Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2010; 6(7):614-626. doi:10.7150/ijbs.6.614 This issue Cite
Research Paper
Antennal expression pattern of two olfactory receptors and an odorant binding protein implicated in host odor detection by the malaria vector Anopheles gambiae
1. University of Hohenheim, Institute of Physiology, Stuttgart, Germany
2. Insect Molecular Genetics and Biotechnology Group, Institute of Biology, National Centre for Scientific Research ”Demokritos”, Athens, Greece
3. Institute of Biology, Faculty of Science, University Neuchâtel, Neuchâtel, Switzerland
4. Present address: Bayer CropScience, Monheim, Germany
Received 2010-8-30; Accepted 2010-10-6; Published 2010-10-8
Abstract
Odor-detection in the malaria mosquito Anopheles gambiae involves large families of diverse proteins, including multiple odorant binding proteins (AgOBPs) and olfactory receptors (AgORs). The receptors AgOR1 and AgOR2, as well as the binding protein AgOBP1, have been implicated in the recognition of human host odors. In this study, we have explored the expression of these olfactory proteins, as well as the ubiquitous odorant receptor heteromerization partner AgOR7, in the thirteen flagellomeres (segments) of female and male antenna. Expressing cells were visualized by adapting a whole mount fluorescence in situ hybridization method. In female mosquitoes, AgOR1-expressing olfactory receptor neurons (ORNs) were almost exclusively segregated in segments 3 to 9, whereas AgOR2-expressing ORNs were distributed over flagellomeres 2 to 13. Different individuals comprised a similar number of cells expressing a distinct AgOR type, although their antennal topography and number per flagellomere varied. AgOBP1-expressing support cells were present in segments 3 to 13 of the female antenna, with increasing numbers towards the distal end. In male mosquitoes, total numbers of AgOR- and AgOBP1-expressing cells were much lower. While AgOR2-expressing cells were found on both terminal flagellomeres, AgOR1 cells were restricted to the most distal segment. High densities of AgOBP1-expressing cells were identified in segment 13, whereas segment 12 comprised very few. Altogether, the results demonstrate that both sexes express the two olfactory receptor types as well as the binding protein AgOBP1 but there is a significant sexual dimorphism concerning the number and distribution of these cells. This may suggest gender-specific differences in the ability to detect distinct odorants, specifically human host-derived volatiles.
Keywords: olfaction, odorant receptor proteins, odorant binding proteins, mRNA expression, sexual dimorphism
Introduction
Millions of people suffer from malaria causing severe illness and death to affected humans. The major vector for transmission of the malaria parasite Plasmodium falciparum is the afrotropical mosquito Anopheles gambiae, causally linked to the requirement of a blood meal by female mosquitoes to complete their gonadotrophic cycle. While female Anopheles rely on their sense of smell to find a blood host in addition to sugar providing plants and appropriate oviposition sites [1], the nectar feeding male mosquitoes mainly use their olfactory system to locate host plant odors [2, 3]. However, males of various blood-sucking mosquito species are known to also respond to odors emanating from hosts of the females [2], which may allow them to find their mating partners at the host location.
Insects detect and discriminate volatile odorants by means of olfactory receptor neurons (ORN) located in sensory structures called olfactory sensilla. In A. gambiae, the majority of olfactory sensilla populate the antennae [4], whereas much lower numbers are located on the other two olfactory appendages, the maxillary palps and the proboscis [5]. Within a sensillum, ORNs are accompanied by several support cells, which secrete odorant binding proteins (OBPs) into the sensillum lymph [6, 7]. OBPs capture odorants upon entering a sensillum through cuticle pores and transfer the signal molecules through the sensillum lymph towards olfactory receptors (ORs) in the dendritic membrane of the ORNs. Large and diverse gene families encoding 57 candidate odorant binding proteins (AgOBPs) [8-11] and 79 putative odorant receptors (AgORs) [12-14] have been annotated from bioinformatic screens of the A. gambiae genome. This high number and diversity of AgOBPs and AgORs suggests specific roles of these proteins in the detection of distinct odorants or odorant classes. In support of this notion, recent functional analysis of the majority of the AgORs revealed a large diversity in their response spectra; while some receptors respond to single or a small number of odorants, others are rather broadly tuned [15-17]. For a few AgORs, it has been indicated that they may play a critical role in host-seeking behaviour and host selection. For example, AgOR1, which responds to the human sweat component 4-methylphenol [15], is selectively expressed in females [13, 18] and down-regulated after a blood meal [13]. Similarly, AgOR2 was found to be predominantly expressed in the female antenna [18] and narrowly tuned to a small set of aromatics including indole [16, 17]. Indoles and phenols are major constituents of the volatile headspace of human sweat [19], but occur also at oviposition sites [20] and in nectar sources [21]. Interestingly, indole and 3-methylindole were recently identified as ligands for the odorant binding protein AgOBP1 and electroantennogram recordings in RNAi knock-down experiments have shown that a proper expression of AgOBP1 is required for an electrophysiological response to the compounds [22].
