10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2010; 6(5):491-498. doi:10.7150/ijbs.6.491 This issue Cite
Research Paper
Identification and Molecular Characterization of a New Member of the Peritrophic Membrane Proteins from the Meadow Moth, Loxostege Sticticalis
1. State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, 100193, China;
2. College of Life Sciences, Agricultural University of Hebei, Baoding, Hebei 071001, China;
3. Department of Biotechnology, Hefei University of Technology, Hefei 230009, China.
Received 2010-6-19; Accepted 2010-8-27; Published 2010-9-6
Abstract
The peritrophic membrane (PM) plays an important role in protecting insects. The PM proteins are important to determinate the formation and function of the PM. A new PM protein, named Lsti99, was identified from the PM of Loxostege sticticalis larvae by cDNA library screening. The full cDNA of Lsti99 is 1392 bp in length, contains an open reading frame (ORF) of 1245 bp that encodes a preprotein of 415 amino acid residues with a 17-amino acid signal peptide. The sequence of Lsti99 showed no homology to other known PM proteins. The recombinant Lsti99 was successfully expressed in insect cells (Sf9) using recombinant baculoviruses and was used to isolate the antibodies to Lsti99 from the polyclonal antiserum. Lsti99 was expressed mainly in the PM, but weaker bands could be detected in the head and integument as well. The Lsti99 protein could be separated from the PM complex by chitinase in vitro, but M2R did not show effect in vitro confirming the chitin-binding activity of Lsti99. The biochemical and physiological functions of Lsti99 in L. sticticalis require further investigation.
Keywords: Peritrophic membrane protein, Chitin-binding activity, Recombinant protein, Loxostege sticticalis, Lsti99.
Introduction
In insects, the midgut is the main organ for digestion and nutrient absorption. In some insects the peritrophic membrane (PM) encircles the gut as a layer and is believed to protect insects from invasion by viruses, bacteria, protozoa, helminthes, and prevents damage to midgut cells by abrasive food particles [1-3]. The PM also appears to protect the midgut epithelium from toxic heme derivatives in mosquitoes [4] associated with digestion of blood meals, and the assimilation and elimination of toxic ammonium ions [5]. The PM is a potential target for insect control [6-7]. Insect PM is mainly composed of proteins and chitin. Four classes of PM proteins have been suggested based on the solubility of those proteins under different extraction conditions [2].
Using molecular methods, 20 PM proteins or putative PM proteins have been identified from several insects, including Lucilia cuprina [8-11], Trichoplusia ni [12-13], Anopheles gambiae [14-15], Chrysomya bezziana [10, 16], Glossina morsitans morsitans [17], Plutella xylostella [18], Mamestra configurata [19], Spodoptera. exigua and Helicoverpa armigera [20]. These proteins have chitin-binding activities as conferred by their characteristic chitin-binding domains or the peritrophin domains [2]. These domains contain 60-75 amino acid residues and are characterized by a conserved registery of cysteine residues and a number of aromatic amino acid residues [2]. It has been suggested that the conserved cysteine residues form intra-domain disulfide bonds contribute to protein stability in the protease-rich gut environment. In the case of TnPM-P42, a chitin deacetylase-like domain is reported instead of a peritrophin domain [2, 12-14]. Identification and characterization of PM from a wide variety of insects will help to develop pest management targets as well as a better understanding of the PM function and development.
In present study, we identified a new gene encoding a PM protein from Loxostege sticticalis larvae by cDNA library screening, which was named as Lsti99. The protein sequence features are different from known PM proteins, but can be liberated from the PM after chitinase treatment.
Materials and methods
Insect
A laboratory colony of L. sticticalis established by our laboratory was used in the present research. The larvae of L. sticticalis were maintained at 22±1 °C, 70-80% RH and L:D 16:8 photoperiod on LB Agar [21]. Tissues for analysis were isolated from fifth instar larval and were stored at -70°C until use.
Cloning and sequencing of the PM protein Lsti99 from L. sticticalis
A cDNA expression library of L. sticticalis midgut was screened by subtractive immunoscreening using antibodies made against a collection of Spodoptera exigua midgut PM proteins [20]. The screening procedure was as described by Wang et al (2004) [22-23]. The clones that positive to the antibodies of PM chitin-binding proteins were selected as candidate clones coding for unidentified PM proteins. These clones were cut and inserted into plasmids using Uni-ZAP XR Gigapack Clonging Kits (Stratagene, La Jolla, CA). The positive clones were sequenced using the dideoxynucleotide chain termination method (Takara Co., Dalian, China).
