10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2010; 6(5):513-524. doi:10.7150/ijbs.6.513 This issue Cite
Research Paper
BP1, an Isoform of DLX4 Homeoprotein, Negatively Regulates BRCA1 in Sporadic Breast Cancer
1. Department of Medicine, Division of Genomic Medicine, George Washington University Medical Center, Washington, DC, USA
2. Center for Vaccine Development, University of Maryland, Baltimore, MD, USA
3. Department of Anesthesiology, George Washington University Medical Center, Washington, DC, USA
4. Armed Forces Institute of Pathology, Washington, DC, USA
5. Department of Medicine, Division of Hematology and Oncology, George Washington University Medical Center, Washington, DC, USA
6. Department of Biochemistry & Molecular Biology, George Washington University Medical Center, Washington, DC, USA
7. Mammalian Genetics Section, NIH, Bethesda, MD, USA
Received 2010-8-25; Accepted 2010-9-10; Published 2010-9-10
Abstract
Introduction: Several lines of evidence point to an important role for BP1, an isoform of DLX4 homeobox gene, in breast carcinogenesis and progression. BRCA1 is a well-known player in the etiology of breast cancer. While familial breast cancer is often marked by BRCA1 mutation and subsequent loss of heterozygosity, sporadic breast cancers exhibit reduced expression of wild type BRCA1, and loss of BRCA1 expression may result in tumor development and progression.
Methods: The Cister algorithm and Genomatix program were used to identify potential BP1 binding sites in BRCA1 gene. Real-time PCR, Western blot and immunohistochemistry analysis were performed to verify the expression of BRCA1 and BP1 in cell lines and breast cancer tissues. Double-stranded siRNA transfection was carried out for silencing BP1 expression. ChIP and EMSA were used to confirm that BP1 specifically binds to BRCA1.
Results: A putative BP1 binding site was identified in the first intron of BRCA1, which was confirmed by chromatin immunoprecipiation and electrophoresis mobility shift assay. BP1 and BRCA1 expression were inversely correlated in breast cancer cell lines and tissues, suggesting that BP1 may suppress BRCA1 transcription through consensus sequence binding.
Conclusions: BP1 homeoprotein represses BRCA1 expression through direct binding to its first intron, which is consistent with a previous study which identified a novel transcriptional repressor element located more than 500 base pairs into the first intron of BRCA1, suggesting that the first intron plays an important role in the negative regulation of BRCA1. Although further functional studies are necessary to confirm its repressor activity towards BRCA1, the elucidation of the role of BP1 in breast tumorigenesis holds great promise in establishing BP1 as a novel target for drug therapy.
Keywords: BP1, DLX4, homeoprotein, BRCA1, breast cancer.
Introduction
Breast cancer is the second leading cause of cancer deaths in women after lung cancer and, excluding cancers of the skin, is the most common form of cancer among women. Breast cancer is one of the most common malignancies affecting women and approximately 207,090 new cases of invasive breast cancer are expected to be diagnosed as well as an estimated 54,010 additional cases of in situ tumor in the United States in 2010, and about 39,840 women are expected to die from the disease [1]. The loss of cellular regulation associated with breast cancer is a multi-step process that involves the coordinated activation or inhibition of specific intact or altered genes, often transcription factors that function as oncogenes or tumor suppressors. BRCA1 is a tumor suppressor gene that is mutated in up to 45% of breast cancer in families with multiple cases of breast cancer [2]. Inherited mutations in the BRCA1 gene are correlated with an increased probability of breast or ovarian cancer, and the normal allele is frequently removed by loss of heterozygosity [3] [4]. In sporadic breast cancer, the incidence of BRCA1 mutation is very low [5]. However, reduced levels of BRCA1 protein and mRNA expression are often observed, suggesting that repression of BRCA1 is involved rather than mutations [6] [7]. This hypothesis was supported by experiments in which the introduction of a normal BRCA1 gene inhibited the growth of breast and ovarian cancer cell lines in vitro and inhibited the tumor growth rate of MCF7 breast cancer cells in nude mice [8]. Growth inhibition was not observed in colon and lung cancer cell lines, further demonstrating that loss of BRCA1 activity plays a special role in breast and ovarian cancers and not other types of cancer.
Other mechanisms have been proposed for BRCA1 inactivation in sporadic cancer including epigenetic silencing. Hypermethylation of the BRCA1 promoter region CpG island was first reported in two out of seven breast tumors and later in two out of six breast tumors and two out of five ovarian tumors [9] [10]. A larger study by Catteau et al. found that BRCA1 promoter is highly methylated in 11% of breast carcinomas and 5% of ovarian carcinomas [11]. Hypermethylation, therefore, appears to not be a significant mechanism for repression of BRCA1 expression in breast tumors. Additional studies have focused on other transcriptional and/or post-transcriptional mechanisms by which BRCA1 inactivation is achieved in breast and ovarian cancer. Baldassarre et al. found that the HMGA1b protein binds directly to the BRCA1 promoter and represses BRCA1 transcription both in vitro and in vivo [12]. In another study, cyclin D1 expression was found to antagonize BRCA1 inhibition of estrogen receptor α (ERα)-dependent gene expression [13].
BRCA1 has been hypothesized to play a role in DNA damage repair, and many studies have reported a reduction in BRCA1 mRNA and protein levels following extended exposure to DNA damaging agents [14] [15] [16]. In one study, repression of p53 by human papilloma virus E6 resulted in an inability to downregulate BRCA1 in response to the DNA-damaging agent adriamycin, while ectopic expression of p53 resulted in the rapid downregulation of BRCA1 mRNA and protein levels [17]. These results, in conjunction with others, suggest a feedback loop where BRCA1 initially induces p53 expression and p53 later acts to negatively regulate BRCA1. The precise mechanism of BRCA1-mediated growth regulation and tumor suppression, however, remains elusive. There is increasing evidence, however, that suggests BRCA1 functions as a regulator of centrosome number. BRCA1 is localized to the centrosome in mitotic cells [18] [19], and interference with BRCA1 function by various methods results in an increase in centrosome number. In one study, mouse fibroblasts derived from exon 11 knockouts were found to have amplified centrosomes [20]. Amino acid residues 504 to 803 of BRCA1 bind γ-tubulin, a major component of centrosomes, and stable overexpression of the γ-tubulin binding domain results in increased centrosome number in tissue culture cells [21]. Acute expression of an inhibitor peptide that binds to the carboxy terminus of BRCA1 causes rapid centrosome amplification in a mammary cell line [22]. In a study by Starita et al., transient inhibition of BRCA1 in mammary tissue-derived cell lines caused rapid accumulation and fragmentation of centrosomes [23]. BRCA1 inhibition in nonmammary cell lines, however, did not result in centrosome amplification.
BP1, a DLX4 isoform, belongs to the homeobox family of master regulatory genes, which are implicated in early development and cell differentiation and are frequently dysregulated in cancer [24]. In normal erythroid cells, BP1 acts as a putative repressor of the β-globin gene [25] [26]. BP1 was expressed in 47% of the adult and 81% of the pediatric acute myeloid leukemia patients, and BP1 overexpression increased the leukemogenic potential of K562 cells in vitro [27]. Furthermore, molecular analysis revealed that BP1 expression is required for cell survival in K562 cells, implicating BP1 in an anti-apoptotic pathway [28]. BP1 was mapped to chromosome 17q21-22, a region of DNA that is frequently amplified in breast cancer and contains BRCA1 and oncogene erbB2 [26]. Since BP1 may be involved in an anti-apoptotic pathway, it was speculated that aberrant expression of BP1 might also promote increased cell survival and growth in other malignancies including breast cancer. BP1 mRNA levels were analyzed in a series of breast cancer cell lines using RT-PCR, and a correlation was found between BP1 expression and tumorigenesis in mice [29]. Cell lines that express little or no BP1 (Hs578T, MDA-MB-435S, and MCF10A) are not tumorigenic, whereas cells that express high levels of BP1 (MCF7-ADR, MDA-MB-468, and MDA-MB-231) are tumorigenic. In the analysis of 46 invasive ductal breast tumors by Fu et al., BP1 expression was detectable in 80% of them as measured by RT-PCR, compared with a lack of expression in six normal breast tissues and low level expression in one normal breast tissue [29]. A correlation was also found in these studies between BP1 expression and estrogen receptor (ER) status as well as between BP1 expression and race. BP1 expression was significantly higher in ER negative (100%) versus ER positive (73%) tumors and in the tumors of African-American women (89%) versus those of Caucasian women (57%). These findings have important scientific and clinical implications. Since ER negative tumors are clinically more aggressive and have a poorer prognosis compared to ER positive tumors, the expression of BP1 might serve as a reliable marker of breast tumor aggressiveness [30]. This was supported by our recent study demonstrating that BP1 enhances cell proliferation and metastatic potential in ER negative Hs578T breast cancer cells [31]. Furthermore, the oncogenic properties of BP1 as revealed in the leukemia study suggest that the high percentage (80%) of BP1 expression in invasive ductal carcinomas contributes to enhanced breast tumor growth rate and invasiveness. We also found that the prevalence of BP1 positive cells and the intensity of BP1 immunostaining increased with the extent of ductal proliferation and carcinogenesis. In addition, BP1 positive cells were observed to have a substantially higher proliferation rate compared to morphologically similar BP1 negative cells, and BP1 co-localized with onco-protein erbB2 in 15 in situ and infiltrating breast ductal carcinomas [32]. We have also identified a number of potential direct BP1 binding targets, including VEGFA, STAT1, ITGA9, etc [33]. BP1 has been shown, therefore, to have a prominent role in breast cancer progression and invasion. However, the precise nature of its role in carcinogenesis remains unclear. In this study, we investigate the role of BP1 in breast tumorigenesis, and evidence will be presented showing how BP1 functions as a transcriptional repressor of BRCA1 in breast cancer.
