF057418 |
| HK1 | hexokinase 1 | 1 | GU324294 |
| HP | haptoglobin | 1 | FN600415 |
| HPS3 | Hermansky-Pudlak syndrome 3 | 1 | DQ885601 |
| HSF1 | heat shock transcription factor 1 | 1 | DQ097507 |
| HSPA1A | heat shock 70kDa protein 1A | 2 | AJ812563 EU099315* |
| IFN@** | interferon cluster | 7 | AY323971-2 AY681350-1 EF503726 AY535404 AY665673 |
| IFNG | interferon, gamma | 5 | AB246350* AF484688* AY466119 EF424252* EF567075* |
| IGF1 | insulin-like growth factor 1 (somatomedin C) | 14 | Y10691 Y16248 Y18832 EU159114-6 FJ032308 AM237594 AY803777 GQ301206* GQ385227 AY743324-5 DQ085612 |
| IGF1R | insulin-like growth factor 1 receptor | 3 | FJ032309 Y12700 AM237595 |
| IGF2 | insulin-like growth factor 2 (somatomedin A) | 2 | FJ032306 DQ085613 |
| IGF2R | insulin-like growth factor 2 receptor | 1 | FJ032307 |
| IGFBP2 | insulin-like growth factor binding protein 2, 36kDa | 3 | AJ223171 Y16352 DQ079594 |
| IGFBP3 | insulin-like growth factor binding protein 3 | 5 | AJ223172 Y16351 AY338972 AY816186 AY304829 |
| IGFBP4 | insulin-like growth factor binding protein 4 | 2 | AJ223170 DQ079592 |
| IGFBP5 | insulin-like growth factor binding protein 5 | 2 | DQ079593 AF045566 |
| IGHG3 | immunoglobulin heavy constant gamma 3 | 1 | GQ140262 |
| IL10 | interleukin 10 | 3 | AB246351* AY325267* EF362779* |
| IL10RA | interleukin 10 receptor, alpha | 1 | AY864730 |
| IL12A | interleukin 12A | 4 | AB246352* EF424253* AY232819* DQ885596 |
| IL12B | interleukin 12B | 3 | AB246353 EF424254 AY198121 |
| IL13 | interleukin 13 | 2 | DQ083523* EF201845 |
| IL15 | interleukin 15 | 3 | AJ891036 DQ083522* EF201846 |
| IL18 | interleukin 18 | 5 | AJ891035 AY394479* EF424256* AY436506* DQ083521 |
| IL1A | interleukin 1, alpha | 2 | AB246786* AY514120* |
| IL1B | interleukin 1, beta | 2 | AB246787* AY514903* |
| IL2 | interleukin 2 | 7 | EF118562^ AB246354* EF397241 EF407852* DQ010150 AF255665 AF363786* |
| IL2RG | interleukin 2 receptor, gamma | 2 | AY864727 AY864729 |
| IL3 | interleukin 3 | 1 | AY553189* |
| IL4 | interleukin 4 | 5 | AJ891034 DQ083520 AY293620* AB246355* EF407850* |
| IL4R | interleukin 4 receptor | 1 | AY864728 |
| IL5 | interleukin 5 | 1 | EF407851* |
| IL6 | interleukin 6 | 3 | AB246788* AY347710* AY601639 |
| IL6R | interleukin 6 receptor | 1 | AY864731 |
| IL8 | interleukin 8 | 3 | FJ595833^ AY952930* AY862415 |
| INHA | inhibin, alpha | 1 | EU884446* |
| INSR | insulin receptor | 1 | Y14017 |
| ISG15 | ISG15 ubiquitin-like modifier | 3 | DQ118136-8^ |
| ITGB1 | integrin, beta 1 | 3 | AH015379 DQ251725-6 |
| ITGB2 | integrin, beta 2 | 11 | AY629299-302 AY737720-1 AY742803 AY821799 AY842449* EU853307 AY252121 |
| KCNMA1 | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | 1 | EU816373 |
| KDR | kinase insert domain receptor | 1 | AM237593 |
| KIT | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | 3 | FJ232938* AY829238 DQ314491* |
| KRT14 | keratin 14 | 1 | DQ846867 |
| LALBA | lactalbumin, alpha- | 20 | EF408609-10 AH014129-32 AY726609-17 EF408824 AF194373^ DQ785796* EF419240-1 |
| LAP | Laryngeal adductor paralysis | 4 | DQ458768* EF418028* EF630358* DQ886701 |