Recombinant Lsti99 expression
A recombinant baculovirus was constructed using the pENTRTM TOPO vector and BaculoDirectTM Linear DNA system (Invitrogen, USA) to over-express Lsti99 in insect cells. Two primers (99-ZFP: 5′-CACCGAGAACTATATTCGCTTCTTAT-3′ and 99-ZRP: 5′-TGGGATCCATTTTCAGTATC-3′) were designed to amplify the Lsti99ORF. The PCR conditions were as follows: after 5 min 94 °C, 30 cycles of 30 sec at 94 °C, 30 sec at 56 °C, and 90 sec at 72 °C, then 10 min at 72 °C. The expected PCR products were excised from the agarose gel and purified using a DNA gel extraction kit (Takara, Japan). Then, the obtained PCR products were cloned into pENTRTM TOPO vector. The LR recombination reaction was performed to transfer the Lsti99 gene into the BaculoDirectTM linear DNA following the protocols provided by the manufacturer. Recombinant Lsti99 was produced as a secreted protein by infecting Sf9 cells with the recombinant baculovirus and the infected culture was maintained in TNM-FH medium supplemented with 10% fetal bovine serum (FBS). The cell culture medium containing the secreted recombinant Lsti99 was collected at 72 h post-infection [24].
Preparation of antibodies reacting to Lsti99
Antibody reacting to the Lsti99 was prepared from an antiserum made against the whole collection of S. exigua midgut PM proteins. The recombinant Lsti99 protein was immobilized onto a piece of supported nitrocellulose membrane (Optitran BA-S85, Schleicher & Schuell, Keene, NH, USA) by incubation of the membrane with the recombinant protein at room temperature for 1 h, followed by extensive washing (5 times) with 0.1 M phosphate-buffered saline (PBS, pH7.4) and incubation in 3% bovine serum albumin (BSA) for 3 h. The nitrocellulose membrane was then incubated with a 100-fold dilution of the S. exigua PM proteins polyclonal antiserum in PBS (0.1M, pH7.4) with 3% BSA at room temperature for 3 h or at 4°C overnight to allow the antibodies in the antiserum to bind to the blotted membrane. Then, the membrane was thoroughly washed five times with 0.1 M PBS (pH7.4) and finally the bound antibodies specific to the Lsti99 were eluted from the membrane by incubation in 5ml of 0.1M glycine buffer (pH 2.5) at room temperature for 10 min, followed by addition of 0.5 ml of 1M Tris-HCl buffer (pH 8.0) to neutralize the pH of the antibody preparation [25].
Treatment of M2R and chitinase to L. sticticalis PM protein Lsti99
The dissected PM were treated with Calcofluor White M2R (M2R, Sigma, Beijing, China) or chitinase (Sigma, Beijing, China). 1.5 g (wet weight) of L. sticticalis PMs were incubated with 2.5 ml of 1% M2R (Sigma, Beijing, China) for 1.5 h at 37°C, followed by centrifugation at 1000 g for 5 min. The supernatant and the remaining PM pellet were stored for further protein extraction. The treatment of L. sticticalis PMs with 0.05 M chitinase (Sigma, Beijing, China) was same as for M2R. The supernatant after M2R or chitinase were directly dried and stored for later use. The pellet were homogenized in 0.1M PBS (pH7.4) and centrifuged at 12,000g for 20 min at 4°C. The supernatants were dried and stored at -70 °C for western blot analysis [26].
Western blot analysis
Proteins for use in Western blotting were extracted from tissues by homogenization in 0.1 M PBS (pH7.4) and centrifugation at 12,000g for 20 min at 4°C. The supernatants were dried and stored at -70 °C. Protein extracts mixed with SDS-PAGE sample buffer were boiled for 10 min, and immediately loaded on the 12% SDS-PAGE gel. After SDS-PAGE, the proteins were blotted on a PVDF membrane (Hybond-P, Amersham), and the membrane was then incubated with Lsti99 antibodies for 2 h at 37 °C. After washing in PBS-Tween (PBST, 0.05% Tween-20 in PBS, pH 7.2), the membrane was incubated in secondary antibodies (HRP-conjugated goat anti-rabbit IgG, dilution 1/2000) for 2 h at 37 °C, and thoroughly washed in PBST (pH7.2). The binding was detected using a DAB stock stain kit (Sino-American Biotechnology Co., China) [27].