Materials and Methods
Cister algorithm (Cis-element Cluster Finder) and Genomatix program. GenBank accession number U37574 for human BRCA1 gene, partial coding sequence was input into the Cister search form (Boston University) to predict regulatory regions in the BRCA1 promoter region. A user-defined cis-element was entered as a TRANSFAC-style matrix for the BP1 consensus binding sequence (A/T)T(A/C)(A/T)ATATG (25). The motif probability threshold was set at 0.01. Genomatix program was also used to predict all potential BP1 homeoprotein binding sites in BRCA1.
Cell lines and cell culture. MCF7 breast cancer cells stably transfected with the pcDNA3.2/BP1 construct or an empty vector were maintained in RPMI 1640 media (Invitrogen) medium with 10% fetal bovine serum plus 500ug/ml of Geneticin (Invitrogen) and 1% penicillin/ streptomycin. BT-20, SKBR3 breast cancer cells were transiently transfected with the pcDNA3.2/BP1 construct or an empty vector and maintained in Eagle's Minimum Essential Medium (ATCC) and McCoy's 5a Medium Modified (ATCC), respectively, with 10% fetal bovine serum plus 500ug/ml of Geneticin (Invitrogen) and 1% penicillin/ streptomycin. Hs578T breast cancer cells stably transfected with the pcDNA3.2/BP1 construct or an empty vector were maintained in DMEM containing high glucose, L-glutamine, and sodium pyruvate (Invitrogen) with 10% fetal bovine serum plus 500ug/ml of G418 [33].
siRNA synthesis and transfection. Double-stranded siRNA targeting BP1 was synthesized using Silencer® siRNA Construction Kit (Ambion). Template oligonucleotides for siRNA synthesis were selected using BP1 (DLX4 Variant 1) accession number NM_138281 by siDirect (University of Tokyo, http://design.rnai.jp/). Two target sequences were selected. An eight nucleotide sequence, 5'-CCTGTCTC-3', was added to the 3' end of the target sequences, as instructed by Silencer protocol. Two target sequences are the following. T1, sense: 5'- GACCTATGGGTAATTTATGCTCCTGTCTC-3', anti-sense: 5'-AAAGCATAAATTACCCATAGGCCTGTCTC-3'; T2, sense: 5'- AAGGAACTGTGCAGATTTAGACCTGTCTC-3', anti-sense: 5'- GTTCTAAATCTGCACAGTTCCCCTGTCTC-3'. The designed sequences were synthesized by Integrated DNA Technologies (IDT). Double-stranded siRNA was then synthesized according to the Silencer protocol, and quantified using the Nanodrop spectrometer (Thermo Scientific). T47D, MDA-MB-231 cells were seeded into two six-well plates with 2.5 x105 cells/well and grown in 2mL RPMI-1640 (Invitrogen), Leibovitz's L-15 Medium (ATCC) respectively, supplemented with 10% fetal bovine serum. After a confluence of 60-70% in each well was reached, the cells were then transfected with T1, T2, Non-targeting Pool siRNA (Thermo Scientific) and no siRNA control using FuGENE® HD Transfection Reagent (Roche Applied Science). The transfection complexes were prepared with 1 µg of siRNA, 200µl of plain medium and 2µl of FuGENEene Reagent. The complexes were applied according to the manufacturers' instructions. After 48h, the transfected cells were collected for verification of BP1 knockdown and analysis of knockdown effects.
Chromatin immunoprecipitation assay. 1.5 x 106 BP1-overexpressing MCF7 or Hs578T cells were crosslinked in either 50 mL RPMI 1640 media or DMEM with 1% formaldehyde for ten minutes at 37ºC. Cells were pelleted by centrifugation and washed twice with cold PBS, resuspended in SDS lysis buffer containing Complete, Mini protease inhibitors (Roche), and incubated for ten minutes on ice. Chromatin was sheared on ice to an average size of 200-500 base pairs and diluted 10 fold with ChIP dilution buffer containing Complete, Mini Protease inhibitors. The lysate was precleared with salmon sperm DNA (Stratagene)/Protein G PLUS/Protein A-Agarose beads (Calbiochem) for one hour at 4ºC with constant rotation. The beads were pelleted, 10% of the supernatant was removed for input, and equal volumes of the remaining supernatant were incubated overnight at 4ºC with constant rotation using 10 μg of BP1 polyclonal antibody (Research Genetics) or no antibody for control. Samples were incubated with 60 μL of salmon sperm DNA/Protein G PLUS/Protein A agarose beads for two hours at 4ºC with constant rotation. Beads were collected by centrifugation and washed once with 1 mL low salt buffer, twice with 1 mL high salt buffer, once with 1 mL LiCl wash buffer, and once with 1 mL Tris-EDTA buffer. Chromatin was eluted twice in 250 μL of freshly made elution buffer at room temperature with constant rotation for 15 minutes. For reverse crosslinking, the eluate and input chromatin were incubated with 20 μL 5 M NaCl for four hours at 65ºC, then stored overnight at -20ºC. Proteins were degraded by incubation with 100 μg/mL proteinase K for one hour at 56ºC. DNA was purified twice by phenol/chloroform/ isoamyl (25:24:1) extraction and precipitated in 1 mL absolute ethanol with 50 μL 3 M sodium acetate at -80ºC for thirty minutes. The precipitated DNA was pelleted by centrifugation and washed with 70% ethanol. After washing, the DNA was pelleted again, allowed to air-dry, and resuspended in Tris-EDTA buffer for use in PCR.
PCR assays. PCR reactions for amplification of chromatin immunoprecipitates and input DNA were performed in a total volume of 25 μL using Taq DNA Polymerase (Invitrogen) following manufacturer's protocol. DNA was initially denatured at 94ºC for 2 minutes, followed by 28 cycles of denaturation at 94ºC for 45 seconds, annealing at 58ºC for 45 seconds, and elongation at 72ºC for 45 seconds, and a final cycle of 94ºC for 45 seconds, 58ºC for 45 seconds, and 72ºC for 7 minutes. Primers were designed to flank the putative BP1 binding site in the first intron of BRCA1 (Forward: 5'-GGACGTTGTCATTAGTTCTTTGG-3'; Reverse: 5'-CGCGAAGAGCAGATAAATCC-3'). PCR products were run on a 2% agarose gel and visualized on a Kodak Image Station 2000MM with Kodak 1D version 6 software.
Electrophoretic mobility shift assay. Electrophoretic mobility shift assay (EMSA) was performed using the DIG Gel Shift Kit, 2nd Generation (Roche) for nonradioactive detection of sequence specific DNA binding proteins following the manufacturer's protocol. Briefly, a 33- base pair double-stranded oligonucleotide probe specific for BP1 binding was 3'-end-labeled using Digoxigenin-11-ddUTP (DIG-11-ddUTP) and terminal transferase. Whole cell extracts from MCF7 BP1 overexpressing cells were incubated with the labeled probe in binding buffer (20 mM HEPES-KOH (pH 7.6), 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM DTT, 0.2% (w/v) Tween 20, and 30 mM KCl) containing 1 µg poly d[I-C] and 0.1 µg poly L-lysine in a final volume of 20 µL for 15 minutes at room temperature. For gel-shift competition, unlabeled specific competitor (50- or 100-fold molar excess) or a non-competitive negative control (25-fold molar excess) was included in the reaction. Sequences of probes were as follows: BP1 binding (Forward: 5'-TTAAAAAGATATATATATATGTTTTTCTAATGT-3'; Reverse: 5'-ACATTAGAAAAACATATATATATATCTTTTTAA-3') and negative control (Forward: 5'-TCTTAGAGGGAGGGCTGAGGGTTTGAAGTCCAACTCCTAAGCC-3'; Reverse: 5'-AGAATCTCCCTCCCGACTCCCAAACTTCAGGTTGAGGATTCGG-3'. DNA-protein complexes were resolved using a pre-run 6% polyacrylamide non-denaturing gel in 0.5% TBE and transferred to a nylon membrane. The oligos were fixed by UV cross-linking and the DIG-labeled probe was visualized by chemiluminescence.