| LBP | lipopolysaccharide binding protein | 1 | FN645646 |
| LDHA | lactate dehydrogenase A | 1 | GU324295 |
| LEP | leptin | 33 | AF387813-4 AH013754^ AY177609 AY338973 AY427959 AY495586-7^ DQ490986 DQ497641 DQ676888-90 DQ831142-3 DQ837736-8 EU030441 EU078405 EU194869 EU199796 EU541447* EU662273-7 EU825672-4 EU888289 GQ385228 |
| LEPR | leptin receptor | 2 | AY177610-1 |
| LHB | luteinizing hormone beta polypeptide | 10 | AY765376* FJ357421* DQ489560^ EF113097-103 |
| LHCGR | luteinizing hormone/choriogonadotropin receptor | 5 | DQ858168-72 |
| LIF | leukemia inhibitory factor | 2 | EU926738 FJ907967 |
| LIPE | lipase, hormone-sensitive | 1 | AY900493 |
| LPO | lactoperoxidase | 1 | EF580919* |
| LTF | lactotransferrin | 25 | Y12775 EU192148 GQ231524 AF281089 DQ522302 DQ838746-51 EF443159 EF488002 EF650854 EU258554 EU263788 EU518482 EU532491 EU581859 EU598149 EU669579 EU683031 EU706286 EU719067 AJ005203 |
| LYZ | lysozyme | 10 | DQ480754-5* GQ497214-6 GQ995483-5 EF535848* AJ225012 |
| MAF1 | MAF1 homolog (S. cerevisiae) | 1 | DQ082847 |
| MAP2K1 | mitogen-activated protein kinase kinase 1 | 1 | EF057415 |
| MEGF11 | multiple EGF-like-domains 11 | 1 | EF057416 |
| MME | membrane metallo-endopeptidase | 1 | DQ885597 |
| MOGAT2 | monoacylglycerol O-acyltransferase 2 | 3 | EF208205 EU239373-4 |
| MPL | myeloproliferative leukemia virus oncogene | 1 | GQ411204 |
| MPO | myeloperoxidase | 1 | EU086095 |
| MSTN | myostatin | 13 | FJ752686-7^ AH014575^ AH013313^ AY254098^ AY363177-8 AY725214 AY854495-7 DQ091762^ DQ159987* |
| MX1 | myxovirus (influenza virus) resistance 1 | 1 | AB462524* |
| MX2 | myxovirus (influenza virus) resistance 2 | 1 | EF052266* |
| MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | 1 | GU296437 |
| MYF5 | myogenic factor 5 | 2 | EF197850^ EF128444* |
| MYF6 | myogenic factor 6 (herculin) | 1 | FJ039849* |
| MYH1 | myosin, heavy chain 1, skeletal muscle, adult | 1 | AB126014 |
| MYOD1 | myogenic differentiation 1 | 1 | FJ194946^ |
| MYOG | myogenin (myogenic factor 4) | 2 | EF636460^ FJ211420 |
| NANOG | Nanog homeobox | 1 | DQ487022 |
| NCAM1 | neural cell adhesion molecule 1 | 1 | AF025988 |
| NLRP5 | NLR family, pyrin domain containing 5 | 2 | AM748273 AM941717 |
| NOS2 | nitric oxide synthase 2, inducible | 2 | EF562453 EF655617 |
| NRAMP1 | natural resistance-associated macrophage protein 1 | 24 | DQ390205 DQ441408 DQ095780-1 DQ376109-10 DQ645386-9 AH015634 DQ658151* AH014155 AY702720 AY707989 AH014681-2 AY860618 AY860620-4 DQ390206 |
| OLR1 | oxidized low density lipoprotein receptor 1 | 53 | GQ415420-51 GQ478023-42 GQ385226 |
| ORM1 | orosomucoid 1 | 1 | FN645647 |
| OVGP1 | oviductal glycoprotein 1, 120kDa | 2 | EU382735* DQ482669* |
| OXT | oxytocin, prepropeptide | 2 | AM234538-9 |
| PAEP | progestagen-associated endometrial protein | 14 | AY775796-802 FN377869 DQ785797* AJ005429 DQ340204-5 AJ492506 AM238696 |
| PAG1 | phosphoprotein associated with glycosphingolipid microdomains 1 | 1 | EU815059* |
| PAG2** | pregnancy-associated glycoprotein 2 | 1 | GU433184 |