Result
Identification and sequence of the cDNA coding for PM protein of L. sticticalis, Lsti99
By screening of the cDNA expression library of the L. sticticalis midgut, 282 positive cDNA clones were obtained and sequenced. Homologous sequences were then identified in the GenBank database using blastp and tblastn programs [28]. The number 99 clone, named Lsti99, showed no similar homologous sequence in GenBank and may encode a previous unidentified PM protein.
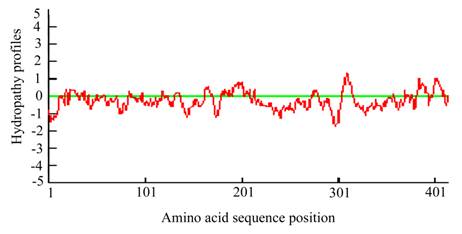
The full cDNA of Lsti99 was 1392 bp, containing an ORF of 1245 bp, followed by an AT-rich untranslated region and a putative polyadenylation signal (AAATAA) located 49 bp upstream of the polyA tail (Fig. 1). The deduced protein sequence showed that Lsti99 being synthesized as a preprotein of 415 amino acid residues with a 17-amino acid signal peptide as predicted by the software SignalP [29]. The predicted molecular weight of mature Lsti99 protein was 45.06 kDa. Prediction of potential O-glycosylation sites using the NetOglyc 3.1 server [30] showed that the proteins had only two potential O-glycosylation sites (Fig. 1) with threonine at positions 200 and 410. Prosite protein pattern search predicted five potential N-glycosylation sites (Fig. 1) [31]. Interestingly, the distribution patterns of these characters did not resemble any of the peritrophin domains from either type I or II PM [2]. The hydropathic nature of Lsti99 was calculated and plotted for each residue in the sequence revealing that most of the residue regions were highly hydrophilic (Fig. 2).
Recombinant protein of Lsti99 and prepare the antibody
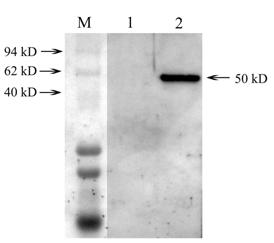
The recombinant Lsti99 were successfully expressed in insect cells (Sf9) as secreted proteins using baculoviruses system (Fig. 3). The apparent molecular weight of the recombinant Lsti99 was ~50 kDa, more than the predicted 45.06 kDa. This can be attributed to the modification to the recombinant protein in the eukaryotic expression system. We isolated the antibodies to the Lsti99 from the polyclonal antiserum using the Lsti99 recombinant protein, according to the methods of Wang and Granados (2003) [25].
Localization of Lsti99 in L. sticticals larvae
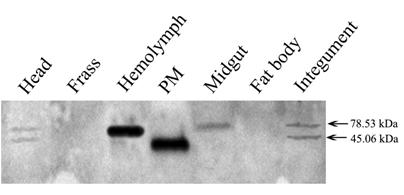
The tissue distribution of Lsti99 in L. sticticals larvae was detected by western blot using the antibody to the recombinant protein. The expected band of ~45.06 kDa was strongest in the PM, but weak band were detected in head and integument also. No bands were found in the hemolymph, midgut, fat body or frass (Fig. 4). Interestingly, there were unexpected clear bands that were co-detected in the hemolymph and midgut, head and integument (Fig. 4).
In vitro effect of M2R and chitinase on Listi99 protein
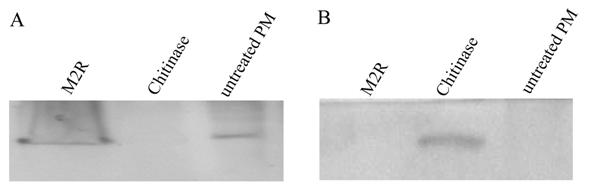
The PM were dissected from larvae and treated with M2R or chitinase. The western blot analysis revealed no band in the PM after the chitinase treatment, but a distinct band in the soluble solutions (Fig. 5). This indicated that the chitinase can dissolve the Lsti99 protein from the PM structure in vitro. When the dissected PMs were treated with M2R or no treatment, the clear band can be detected in PM, not in the soluble solutions (Fig. 5), indicating that M2R has no effect on the Lsti99 in vitro.