RNA extraction and Real-time RT-PCR assays. Total RNA was extracted from human breast cancer cell lines with Trizol reagent (Invitrogen) according to the manufacturer's protocol. cDNA synthesis was carried out with 1 μg of total RNA using the First-Strand cDNA Synthesis Kit (Invitrogen). Templates for real-time PCR standards were prepared by amplification of the target genes using cDNA in a total volume of 25 μL using Taq DNA Polymerase (Invitrogen) following the manufacturer's protocol. Specifically, PCR was performed at the following conditions: denaturing at 94ºC for 2 minutes, followed by 28 cycles of denaturation at 94ºC for 45 seconds, annealing at 60ºC for 45 seconds, and elongation at 72ºC for 45 seconds, with an additional elongation step at 72ºC for 5 minutes. The primers used were specific for BP1 (Forward: 5'-CAAAGCTGTCTTCCCAGACC-3'; Reverse: 5'-GTTGTAGGGGACAAGCCAAG-3'), BRCA1 (Forward 5'-TGTGAGGCACCTGTGGTGA-3'; Reverse: 5'-CAGCTCCTGGCACTGGTAGAG-3'), or 18S (Forward 5'-CCGCAGCTAGGAATAATGGA-3'; Reverse: 5'-CCCTCTTAATCATGGCCTCA-3'). Real-time PCR standards comprised a series of 5 dilutions of the respective PCR product at 1, 1:10, 1:100, 1:1,000, and 1:10,000 for relative quantitation of target expression levels.
Real-time PCR reactions with the above primers were performed in a total volume of 25 μL using Platinum SYBR Green qPCR SuperMix UDG with ROX (Invitrogen) according to the manufacturer's protocol. Thermocycling was performed using a 7300 Real Time PCR System (Applied Biosystems) with the following conditions: 1 cycle of 50ºC for 2 minutes, 1 cycle of 95ºC for 10 minutes, 40 cycles of 95ºC for 15 seconds and 60ºC for 1 minute, and a final cycle of 95ºC for 15 seconds, 60ºC for 30 seconds, and 95ºC for 15 seconds. Data was analyzed using 7300 System Sequence Detection Software version 1.2 (Applied Biosystems). The relative expression levels of BP1 and BRCA1 were calculated using the delta Ct value and normalized against ribosomal 18S expression.
Protein extraction and Western blot analysis. Proteins were extracted from cell lines using T-PER Tissue Protein Extraction Reagent (Pierce) according to the manufacturer's protocol. Complete, Mini Protease Inhibitor Cocktail (Roche) was added to the extraction solution prior to lysis to prevent proteolytic activity. Cell protein lysate was prepared with SDS gel-loading buffer containing β-mercaptoethanol and heated at 98ºC for 15 minutes. Proteins were separated by SDS-PAGE using a 12% Ready Gel Tris-HCl pre-cast polyacrylamide gel (Bio-Rad) and transferred overnight. The membrane was blocked prior to the addition of the primary antibody with 5% milk in Tris/NaCl/EDTA (TNE) with 0.05% Tween. The membrane was incubated overnight with either BP1 (NB 100-481) rabbit polyclonal antibody (Novus Biologicals) at a dilution of 1:5,000 in TNE buffer with 0.05% Tween, BRCA1 monoclonal antibody (Cat# NB100-598) (Novus Biologicals), which recognizes the epitope amino acid 1314-1600, at a dilution of 1:500 in TNE buffer with 0.05% Tween and 5% milk, or β-actin (A 5441) mouse monoclonal antibody (Sigma) at a dilution of 1:10,000 in TNE buffer with 0.05% Tween. The membrane was washed 3 times with TNE/0.05% Tween and incubated with a goat-anti-rabbit IgG conjugated to horse radish peroxidase (Santa Cruz) for BP1 or a goat-anti-mouse IgG (Amersham) for BRCA1 and β-actin at a 1:10,000 dilution in TNE/0.05% Tween (and 5% milk for BRCA1). Enhanced chemiluminescence with Western Blotting Luminol Reagent (Santa Cruz) was used according to the manufacturer's protocol to visualize proteins and quantify band intensity.
Immunhistochemistry assays (IHC). Consecutive tissue sections at 8-10 μm thickness were made from fresh frozen human breast tumors (n=15) with co-existing normal, pre-invasive, and invasive components. Sections were placed on positively charged microscopic slides and fixed in 10% buffered formalin for 10 minutes at room temperature. Next, sections were washed with PBS (pH 7.0) containing 0.2% Tween 20 three times each for 10-15 minutes at room temperature and subjected to morphological and immunohistochemical assessment using previously published criteria and protocols [32].
To assess the potential correlation between BRCA1 and BP1 expression, sets of two immediate adjacent sections from each case were immunostained with affinity purified rabbit polyclonal antibodies raised against a peptide mapping at the C-terminus of human BRCA1 (I-20) (Cat# sc-646) (Santa Cruz) and against a peptide mapping at the N-terminus of BP1 of human origin (Research Genetics). Immunostaining was carried out using previously published protocols [34] [35]. Briefly, sections were incubated with the primary antibody and different control solutions, including the substitution of pre-immune IgG for the primary antibody, substitution of PBS for the primary or secondary antibody, and pre-absorption of the primary antibody with the corresponding peptide (done only for BP1), overnight at room temperature. After the incubation, sections were washed with PBS containing 0.2% Tween 20 three times each for 5-7 minutes. The antigen/antibody complexes were detected using an ABC detection kit (Vector) and an AP-red chromogen kit (Zymed) according to the manufacturers' protocols.
The expression status of BP1 and BRCA1 at the same cell population in two different sections was compared. A given cell was considered to be positive if distinct chromogen coloration was seen in all three duplicated procedures and no chromogen coloration was seen in any of the controls.
Results
Identification of potential BP1 homeoprotein binding sites in BRCA1
Analysis of a partial coding sequence of the human BRCA1 gene (GenBank accession U37574) using the Cister algorithm and Genomatix program identified 21 putative BP1 binding sites (ATATATATG) within the first intron. We focused on the one with strongest potential from nucleotide positions 2,830 to 2,838, which is -47 to -39 base pairs upstream of the translational start site, suggesting that BP1 might transcriptionally suppress BRCA1 through binding downstream of its promoter (Fig. 1A, Fig. 6). The partial coding sequence is 3,798 nucleotides long and includes the promoter and 5' UTR, which consists of exons 1a and 1b as well as exon 2.
Cister search results and PCR amplification of ChIP DNA samples from MCF7 and Hs578T cells. A. Cister predicts regulatory regions in DNA sequences by searching for clusters of cis-elements. The red line adjacent to the pink protein coding region corresponds to the putative BP1 binding site located on the positive strand of the human BRCA1 gene and indicates the probability that a regulatory factor binds at this position. B. ChIP assays were conducted with a BP1 antibody and a no antibody control. A 186 base pair PCR fragment was amplified with BP1 immunoprecipitated and input DNA from BP1 overexpressing MCF7 and Hs578T cells, but not with the no antibody control, using primers flanking the putative BP1 binding site. C. Correlation of BP1 mRNA expression with the level of ChIPed BRCA1 genomic DNA analysis. ChIP assays were performed on a series of MCF7 cell lines with different levels of BP1 expression. Various levels of BRCA1 genomic DNA were amplified from the ChIP DNA by SYBR real-time PCR, in correlation with the mRNA expression of BP1 in each cell line.