ORs are expressed by ORNs and OBPs by support cells. Knowledge on the number and topography of these cells in the antennae may give first valuable information on the importance of candidate recognition proteins for the detection of behaviourally relevant odorants. The number and topography of cells expressing pheromone receptors or binding proteins involved in detection of distinct pheromone components have recently been determined by in situ hybridization studies on moth antennae [23-26, 26, 27]. In view of the general lack of established in situ hybridization protocols in A. gambiae, the expression of the different AgORs or AgOBPs in the antenna has not been assessed yet. In this study we set out to adapt a whole mount fluorescence in situ hybridization (WM-FISH) method successfully used previously with moth antennae [24] to the antenna of A. gambiae. Focussing on olfactory proteins possibly involved in the detection of human host odors, we visualized the cells with transcripts for AgOR1, AgOR2 and AgOBP1 and determined their topographic distribution in the antennae of female and male A. gambiae. In addition, the cells bearing transcripts for AgOR7, representing the ubiquitous heteromerization partner of A. gambiae ORs [28, 29] were localized.
Materials and Methods
Animal rearing and tissue preparation
Eggs and larvae of the Anopheles gambiae (Giles) s.s. strain Kisumu were kindly provided by Bayer CropScience, Monheim, Germany. The laboratory strain was originally derived from the region of Kisumu, Kenya. Animals were reared to adults at the University of Hohenheim at 28°C with a day-night cycle of 12:12. Anopheles gambiae (Giles) s.s. strain 16CSS was originally derived from Lagos, Nigeria. Animals were reared at the University of Neuchâtel as described earlier [30] and were transferred to Hohenheim for in situ hybridization experiments. After emergence, animals had access to 10% sucrose ad libitum. For in situ hybridization, one to 3-days old mosquitoes of A. gambiae strain Kisumu were used. Animals of strain 16CSS were 8 - 11 days old.
Whole mount fluorescence in situ hybridization (WM-FISH)
All incubations and washes were made in a volume of 0.25 ml in thin walled PCR tubes (Kisker, Germany) with slow rotation on an overhead shaker, in a hybridization oven or by shaking moderately on a heating block. Antennae were dissected from cold anesthetized animals and transferred to 4% paraformaldehyde in 0.1 M NaCO3, pH 9.5, 0.03% Triton X-100. Antennae were fixed for 20 - 24 hours at 6°C followed by a wash at room temperature for 1 min in PBS (phosphate-buffered saline = 145 mM NaCl, 1.4 mM KH2PO4, 8 mM Na2HPO4, pH 7.1) containing 0.03% Triton X-100. Subsequently, antennae were incubated for 10 min in 0.2 M HCl, 0.03% Triton X-100 and washed for 2 min in PBS with 1% Triton X-100. Antennae were transferred to whole mount in situ hybridization solution (50% formamide, 5xSSC, 1xDenhardt's reagent, 50 µg/ml yeast RNA, 1% Tween 20, 0.1% Chaps, 5 mM EDTA pH 8.0) and directly subjected to prehybridization or stored for up to 11 days at 6°C in the solution. Storage did not cause any obvious loss of signal intensity in the following WM-FISH procedure. Prehybridization was performed at 55°C for 6 hours. After this, antennae were incubated for at least 48 hours at the same temperature in hybridization solution containing the digoxigenin (DIG)-labelled antisense RNA probe. Post-hybridization, the antennae were washed four times for 15 min each in 0.1xSSC, 0.03% Triton X-100 at 60°C and then treated with 1% blocking reagent (Roche) in TBS (100 mM Tris, 150 mM NaCl, pH 7.5), 0.03% Triton X-100 for 5 hours at 6°C. DIG-labelled probes were detected by incubation for at least 48 hours with an anti-DIG AP-conjugated antibody (Roche) diluted 1:500 in TBS, 0.03% Triton X-100 with 1% blocking reagent. After washing five times for 10 min in TBS with 0.05% Tween at room temperature, DIG-labelled probes were visualized by incubation in the dark for 5 hours with HNPP (2-hydroxy-3-naphtoic acid-2'-phenylanilide phosphate, Roche) 1:100 in DAP-buffer (100 mM Tris, 100 mM NaCl, 50 mM MgCl2, pH 8.0) at 6°C. After a short wash in PBS, antennae were mounted in Moviol (10% polyvinylalcohol 4-88, 20% glycerol in PBS).