Nucleotide sequence and deduced amino acid sequence of the Lsti99 cDNA in L. sticticalis. The suggested start codon ATG and stop codon TAG are indicated in boxes. A putative polyadenylation signal is shaded with grey. The predicted signal peptide is underlined. Arrows under the nucleotide sequences represent the position of the different synthetic primers used in recombinant expression. The N-glycosylation sites in Lsti99 were predicted using Expert Protein Analysis System, and shaded in black. The O-glycosylation sites in Lsti99 were predicted using the NetOglyc 3.1 server and showed with dotted line. The nucleotide sequence reported here is deposited in the GenBank database under the accession number FJ798745.

The predicted hydropathy profiles of the deduced amino acid sequences of the Lsti99. The hydropathy profiles was predicted using DNAman 6.3.0.99.
Western blotting analysis of expressed Lsti99 in Sf9 cells. M, represent the protein marker; lane 1, Western-blotting of the normal Sf9; lane 2, Western-blotting of the Sf9 being infected by recombinant virus.
Tissue distribution of the Lsti99 based on the western blot. 20 µg of protein samples extracted from several different laval tissues were loaded on each lane for immunoblot analysis.
Western blot analysis of the PM proteins treated with M2R and chitinase. A, the proteins from the pellet after treatment were used for analysis; B, the soluble solutions were used for analysis.
Discussion
To date, about 20 PM proteins have been identified and characterized, which posses invariably chitin-binding activities as conferred by their characteristic peritrophin domains [2]. The peritrophin domains contain 60-75 amino acid residues and are characterized by a conserved register of cysteine residues and a number of aromatic amino acid residues [32]. In this study, we identified a novel PM chitin-binding protein from L. sticiticals, Lsti99, distinctly different from the peritrophin-type chitin-binding proteins. Lsti99 lacks the peritrophin domain, but could be liberated from the PM after chitinase treatment (Fig. 5). The mature protein of Lsti99 was predicted to be 45.06 kDa. The baculovirus system was used to express the recombinant Lsti99 protein and the expressed protein was measured to be 50 kDa. The higher molecular weight can be attributed to the post-translational modifications in the eukaryotic expression system.
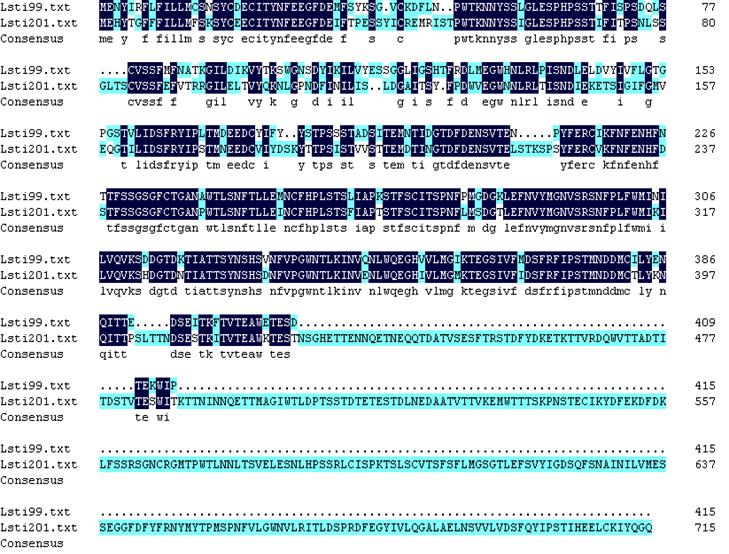
The antibodies to Lsti99 were used to characterize the tissue localization of the protein. The Lsti99 was mainly expressed in the PM, and also could be detected in the head and integument. The expression showed some differences to that of TnPM-P42 protein, another PM protein without peritrophin domains in Trichoplusia ni [23]. The TnPM-P42 protein was only expressed in the PM of midgut, not in hemolymph, malpighian tubules, salivary glands, fat body, or integument [23]. It is interesting that another bigger band also appeared in the head, hemolymph and integument (Fig. 4), which can be attributed to the cross activity of the Lsti99 antibodies to other proteins. Recently, another clone, nameed Lsti201 (GenBank FJ798746) from the L. sticiticals midgut cDNA library of was sequenced and compared with Lsti99. The ORF of Lsti201 encodes a preprotein of 715aa, which is longer than Lsti99 (415aa). The two genes showed higher than 70% similarity at the N terminal and middle regions (Fig. S1). The antibody to Lsti99 showed cross activity to the recombinant protein of Lsti201 with E.coli expression system. However, the Lsti201 did not express in the PM (Fig 4), and showed no chitin binding activities.