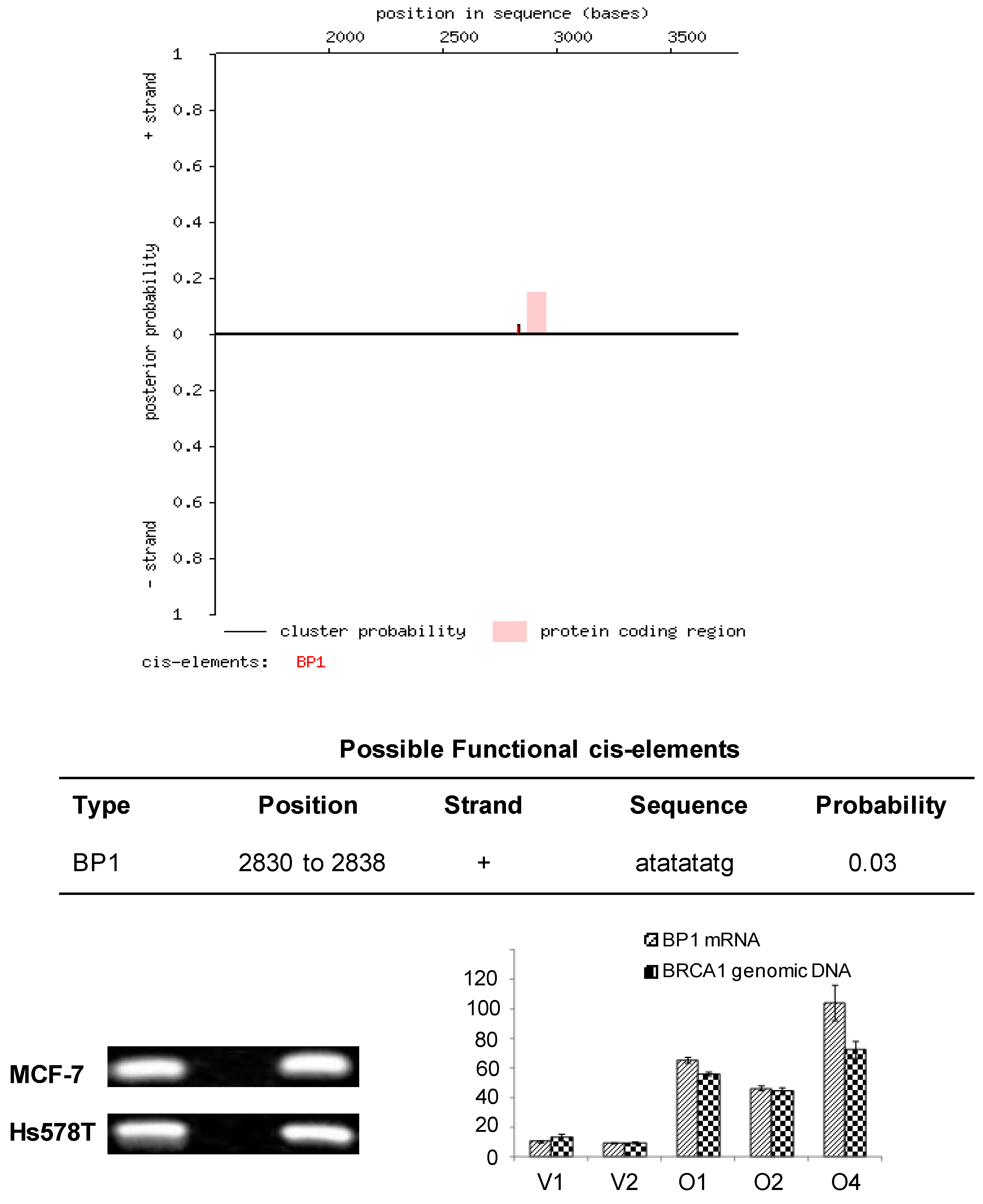
Confirmation of BP1 binding to the first intron of BRCA1
Based on the sequence analysis, primers were designed flanking the putative BP1 binding site to be used in PCR analysis of immunoprecipitates isolated by ChIP. To determine whether BP1 binds to the putative consensus sequence identified by Cister, ChIP was performed with BP1 overexpressing MCF7 and Hs578T cell lines. Chromatin immunoprecipitates were analyzed by PCR and a 186 base pair product was amplified from BP1 immunoprecipitated chromatin but no product was formed from the no antibody control for both cell lines analyzed (Fig. 1B). To evaluate the binding efficiency, we performed the ChIP on cell lines overexpressing BP1 and their controls for both MCF7 and Hs578T cells. The real-time PCR data showed that the amount of pulled-down complex was correlated with the level of BP1 expression in those cells (Fig. 1C).
Electrophoretic mobility shift assay (EMSA). Protein extract isolated from BP1 overexpressing MCF7 cells binds specifically to a 33- nucleotide double-stranded sequence containing the putative BP1 binding site in the first intron of BRCA1. Labeled probe specific for the BP1 binding site was used in the binding reactions with the following additions: Lane 1, free probe; lane 2, protein extract; lane 3, protein extract plus 50-fold molar excess of unlabeled BP1 specific double-stranded probe; lane 4, protein extract plus 100-fold molar excess of unlabeled BP1 specific double-stranded probe; lane 5, protein extract plus 100-fold molar excess of unlabeled scrambled probe.
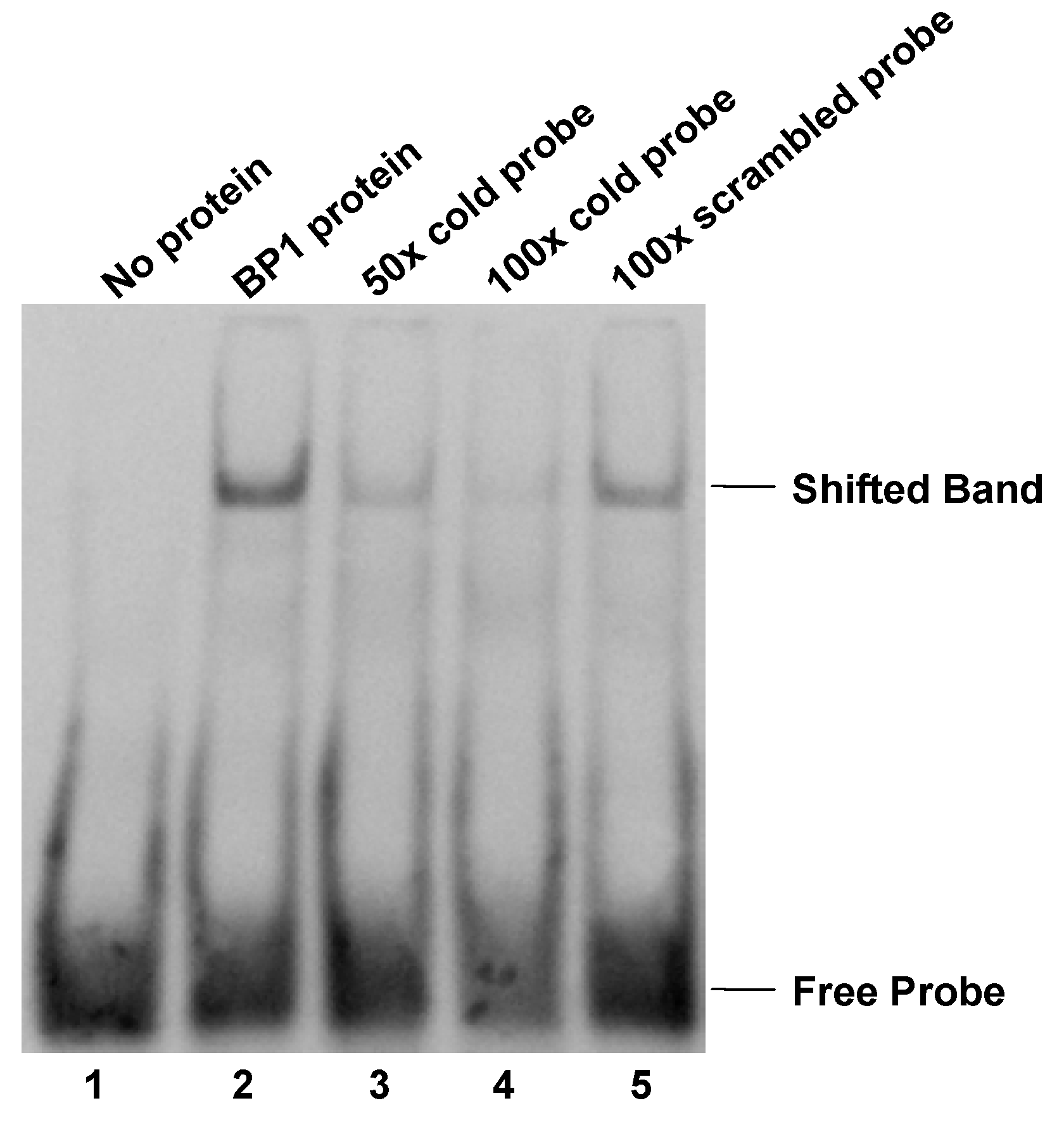
To further evaluate the possibility of protein binding to the putative BP1 binding site, EMSA was performed using a DIG-labeled 33-mer specific for the BP1 binding site in the first intron of BRCA1. Binding to the DIG-labeled probe was observed as a shifted band in protein extracts isolated from BP1 overexpressing MCF7 cells (Fig. 2 lane 2). Specificity of protein binding to the DIG-labeled oligomer was tested by competitive inhibition of binding by the addition of 50- and 100-fold molar excess of unlabeled specific probe (Fig. 2 lanes 3 and 4). When 50-fold molar excess unlabeled probe specific for BP1 binding was added to the protein extract, binding to the DIG-labeled oligomer was reduced by more than half. The addition of 100-fold molar excess of unlabeled specific probe reduced binding to the DIG-labeled probe even further, almost entirely eliminating the observed DNA shift. Furthermore, the addition of 100-fold molar excess of an unlabeled non-competitive oligomer to the protein extract did not affect the gel mobility shift of the DIG-labeled probe (Fig. 2 lane 5). To test the specificity of the binding, super shift assay with addition of BP1 antibody was performed and a super shifted band was observed (data not shown).