| PAPOLA | poly(A) polymerase alpha | 2 | FJ386493 GQ896282 |
| PAPPA | pregnancy-associated plasma protein A, pappalysin 1 | 1 | DQ079591 |
| PDCD1 | programmed cell death 1 | 2 | FJ827145* FJ827147* |
| PDHB | pyruvate dehydrogenase (lipoamide) beta | 1 | GU324296 |
| PFKM | phosphofructokinase, muscle | 1 | GU324297 |
| PGR | progesterone receptor | 6 | EU662278-82 AM237584 |
| PKD2 | polycystic kidney disease 2 | 1 | DQ230325 |
| PKM2 | pyruvate kinase, muscle | 1 | EF057417 |
| PLOD2 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | 1 | DQ418774 |
| PLS1 | plastin 1 | 1 | DQ418775 |
| PLZF | Promyelocytic leukemia zinc finger | 1 | EU195081 |
| POU1F1 | POU class 1 homeobox 1 | 2 | AY971145 GQ385224 |
| POU5F1 | POU class 5 homeobox 1 | 2 | DQ487023 EU926737 |
| PPARG | peroxisome proliferator-activated receptor gamma | 2 | GU066311 EU887290^ |
| PRDX1 | peroxiredoxin 1 | 1 | DQ082848 |
| PRKAG3 | protein kinase, AMP-activated, gamma 3 non-catalytic subunit | 2 | DQ646431 DQ525861 |
| PRL | prolactin | 4 | EF054878* EU352822 DQ287249 EU340420 |
| PRLR | prolactin receptor | 1 | GQ339914 |
| PRNP | prion protein | 6 | AY720689-90 AY768533-4 AY320365 AY769894 |
| PROS1 | protein S (alpha) | 1 | DQ885599 |
| PRSS7 | protease, serine, 7 (enterokinase) | 1 | DQ518426 |
| PTGES | prostaglandin E synthase | 1 | DQ167808 |
| PTGFR | prostaglandin F receptor (FP) | 2 | DQ100351* AY346134* |
| PTGS2 | prostaglandin-endoperoxide synthase 2 | 1 | EF028069 |
| PXMP4 | peroxisomal membrane protein 4, 24kDa | 1 | EU714054 |
| REXO1 | REX1, RNA exonuclease 1 homolog (S. cerevisiae) | 1 | FJ269033 |
| RN18S1 | RNA, 18S ribosomal 1 | 2 | GU324290 Y12516 |
| RN28S1 | RNA, 28S ribosomal 1 | 1 | AJ621496 |
| RNASE1 | ribonuclease, RNase A family, 1 (pancreatic) | 3 | AF259554 AJ011842-3 |
| RNF13 | ring finger protein 13 | 1 | DQ885600 |
| RPSA | ribosomal protein SA | 1 | GQ452383^# |
| SAA@ | serum amyloid A1 cluster | 1 | FN599527 |
| SCD | stearoyl-CoA desaturase (delta-9-desaturase) | 10 | FN395259 FM876222 AM600640 AH015836 DQ088625 DQ646700-1 GQ336861-2 EF122150 |
| SDHA | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 1 | GU324298 |
| SELT | selenoprotein T | 1 | DQ899756 |
| SERPINA1 | serpin peptidase inhibitor, clade A, member 1 | 1 | GQ385225 |
| SERPINA14** | serpin peptidase inhibitor, clade A, member 14 | 1 | DQ661648* |
| SFTPD | surfactant protein D | 1 | GQ231525 |
| SLAMF1 | signaling lymphocytic activation molecule family member 1 | 1 | DQ228868* |
| SLC11A1 | solute carrier family 11, member 1 | 5 | FJ827149* U27105* AY702721 DQ855416-7 |
| SLC11A2 | solute carrier family 11, member 2 | 1 | FJ827151* |
| SLC24A1 | solute carrier family 24, member 1 | 1 | EF121559 |
| SLC26A2 | solute carrier family 26, member 2 | 1 | AY350740^ |
| SLC2A1 | solute carrier family 2, member 1 | 3 | GU324293 FJ468009 AJ812564 |
| SLC5A4 | solute carrier family 5, member 4 | 1 | AF254423 |
| SLC5A5 | solute carrier family 5, member 5 | 1 | FJ468010 |
| SLC5A8 | solute carrier family 5, member 8 | 1 | FJ468011 |
| SLPI | secretory leukocyte peptidase inhibitor | 1 | EU700055* |