Alignment of the amino acid sequences of Lsti99 and Lsti201. The conserved amino acids were shaded. Lsti99, FJ798745; Lsti201 FJ798746.
When the dissected PMs were treated with chitinase, no band was detected after western blot, but the soluble solutions showed a distinct band. When incubated with M2R, the Lsti99 band showed no difference to the control, which is in accordance to the results of M2R to S. exigua in vitro [26]. The above result indicated that the Lsti99 protein is one of the PM proteins, which can bind to chitin and be separated from the complex by chitinase. However, the biochemical and physiological functions of Lsti99 need to be further elucidated.
Acknowledgements
We appreciate helpful comments provided by the anonymous reviewers. We thank to Dr. Carl W. Doud and Dr. Murugan (Department of Entomology, Kansas State University) for helping revise of the manuscript. This work was supported by National Program on Key Basic Research Projects (“973” program, 2006CB102007), the National Key Technologies R&D Program the People's Republic of China. (2006BADA01, 2006BADA05), and the Program for New Century Excellent Talents in University (NCET-07-0251).
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Lehane MJ. Peritrophic matrix structure and function. Annu Rev Entomol. 1997;42:525-50
2. Tellam RL, Wijffels G, Willadsen P. Peritrophic membrane proteins. Insect Biochem Mol Biol. 1999;29:87-101
3. Bolognesi R, Ribeiro AF, Terra WR. et al. The peritrophic membrane of Spodoptera frugiperda: secretion of peritrophins and role in immobilization and recycling digestive enzymes. Arch Insect Biochem Physiol. 2001;47:62-75
4. Pascoa V, Oliveira PL, Dansa-Petretski M. et al. Aedes aegypti peritrophic matrix and its interaction with heme during blood digestion. Insect Biochem Mol Biol. 2002;32:517-23
5. Kato N, Dasgupta R, Smartt C. et al. Glucosamine: fructose-6-phosphate aminotransferase: gene characterization, chitin biosynthesis and peritrophic matrix formation in Aedes aegypti. Insect Mol Biol. 2002;11:207-16
6. Casu R, Eisemann C, Pearson R. et al. Antibody-mediated inhibition of the growth of larvae from an insect causing sutaneous myiasis in a mammalian host. Proc Natl Acad Sci USA. 1997;94:8939-44
7. Shao L, Devenport M, Fujioka H. et al. Identification and characterization of a novel peritrophic matrix protein, Ae-Aper50, and the microvillar membrane protein, AEG12, from the mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2005;35:947-59
8. Elvin C, Vuocolo T, Pearson R. et al. Characterization of a major peritrophic membrane protein, peritrophin-44, from the larvae of Lucilia cuprina: cDNA and deduced amino acid sequences. J Biol Chem. 1996;271:8925-35
9. Schorderet S, Pearson RD, Vuocolo T. et al. cDNA and deduced amino acid sequences of a peritrophic membrane glycoprotein, 'peritrophin-48', from the larvae of Lucilia cuprina. Insect Biochem Mol Biol. 1998;28:99-111
10. Wijffels G, Eisemann C, Riding G. et al. A novel family of chitin-binding proteins from insect type 2 peritrophic matrix. cDNA sequences, chitin binding activity, and cellular localization. J Biol Chem. 2001;276:15527-36
11. Tellam RL, Vuocolo T, Eisemann C. et al. Identification of an immunoprotective mucin-like protein, peritrophin-55, from the peritrophic matrix of Lucilia cuprina larvae. Insect Biochem Mol Biol. 2003;33:239-52
12. Wang P, Granados RR. Molecular cloning and sequencing of a novel invertebrate intestinal mucin cDNA. J Biol Chem. 1997;272:16663-9
13. Wang P, Granados RR. Molecular structure of the peritrophic membrane (PM): identification of potential PM target sites for insect control. Arch Insect Biochem Physiol. 2001;47:110-8
14. Shen Z, Jacobs-Lorena M. A type I peritrophic matrix protein from the malaria vector Anopheles gambiae binds to chitin. Cloning, expression and characterization. J Biol Chem. 1998;273:17665-70