Overexpression of BP1 in breast cancer cells down-regulates BRCA1 expression
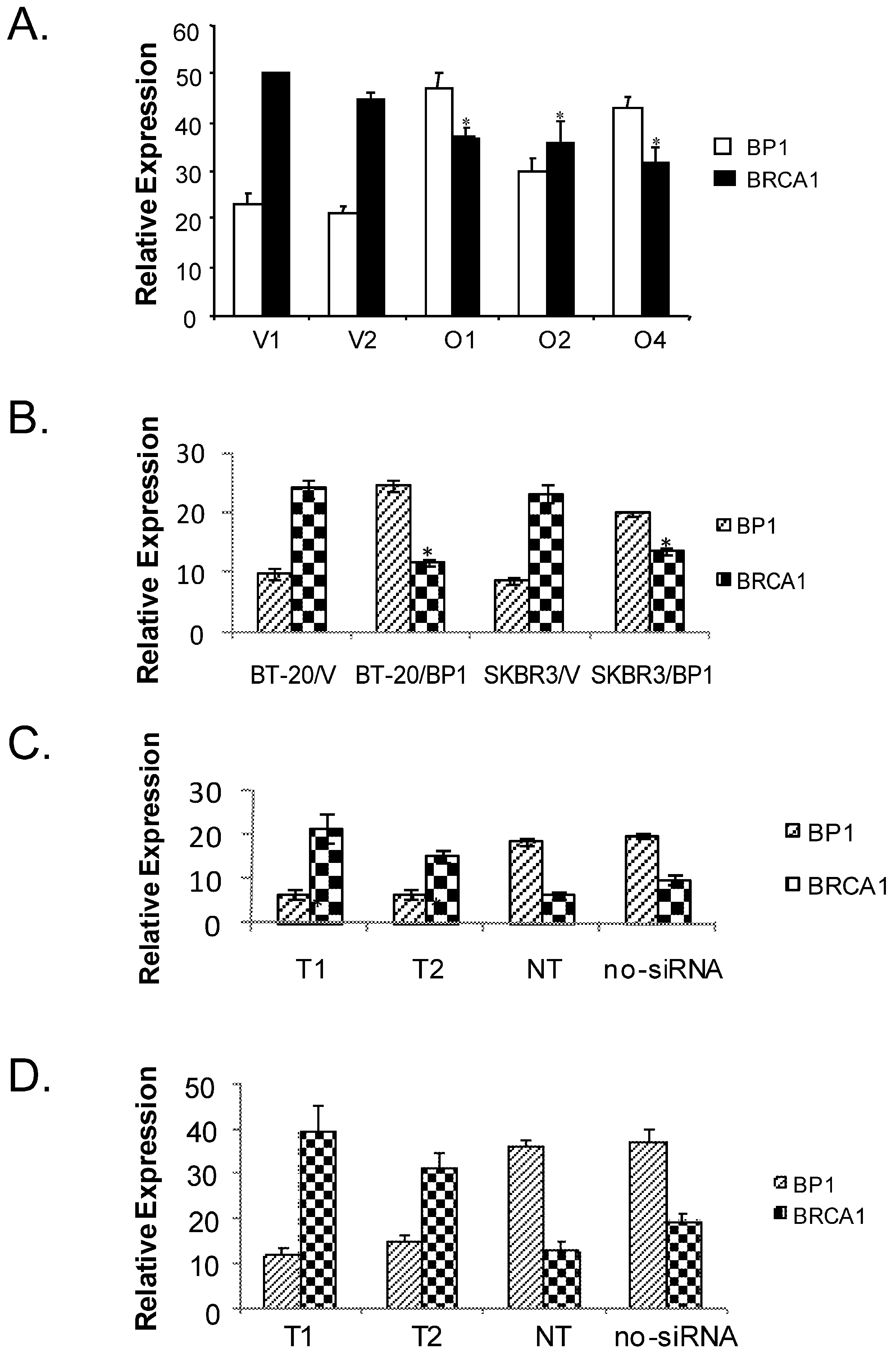
To verify BP1 expression represses BRCA1 in breast cancer cells, real-time RT-PCR was performed to quantitatively determine the transcription of BP1 and BRCA1 mRNA levels in BP1-overexpressing MCF7 cell lines compared to vector controls (Fig. 3A), as well as in transiently transfected BT-20 and SKBR3 cell lines (Fig. 3B). Real-time PCR reactions were performed in duplicate three times and normalized against 18S rRNA expression. For BP1, the normalized expression levels for BP1-overexpressing lines O1, O2, and O4 were 94.6, 60.0, and 85.7. Vector control lines V1 and V2 exhibited levels of 46.0 and 41.8 for an average of 43.9. BP1 overexpressors, therefore, had levels of BP1 mRNA expression 1.4 to 2.2 fold greater on average compared to vector controls. For BRCA1, the normalized expression levels for vector control cell lines V1 and V2 were 100.0 and 89.0 for an average of 94.5. Expression levels for overexpressing lines O1, O2, and O4 were 74.0, 71.6, and 63.4, a decrease of 21.7%, 24.2%, and 32.9% respectively relative to vector controls. In transiently transfected BT-20 and SKBR3 breast cancer cells, the expression of BP1were 2.1 and 1.6 fold higher respectively compared to their vector controls, while BRCA1 were down-regulated by 2.1 and 1.7 fold respectively (Fig. 3B). The mRNA expression of BP1 and BRCA1 were consistently inverse-correlated.
Real-time RT-PCR analysis of BP1 and BRCA1 expression. Relative BP1 and BRCA1 mRNA expression levels were quantified by real-time RT-PCR and normalized against 18S expression levels. Data are the mean ± standard error for three experiments for BRCA1 and BP1 with duplicates each. Student's t test was done for statistical significance analysis (*P < 0.05). A. Stable MCF7 BP1 overexpressors (O1, O2, and O4) vs. vector controls (V1 and V2). B. Transient BP1 overexpressor (BT-20/BP1) vs. vector control (BT-20/V); and transient BP1 overexpressor (SKBR3/BP1) vs. vector control (SKBR3/V). C. T47D cells treated with BP1 siRNA target T1 and T2 respectively comparing non-targeting (NT) and no siRNA controls. D. MDA-MB-231 cells treated with BP1 siRNA target T1 and T2 respectively comparing non-targeting (NT) and no siRNA controls.
BP1 siRNA knock-down in breast cancer cells up-regulates BRCA1 expression
To verify if BP1 siRNA knock-down can potentially activate BRCA1 expression, we transiently transfected T47D and MDA-MB-231 cells respectively with BP1 siRNA targets and controls. Real-time PCR reactions were performed in duplicate three times and normalized against 18S rRNA expression. The normalized values for the expression of BP1 and BRCA1 for the following transfections: T1, T2, non-targetting (NT) and no siRNA control were shown for T47D (Figure 3C), and MDA-MB-231 (Figure 3D). Both T1 and T2 siRNA knocked down BP1 expression by more than 2 fold, while BRCA1 expression increased by 2.5-3 fold. The purpose of NT was to ensure that the BP1 knockdown is specific by its targets, not by random siRNA targets.
Western blot analysis of BP1 and BRCA1 protein levels. Whole-cell extracts from MCF7 vector controls (V1 and V2) and BP1 overexpressors (O1, O2, and O4) were analyzed by Western blotting using antibodies for BP1 or BRCA1 (4A). Protein expression levels were normalized against β-actin. BRCA1 protein expression is decreased by 72-79% in BP1 overexpressors relative to vector controls. Relative BP1 and BRCA1 expression levels are presented based on results from one experiment each (4B.).