| SMOC1 | SPARC related modular calcium binding 1 | 4 | AY947405 EU370565* DQ159955* EF446167* |
| SOX2 | SRY (sex determining region Y)-box 2 | 4 | GQ451841* GQ853881^ DQ487021 EU661361 |
| SPP1 | secreted phosphoprotein 1 | 3 | DQ295062* DQ415660* DQ899755^ |
| SRY | sex determining region Y | 15 | GQ259332^ AY323859 DQ119747^ EF465091 EF465093-4 AY341337^ DQ417872^ EF465092 EF465095-6 EU016229^ DQ336535^ FJ546413-4^ |
| SST | somatostatin | 1 | GQ864014^ |
| STAR | steroidogenic acute regulatory protein | 1 | DQ062682 |
| STAT3 | signal transducer and activator of transcription 3 | 1 | DQ487026 |
| STAT5A | signal transducer and activator of transcription 5A | 2 | EU887291 EU030440 |
| TAP** | tracheal antimicrobial peptide | 2 | AB299981 AB299970 |
| TEK | TEK tyrosine kinase, endothelial | 1 | AM237587 |
| TFDP2 | transcription factor Dp-2 | 1 | DQ418776 |
| THPO | thrombopoietin | 1 | GQ411203 |
| TIE1 | tyrosine kinase with immunoglobulin-like and EGF-like domains 1 | 1 | AM237588 |
| TLR1 | toll-like receptor 1 | 2 | EU005235 GU451251^ |
| TLR10 | toll-like receptor 10 | 1 | EU005244 |
| TLR2 | toll-like receptor 2 | 5 | DQ131599 DQ288130* EU005236 EU178742^ GU441859 |
| TLR3 | toll-like receptor 3 | 3 | DQ508811* EU005237 FJ606788^ |
| TLR4 | toll-like receptor 4 | 10 | GQ851932 EF609240 EF562451-2 EU005238 GU002362 GU176310 DQ393127 EU386358^ DQ857349* |
| TLR5 | toll-like receptor 5 | 2 | EU005239 GQ866978^ |
| TLR6 | toll-like receptor 6 | 2 | EU005240 GQ866885 |
| TLR7 | toll-like receptor 7 | 3 | EU005241 GQ925364^ GU214059^ |
| TLR8 | toll-like receptor 8 | 2 | EU005242 GQ922103^ |
| TLR9 | toll-like receptor 9 | 3 | EU005243 EU747827 FJ606787^ |
| TNF | tumor necrosis factor | 5 | EF562449* EF562450* AB246789* EF424255* AY221123* |
| TNP1 | transition protein 1 | 1 | EU979632 |
| TNP2 | transition protein 2 | 3 | EU979633 EU999142-3 |
| TSHB | thyroid stimulating hormone, beta | 1 | FJ357420* |
| TSPY** | testis-specific protein | 4 | EU386184 EU386187-8 EU032586 |
| TUBB3 | tubulin, beta 3 | 1 | DQ487030 |
| TYROBP | TYRO protein tyrosine kinase binding protein | 1 | EF118738^ |
| U2AF1 | U2 small nuclear RNA auxiliary factor 1 | 1 | FJ415609 |
| UACA | uveal autoantigen with coiled-coil domains and ankyrin repeats | 1 | EF088289 |
| UBA52 | ubiquitin A-52 residue ribosomal protein fusion product 1 | 1 | AY842446* |
| USP25 | ubiquitin specific peptidase 25 | 1 | DQ418777 |
| UTY | ubiquitously transcribed tetratricopeptide repeat gene, Y-linked | 1 | AY936541 |
| VEGFA | vascular endothelial growth factor A | 1 | AM237592 |
| VIM | vimentin | 1 | DQ487027 |
| YWHAZ | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 1 | GU324299 |
| ZFX | zinc finger protein, X-linked | 2 | Y08942 X99827# |
| ZFY | zinc finger protein, Y-linked | 5 | DQ336545 DQ336555 AY928827 X99826 GQ259330 |
Notes:
* Entry is mRNA with complete coding sequence for the gene;
^ Entry is genomic DNA with complete coding sequence for the gene;
# Entry is a pseudogene;
**Specific gene symbols are used in water buffalo;
@Entry belongs a gene cluster.