15. Devenport M, Fujioka H, Donnelly M. et al. Storage and secretion of Ag-Aper14, a novel peritrophic matrix protein, and Ag-Muc1 from the mosquito Anopheles gambiae. Cell Tissue Res. 2005;320:175-85
16. Vuocolo T, Eisemann CH, Pearson RD. et al. Identification and molecular characterization of a peritrophin gene, peritrophin-48, from the myiasis fly Chrysomya bezziana. Insect Biochem Mol Biol. 2001;31:919-32
17. Hao Z, Aksoy S. Proventriculus-specific cDNAs characterized from the tsetse, Glossina morsitans morsitans. Insect Biochem Mol Biol. 2002;32:1663-71
18. Sarauer BL, Gillott C, Hegedust D. Characterization of an intestinal mucin from the peritrophic matrix of the diamondback moth, Plutella xylostella. Insect Mol Biol. 2003;12:333-43
19. Shi X, Chamankhah M, Visal-Shah S. et al. Modeling the structure of the type I peritrophic matrix: characterization of a Mamestra configurata intestinal mucin and a novel peritrophin containing 19 chitin binding domains. Insect Biochem Mol Biol. 2004;34:1101-15
20. Zhang X, Li J, Guo W. et al. A Method of rapid and efficient isolation of highly specific antibodies from an antiserum against a pool of proteins. Acta Agri boreali-sinica. 2008;23:111-3
21. Jiang XF, Huang SH, Luo LZ. et al. Diapause termination, post-diapause development and reproduction in the beet webworm, Loxostege sticticalis (Lepidoptera: Pyralidae). J Insect Physiol. 2010;56:1325-31
22. Wang P, Li G, Granados RR. Identification of two new peritrophic membrane proteins from larval Trichoplusia ni: structural characteristics and their functions in the protease rich insect gut. Insect Biochem Mol Biol. 2004;34:215-27
23. Guo W, Li G, Pang Y. et al. A novel chitin-binding protein identified from the peritrophic membrane of the cabbage looper, Trichoplusia ni. Insect Biochem Mol Biol. 2005;35:1224-34
24. Zhang Y, Hu J, Miao Y. et al. Expression of EGFP-spider dragline silk fusion protein in BmN cells and larvae of silkworm showed the solubility is primary limit for dragline proteins yield. Mol Biol Rep. 2008;35:329-35
25. Wang P, Granados RR. Rapid and efficient isolation of highly specific antibodies from an antiserum against a pool of proteins. Biotech Histochem. 2003;78:201-5
26. Wang P, Granados RR. Calcofluor disrupts the midgut defense system in insects. Insect Biochem Mol Biol. 2000;30:135-43
27. Wei ZJ, Zhang QR, Kang L. et al. Molecular characterization and expression of prothoracicotropic hormone during development and pupal diapause in the cotton bollworm, Helicoverpa armigera. J Insect Physiol. 2005;51:691-700
28. Altschul SF, Gish W, Miller W. et al. Basic local alignment search tool. J Mol Biol. 1990;215:403-10
29. Bendtsen JD, Nielsen H, von Heijne G. et al. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783-95
30. Hansen JE, Lund O, Tolstrup N. et al. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconjugate J. 1998;15:115-30
31. Falquet L, Pagni M, Bucher P. et al. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235-8
32. Wang P, Granados RR. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc Natl Acad Sci USA. 1997;94:6977-82
Author contact
![]() Corresponding author: Ya-Zhong Cao, State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, 100193, People's Republic of China; Wei Guo, College of Life Sciences, Agricultural University of Hebei, Baoding, Hebei 071001, People's Republic of China. Tel: 86-010-62815619. Fax: 86-01062815619. E-mail: ajiaozicom; jyincn.
Corresponding author: Ya-Zhong Cao, State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, 100193, People's Republic of China; Wei Guo, College of Life Sciences, Agricultural University of Hebei, Baoding, Hebei 071001, People's Republic of China. Tel: 86-010-62815619. Fax: 86-01062815619. E-mail: ajiaozicom; jyincn.

 Global reach, higher impact
Global reach, higher impact