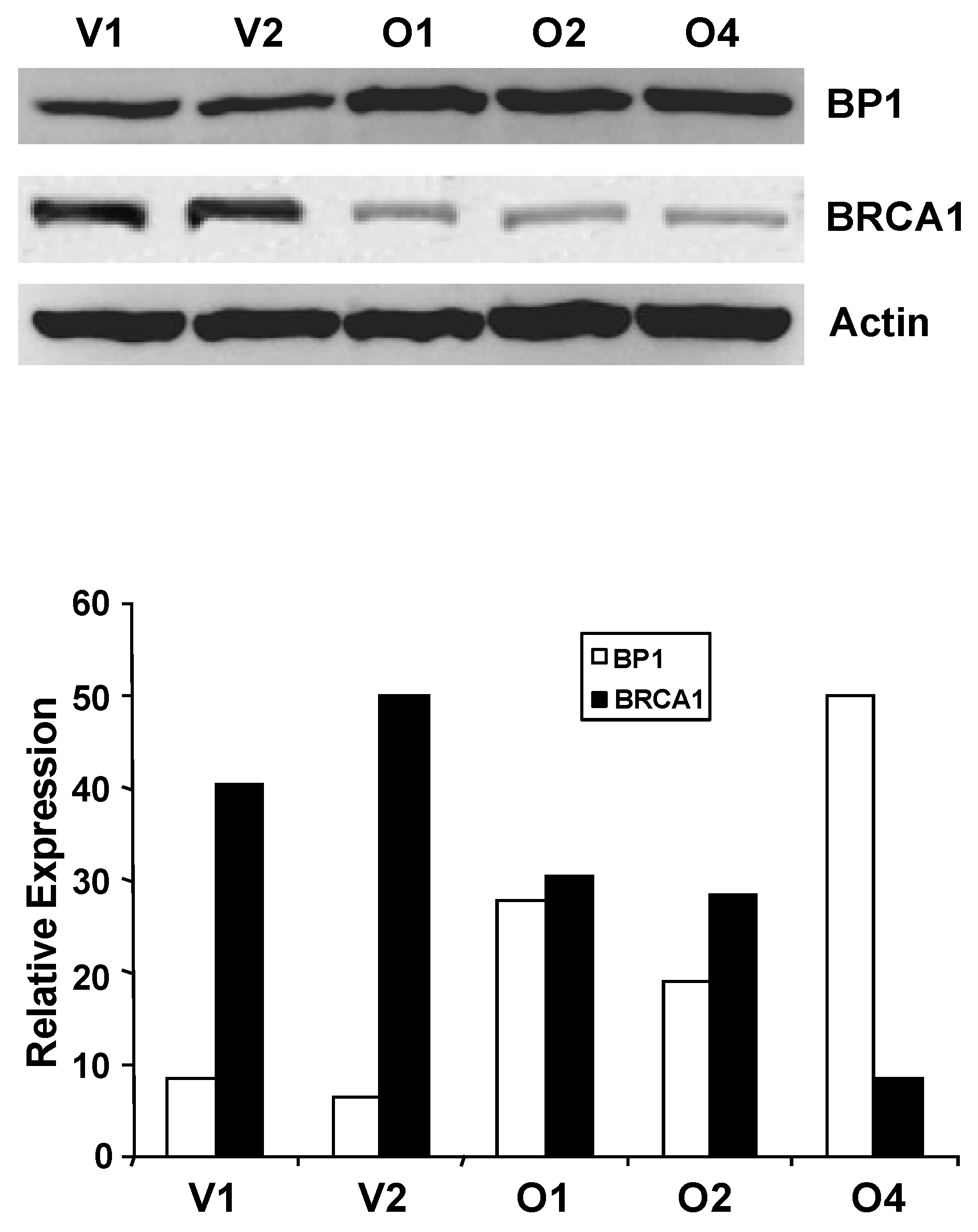
We next questioned whether altered BRCA1 mRNA expression in BP1 overexpressors was associated with altered BRCA1 expression at the protein level. Western blot analysis was performed using whole-cell extracts isolated from MCF7 BP1 overexpressing and vector control cell lines (Fig. 4). Band intensity for BP1 and BRCA1 was quantified and normalized against actin using NIH ImageJ (Version 1.43). Relative protein expression levels for BP1 overexpressing cell lines O1, O2, and O4 were 100.0, 82.5, and 87.4, while expression levels for vector control cell lines V1 and V2 were 42.9 and 42.9. BP1 protein levels, therefore, were on average increased by 2.0 to 2.3 fold in BP1 overexpressing cell lines compared to vector controls. For BRCA1, the normalized expression levels for vector control cell lines V1 and V2 were 100.0 and 93.5 for an average of 96.8, while BP1 overexpressing lines O1, O2, and O4 were 27.0, 25.0, and 19.9, a decrease of 72.1%, 74.2%, and 79.4% respectively relative to vector controls.
Inverse correlation between BP1 and BRCA1 immunostaining in patient tissue
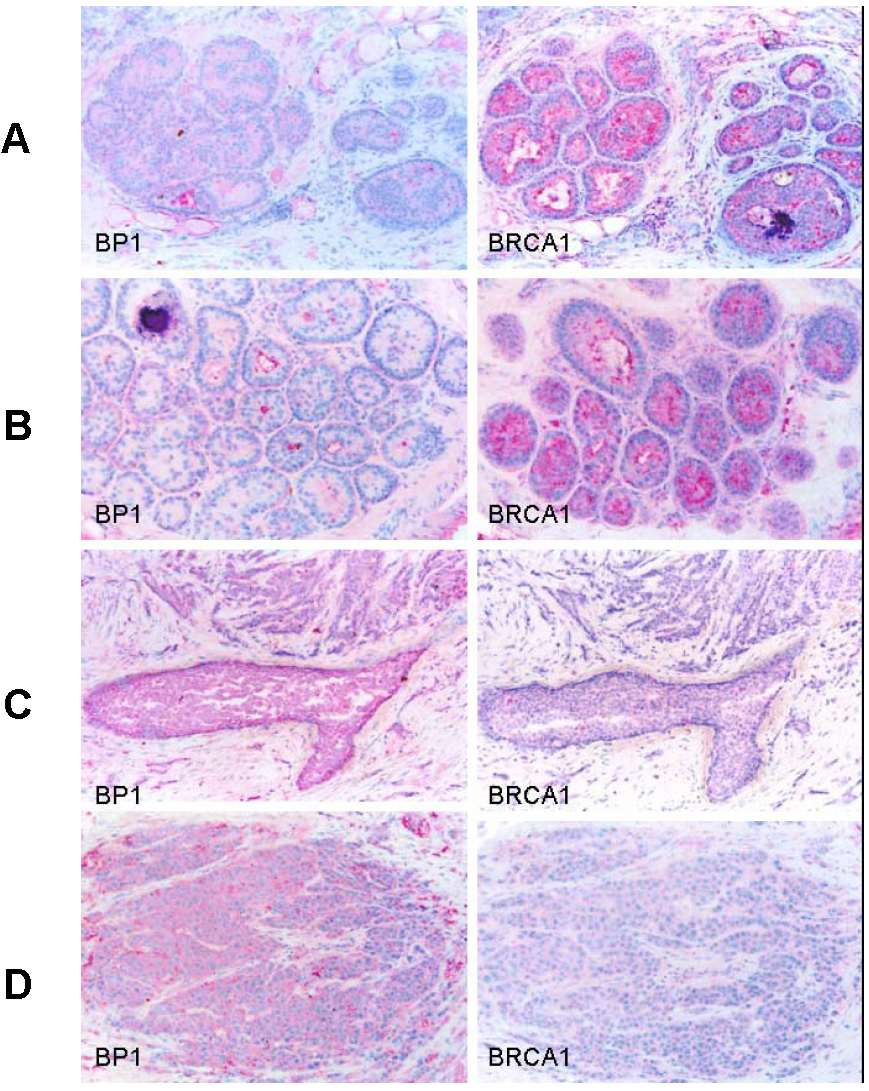
Immunostaining was also performed using antibodies for BP1 and BRCA1 on consecutive human breast tumor tissue sections (n=15) with co-existing normal, hyperplasia, in situ, and invasive components, and distinct BRCA1 and BP1 immuno-reactivity was seen in a subset of epithelial cells. The expression of BRCA1 and BP1 appeared to be inversely correlated. Distinct BRCA1 expression was consistently seen in most normal and hyperplastic cells (Fig. 5A and 5B), whereas its expression appeared to decrease with tumor progression, with no or substantially reduced expression in most in situ and invasive tumor cells (Fig. 5C and 5D). In contrast, no distinct BP1 expression was seen in most normal and hyperplastic cells (Fig. 5A and 5B), whereas intense and uniform BP1 immunoreactivity was consistently present in most in situ and invasive cancer cells (Fig. 5C and 5D).