5. Conclusions
- As described above, the water buffalo community has been working hard to generate genome resources in terms of cytogenetic characterization, whole genome mapping and whole genome sequencing. Certainly, genome science comes of age in this important animal species.
- The vast amount of genetic/genomic resources for cattle research has served as shortcuts for the water buffalo community to initiate genome science in the species. As bovine genome resources have advanced significantly during the last thirty years, so has that of water buffalo.
- Completion of the studies reported herein will help develop essential genome technologies for breeders and farmers to improve reproductive performance and production potential in water buffalo.
Acknowledgements
This work was supported by USDA/FAS grant BIO12-001-009 to Z.J.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Kierstein G. et al. Analysis of mitochondrial D-loop region casts new light on domestic water buffalo (Bubalus bubalis) phylogeny. Mol Phylogenet Evol. 2004;30(2):308-24
2. Bubalus bubalis 2004. Roth J and Myers P. http://animaldiversity.ummz.umich.edu/site/accounts/information/Bubalus_bubalis.html
3. Yindee M. et al. Y-chromosomal variation confirms independent domestications of swamp and river buffalo. Anim Genet. 2010 [Epub ahead of print]
4. FAO. Water Buffalo: an asset undervalued, F.R.O.f.A.a. Bangkok, Thailand: Pacific Editor. 2000:1-6
5. Iannuzzi L, Di Meo G. Water Buffalo. In: (ed.) Cockett NE, Kole C. Genome Mapping and Genomics in Domestic Animals. Berlin Herdelberg, Germany: Springer-Verlag. 2009
6. Council N.R. The water buffalo: new prospects for an underutilized animal. Washington DC: National Academy Press. 1981
7. Cockrill W.R. The water buffalo: a review. Br Vet J. 1981;137(1):8-16
8. Chen Y.C, Li X.H. New evidence of the origin and domestication of the Chinese swamp buffalo (Bubalus bubalis). Buffalo Journal. 1989;1:51-55
9. Kumar S. et al. Phylogeography and domestication of Indian river buffalo. BMC Evol Biol. 2007;7:186
10. Kumar S. et al. Mitochondrial DNA analyses of Indian water buffalo support a distinct genetic origin of river and swamp buffalo. Anim Genet. 2007;38(3):227-32
11. Buchholz C. Cattle. In: (ed.) Parker S.P. Grzimek's Encyclopedia of Mammals. New York: McGraw-Hill Publishing Company. 1990:360-417
12. Amaral M.E. et al. A first generation whole genome RH map of the river buffalo with comparison to domestic cattle. Genomics BMC. 2008;9:631
13. Gallagher DSJr, Womack JE. Chromosome conservation in the Bovidae. Hered J. 1992;83(4):287-98
14. Iannuzzi L. et al. Comparative FISH mapping in river buffalo and sheep chromosomes: assignment of forty autosomal type I loci from sixteen human chromosomes. Cytogenet Cell Genet. 2001;94(1-2):43-8
15. Iannuzzi L, King W.A, Di Berardino D. Chromosome evolution in domestic bovids as revealed by chromosome banding and FISH-mapping techniques. Cytogenet Genome Res. 2009;126(1-2):49-62
16. Gallagher DSJr. et al. Applications of chromosomal fish in the Bovidae with emphases on physical mapping in domestic cattle and comparative cytogenetic analyses of the tribe Bovini. Anim Biotechnol. 1999;10(3):105-8