Preparation of in situ hybridization probes
DIG-labelled antisense or sense riboprobes for AgOR1, AgOR2, AgOR7 [13, 28, 29] and AgOBP1 [11] were transcribed from linearized recombinant pGem-T Easy and Bluescript plasmids containing the coding regions of the OBP and OR genes. RNAs were transcribed using a T3/T7 RNA transcription system (Roche) following recommended protocols. Receptor RNA probes were subsequently fragmented to an average length of about 600 bp by incubation in carbonate buffer (80 mM NaHCO3 120 mM Na2CO3, pH 10.2) following the protocol of [31].
Data analysis
Antennae were analyzed on a Zeiss LSM510 Meta laser scanning microscope (Zeiss, Oberkochen, Germany). Figures were arranged using appropriate graphic programs. Images were not altered except to adjust the brightness or contrast for uniform tone within a single figure. For quantifying the number of AgOR1- and AgOR2-expressing cells along the antennae, the labelled cells in each segment were counted under confocal microscope inspection.
Results
In A. gambiae the antennae of both sexes consist of 13 flagellomeres (segments) but otherwise are sexually dimorphic (Fig. 1A and Fig. 3A). In female mosquitoes, each antenna carries around 750 chemosensory sensilla distributed over all 13 segments. In contrast, the antenna of males comprises only approximately 250 chemosensory sensilla restricted to the distal two segments (12 and 13) [5]. Segments 1 - 11 are populated by long bristles (fibrillae) (Fig. 3A) supposed to function as auditory sensors tuned to detect wing beat sounds of conspecific females [32]. In this study, we set out to determine the number and topographic distribution of the cells, which express the olfactory receptor types AgOR1, AgOR2, AgOR7 and the odorant binding protein AgOBP1, in the antenna of female and male A. gambiae. Towards this goal, we adapted a whole mount fluorescent in situ hybridization (WM-FISH) protocol, which allowed us to visualize the expressing cells by means of confocal laser scanning microscopy (LSM) and to analyze their localization within the antenna.