Expression status of BP1 and BRCA1 in normal and different breast lesions. Consecutive tissue sections from human breast tumors (n=15) with co-existing normal, pre-invasive, and invasive components were immunostained using antibodies for BP1 and BRCA1, and the expression status of BP1 and BRCA1 at the same cell population in two different sections was compared. Distinct BRCA1 expression but no distinct BP1 expression was consistently seen in most normal (A) and hyperplastic (B) cells. BRCA1 expression appeared to decrease with the extent of tumor progression, with no or substantially reduced expression in most in situ (C) and invasive (D) tumor cells, while intense and uniform BP1 immunoreactivity was consistently present.
Discussion
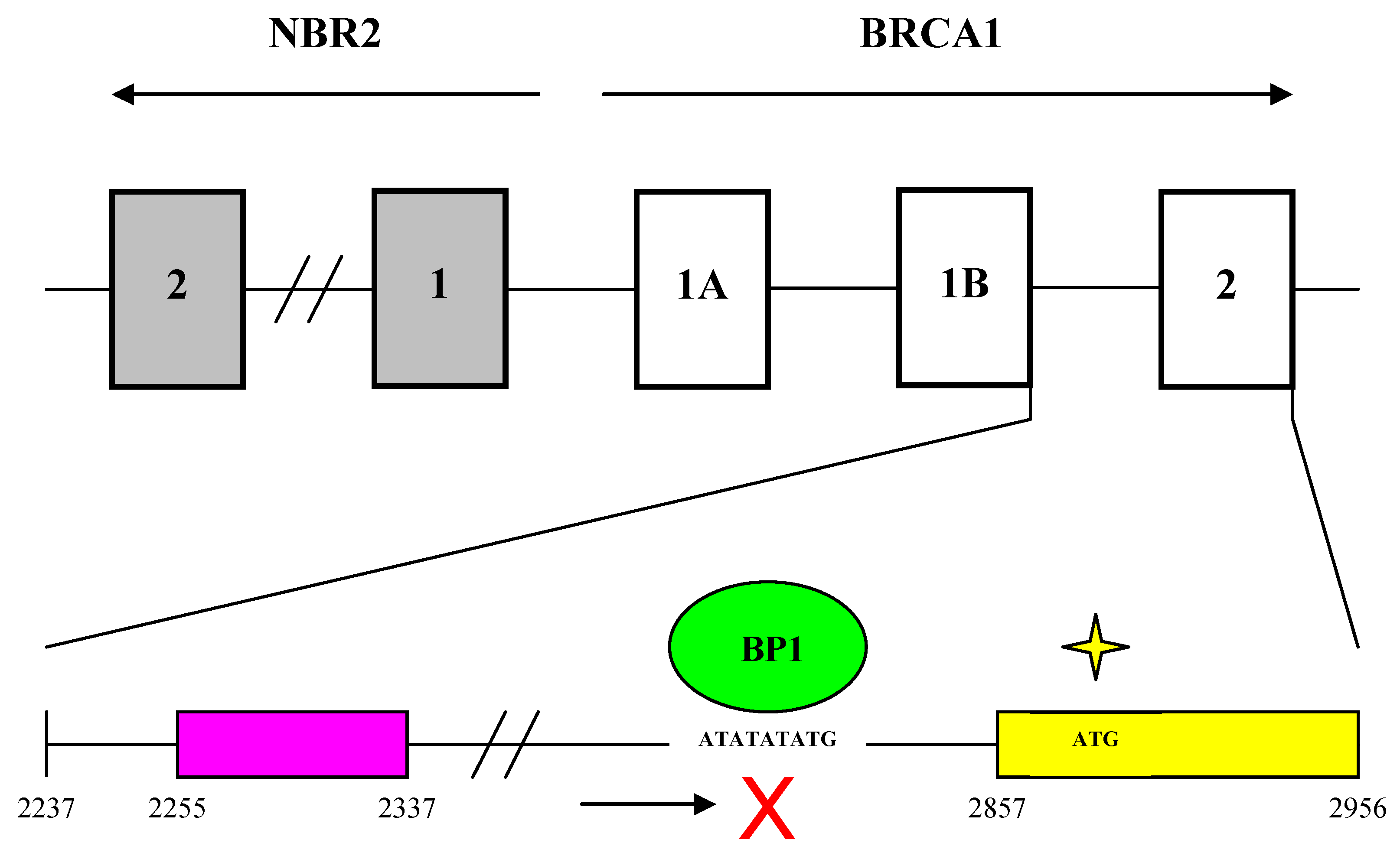
These results help elucidate the molecular mechanisms controlling BRCA1 expression and provide an updated picture of the genomic organization of the BRCA1 promoter region (Fig. 6). BRCA1 transcription is dependent on a bi-directional promoter located within a 229 base pair intergenic region between BRCA1 and a neighboring unrelated gene called NBR2 [36]. Deletion analysis identified a minimal 56 base pair EcoRI-HaeIII fragment within the intergenic region that retained bi-directional promoter activity. In addition, a number of sequence elements have been identified that potentially regulate the BRCA1 promoter. These include a CREB/ATF-1 site that likely functions as a transcriptional enhancer [10] [37] and an E2F site that potentially mediates estrogen-dependent BRCA1 expression [38]. Regulatory elements located within noncoding regions of the BRCA1 gene have also been identified. Suen and Goss used deletion analysis to localize a 36 base pair repressor element located more than 500 base pairs into the first intron of BRCA1 [39]. The region was found to contain several GA-rich sequences that may serve as potential binding sites for the ets or Sp1 families of transcription factors. More recently, cross-species comparative sequence analysis of human and mouse intronic sequences in the BRCA1 gene identified two evolutionarily conserved noncoding sequences in intron 2, 5 kb downstream of the BRCA1 promoter [40]. The finding that BP1 binds to the first intron of BRCA1, therefore, is consistent with the results of the previous two studies that identified regulatory elements located within noncoding regions of the BRCA1 gene.
The results presented in this paper for the first time suggest that BP1 binds to the first intron of BRCA1 and negatively regulates its transcription. Since BRCA1 is developmentally expressed and is involved in cell proliferation and differentiation, it is logical that its transcription is in part regulated by a homeoprotein such as BP1.
Genomic organization of the BRCA1 promoter region and localization of the BP1 consensus binding site. The top part of the diagram shows the organization of introns and exons for the BRCA1 and NBR2 genes (marked by open and gray boxes respectively). Transcription of BRCA1 and its neighboring gene NBR2 proceeds toward the right and left sides respectively. A 56 base pair minimal region located within a 229 base pair intergenic region between BRCA1 and NBR2 functions as a minimal bi-directional promoter for the two divergently transcribed genes. The bottom part of the diagram shows the region from the beginning of intron 1 to the end of exon 2 of BRCA1. Numbering of nucleotide positions is the same as for GenBank accession number U37574. A putative repressor region located between nucleotides 2255 and 2337 is marked by a purple box and contains several GA-rich sequences that may serve as potential binding sites for the ets or Sp1 families of transcription factors. A yellow box represents exon 2 from nucleotide positions 2857 to 2956. BP1 is shown as a green oval binding to the consensus sequence ATATATATG within the first intron of BRCA1 from nucleotide positions 2830 to 2838, which is -47 to -39 base pairs upstream of the translational start site in exon 2 (marked by a yellow star).
Conclusions
Our results demonstrated that BP1, an isoform of DLX4 homeoprotein, negatively regulates the expression of BRCA1 through binding to its intron, which suggests that overexpression of BP1 might be a potential initiator to inactivate BRCA1. Therefore, targeting BP1 may provide a new avenue for breast cancer prevention and treatment.
Abbreviations
DLX4: Distal Less 4; BRCA1: Breast Cancer 1; BP1: Beta Protein 1; EMSA: Electrophoretic Mobility Shift Assay; ChIP: Chromatin Immunoprecipitation.
Competing interests
The authors declare that they have no competing interests.
Acknowledgements
We would like to thank the support from Dr. Alan Wasserman, Chairman of the Department of Medicine and Dr. Allan Goldstein, former Chairman of the Department of Biochemistry and Molecular Biology. This work was supported by NIH grant CA102928 (SWF) and the McCormick Genomics Grant (SWF).