17. Barker J.S. et al. Genetic diversity of Asian water buffalo (Bubalus bubalis): microsatellite variation and a comparison with protein-coding loci. Anim Genet. 1997;28(2):103-15
18. Ritz L.R. et al. Phylogenetic analysis of the tribe Bovini using microsatellites. Anim Genet. 2000;31(3):178-85
19. Kumar S. et al. Genetic variation and relationships among eight Indian riverine buffalo breeds. Mol Ecol. 2006;15(3):593-600
20. Lei C.Z. et al. Independent maternal origin of Chinese swamp buffalo (Bubalus bubalis). Anim Genet. 2007;38(2):97-102
21. Di Meo G.P. et al. An extended river buffalo (Bubalus bubalis, 2n = 50) cytogenetic map: assignment of 68 autosomal loci by FISH-mapping and R-banding and comparison with human chromosomes. Chromosome Res. 2008;16(6):827-37
22. Amaral M.E. et al. Construction of a river buffalo (Bubalus bubalis) whole-genome radiation hybrid panel and preliminary RH mapping of chromosomes 3 and 10. Anim Genet. 2007;38(3):311-4
23. Goldammer T. et al. A radiation hybrid map of river buffalo (Bubalus bubalis) chromosome 7 and comparative mapping to the cattle and human genomes. Cytogenet Genome Res. 2007;119(3-4):235-41
24. Ianella P. et al. First radiation hybrid map of the river buffalo X chromosome (BBUX) and comparison with BTAX. Anim Genet. 2008;39(2):196-200
25. Miziara M.N. et al. A radiation hybrid map of river buffalo (Bubalus bubalis) chromosome 1 (BBU1). Cytogenet Genome Res. 2007;119(1-2):100-4
26. Stafuzza N.B. et al. Preliminary radiation hybrid map for river buffalo chromosome 6 and comparison to bovine chromosome 3. Anim Genet. 2007;38(4):406-9
27. Stafuzza N.B. et al. Comparative RH maps of the river buffalo and bovine Y chromosomes. Cytogenet Genome Res. 2009;126(1-2):132-8
28. Tanaka K. et al. Characterization and chromosomal distribution of satellite DNA sequences of the water buffalo (Bubalus bubalis). Hered J. 1999;90(3):418-22
29. Pathak D. et al. Chromosomal localization, copy number assessment, and transcriptional status of BamHI repeat fractions in water buffalo Bubalus bubalis. DNA Cell Biol. 2006;25(4):206-14
30. Srivastava J. et al. Transcriptional status of known and novel genes tagged with consensus of 33.15 repeat loci employing minisatellite-associated sequence amplification (MASA) and real-time PCR in water buffalo, Bubalus bubalis. DNA Cell Biol. 2006;25(1):31-48
31. Metzker M.L. Emerging technologies in DNA sequencing. Genome Res. 2005;15(12):1767-76
32. Jiang Z, Rokhsar D.S, Harland R.M. Old can be new again: HAPPY whole genome sequencing, mapping and assembly. Int J Biol Sci. 2009;5(4):298-303
33. Voelkerding K.V, Dames S.A, Durtschi J.D. Next-generation sequencing: from basic research to diagnostics. Clin Chem. 2009;55(4):641-58
34. Jayaraman K.S. Indian Buffalo at Crossroads of Science. Boloji.com. 2009
Author contact
![]() Corresponding author: Dr. Zhihua Jiang, Department of Animal Sciences, Washington State University, Pullman, WA 99164 - 6351. Tel: +509 335 8761; Fax: +509 335 4246; E-mail: jiangzedu
Corresponding author: Dr. Zhihua Jiang, Department of Animal Sciences, Washington State University, Pullman, WA 99164 - 6351. Tel: +509 335 8761; Fax: +509 335 4246; E-mail: jiangzedu

 Global reach, higher impact
Global reach, higher impact