References
1. Atlanta: American Cancer Society Inc. Society AC. http://www.cancer.org
2. Institute NC. Genetics of breast and ovarian cancer. Bethesda, MD: Institute NC. 2005
3. Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W. et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66-71
4. Smith SA, Easton DF, Evans DG, Ponder BA. Allele losses in the region 17q12-21 in familial breast and ovarian cancer involve the wild-type chromosome. Nat Genet. 1992;2(2):128-131
5. Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y. et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266(5182):120-122
6. Thompson ME, Jensen RA, Obermiller PS, Page DL, Holt JT. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat Genet. 1995;9(4):444-450
7. Wilson CA, Ramos L, Villasenor MR, Anders KH, Press MF, Clarke K, Karlan B, Chen JJ, Scully R, Livingston D. et al. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat Genet. 1999;21(2):236-240
8. Holt JT, Thompson ME, Szabo C, Robinson-Benion C, Arteaga CL, King MC, Jensen RA. Growth retardation and tumour inhibition by BRCA1. Nat Genet. 1996;12(3):298-302
9. Dobrovic A, Simpfendorfer D. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 1997;57(16):3347-3350
10. Mancini DN, Rodenhiser DI, Ainsworth PJ, O'Malley FP, Singh SM, Xing W, Archer TK. CpG methylation within the 5' regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene. 1998;16(9):1161-1169
11. Catteau A, Harris WH, Xu CF, Solomon E. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene. 1999;18(11):1957-1965
12. Baldassarre G, Battista S, Belletti B, Thakur S, Pentimalli F, Trapasso F, Fedele M, Pierantoni G, Croce CM, Fusco A. Negative regulation of BRCA1 gene expression by HMGA1 proteins accounts for the reduced BRCA1 protein levels in sporadic breast carcinoma. Mol Cell Biol. 2003;23(7):2225-2238
13. Wang C, Fan S, Li Z, Fu M, Rao M, Ma Y, Lisanti MP, Albanese C, Katzenellenbogen BS, Kushner PJ. et al. Cyclin D1 antagonizes BRCA1 repression of estrogen receptor alpha activity. Cancer Res. 2005;65(15):6557-6567
14. Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90(3):425-435
15. Andres JL, Fan S, Turkel GJ, Wang JA, Twu NF, Yuan RQ, Lamszus K, Goldberg ID, Rosen EM. Regulation of BRCA1 and BRCA2 expression in human breast cancer cells by DNA-damaging agents. Oncogene. 1998;16(17):2229-2241
16. Fan S, Twu NF, Wang JA, Yuan RQ, Andres J, Goldberg ID, Rosen EM. Down-regulation of BRCA1 and BRCA2 in human ovarian cancer cells exposed to adriamycin and ultraviolet radiation. Int J Cancer. 1998;77(4):600-609
17. MacLachlan TK, Dash BC, Dicker DT, El-Deiry WS. Repression of BRCA1 through a feedback loop involving p53. J Biol Chem. 2000;275(41):31869-31875
18. Hsu LC, White RL. BRCA1 is associated with the centrosome during mitosis. Proc Natl Acad Sci U S A. 1998;95(22):12983-12988
19. Lotti LV, Ottini L, D'Amico C, Gradini R, Cama A, Belleudi F, Frati L, Torrisi MR, Mariani-Costantini R. Subcellular localization of the BRCA1 gene product in mitotic cells. Genes Chromosomes Cancer. 2002;35(3):193-203
20. Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, Harris CC, Ried T, Deng CX. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999;3(3):389-395
21. Hsu LC, Doan TP, White RL. Identification of a gamma-tubulin-binding domain in BRCA1. Cancer Res. 2001;61(21):7713-7718
22. Schlegel BP, Starita LM, Parvin JD. Overexpression of a protein fragment of RNA helicase A causes inhibition of endogenous BRCA1 function and defects in ploidy and cytokinesis in mammary epithelial cells. Oncogene. 2003;22(7):983-991
23. Starita LM, Machida Y, Sankaran S, Elias JE, Griffin K, Schlegel BP, Gygi SP, Parvin JD. BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol Cell Biol. 2004;24(19):8457-8466
24. Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2(10):777-785
25. Chase MB, Fu S, Haga SB, Davenport G, Stevenson H, Do K, Morgan D, Mah AL, Berg PE. BP1, a homeodomain-containing isoform of DLX4, represses the beta-globin gene. Mol Cell Biol. 2002;22(8):2505-2514
26. Fu S, Stevenson H, Strovel JW, Haga SB, Stamberg J, Do K, Berg PE. Distinct functions of two isoforms of a homeobox gene, BP1 and DLX7, in the regulation of the beta-globin gene. Gene. 2001;278(1-2):131-139
27. Haga SB, Fu S, Karp JE, Ross DD, Williams DM, Hankins WD, Behm F, Ruscetti FW, Chang M, Smith BD. et al. BP1, a new homeobox gene, is frequently expressed in acute leukemias. Leukemia. 2000;14(11):1867-1875
28. Stevenson HS, Fu SW, Pinzone JJ, Rheey J, Simmens SJ, Berg PE. BP1 transcriptionally activates bcl-2 and inhibits TNFalpha-induced cell death in MCF7 breast cancer cells. Breast Cancer Res. 2007;9(5):R60
29. Fu SW, Schwartz A, Stevenson H, Pinzone JJ, Davenport GJ, Orenstein JM, Gutierrez P, Simmens SJ, Abraham J, Poola I. et al. Correlation of expression of BP1, a homeobox gene, with estrogen receptor status in breast cancer. Breast Cancer Res. 2003;5(4):R82-87
30. Sheikh MS, Garcia M, Pujol P, Fontana JA, Rochefort H. Why are estrogen-receptor-negative breast cancers more aggressive than the estrogen-receptor-positive breast cancers? Invasion Metastasis. 1994;14(1-6):329-336
31. Fu Y, Lian Y, Kim KS, Zhang L, Hindle AK, Brody F, Siegel RS, McCaffrey TA, Fu SW. BP1 Homeoprotein Enhances Metastatic Potential in ER-negative Breast Cancer. J Cancer. 2010;1(1):54-62
32. Man YG, Fu SW, Schwartz A, Pinzone JJ, Simmens SJ, Berg PE. Expression of BP1, a novel homeobox gene, correlates with breast cancer progression and invasion. Breast Cancer Res Treat. 2005;90(3):241-247
33. Song Y, Dang C, Fu Y, Lian Y, Hottel J, Li X, McCaffrey T, Fu SW. Genome-wide analysis of BP1 transcriptional targets in breast cancer cell line Hs578T. Int J Biol Sci. 2009;5(1):1-12
34. Man YG, Ball WD, Culp DJ, Hand AR, Moreira JE. Persistence of a perinatal cellular phenotype in submandibular glands of adult rat. J Histochem Cytochem. 1995;43(12):1203-1215
35. Man YG. A simple epitope retrieval method without the use of microwave oven or enzyme digestion. Applied Immunohistochemistry. 1996;4:139-141
36. Suen TC, Goss PE. Transcription of BRCA1 is dependent on the formation of a specific protein-DNA complex on the minimal BRCA1 Bi-directional promoter. J Biol Chem. 1999;274(44):31297-31304
37. Thakur S, Croce CM. Positive regulation of the BRCA1 promoter. J Biol Chem. 1999;274(13):8837-8843
38. Wang A, Schneider-Broussard R, Kumar AP, MacLeod MC, Johnson DG. Regulation of BRCA1 expression by the Rb-E2F pathway. J Biol Chem. 2000;275(6):4532-4536
39. Suen TC, Goss PE. Identification of a novel transcriptional repressor element located in the first intron of the human BRCA1 gene. Oncogene. 2001;20(4):440-450
40. Wardrop SL, Brown MA. Identification of two evolutionarily conserved and functional regulatory elements in intron 2 of the human BRCA1 gene. Genomics. 2005;86(3):316-328
Author contact
![]() Corresponding author: Sidney W. Fu, M.D., Ph.D., Associate Professor, Dept. of Medicine, Division of Genomic Medicine, The George Washington University Medical Center, 2300 I Street, N.W. Room 523A, Washington, DC 20037. Email: sfuedu; 202-994-4767 (Office), 202-994-0691 (Lab); Fax: 202-994-2487
Corresponding author: Sidney W. Fu, M.D., Ph.D., Associate Professor, Dept. of Medicine, Division of Genomic Medicine, The George Washington University Medical Center, 2300 I Street, N.W. Room 523A, Washington, DC 20037. Email: sfuedu; 202-994-4767 (Office), 202-994-0691 (Lab); Fax: 202-994-2487

 Global reach, higher impact
Global reach, higher impact