Expression of AgOR1 in the antenna of female mosquitoes
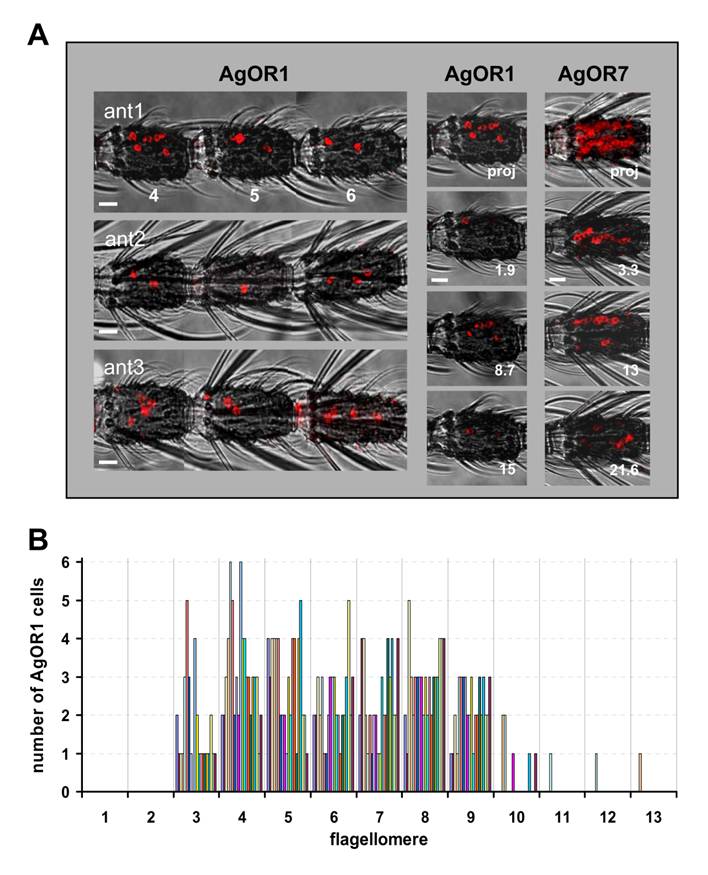
In the WM-FISH experiments AgOR1-positive cells were very rarely observed in segments 1, 2 and 10 to 13. Figures 1B and 1C illustrate segments 3 - 9, in which AgOR1-expressing cells in female antennae were regularly visualized. No differences were found between strains (Fig. 1B: Kisumu, Fig. 1C: 16CSS). At higher magnification figure 1D shows that the soma region of AgOR1-positive cells is stained, whereas the nucleus appears unstained. This cellular labelling is typical for FISH-experiments with olfactory receptor probes [23, 24]. AgOR1 expression was variable within a segment, both longitudinally as well as on the circumference (Fig. 2A, left panel). In addition, the number of AgOR1-expressing cells varied between segments and between individual antennae (Table 1 and Fig. 2B). This variability was seen in both A. gambiae strains tested. Expression was restricted mostly to segments 3 - 9 with generally between 1 - 3 stained cells per segment, although sometimes up to 6 stained cells were found. Segment 10 occasionally expressed 1 - 2 AgOR1 cells, whereas only rarely did segments 11 - 13 contain a single stained cell (Fig. 2B). One antenna contained on average 18.1 (±3.5, n=20) labelled cells (Table 1). AgOR1-expressing cells were found at different optical planes of a confocal LSM image stack (Fig. 2A, middle panel), indicating different positions on the circumference of the antenna. Occasionally, labelled cells appeared on opposite sites of the antennal segments (not shown), demonstrating that the applied method allowed detection of labelled cells in all the width of the antenna.
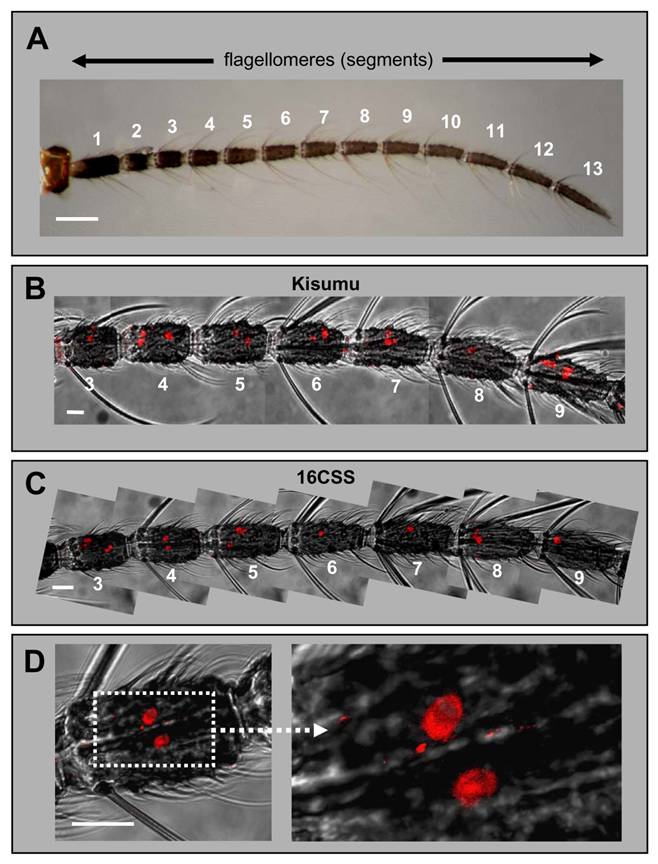
Expression of AgOR1 in the female antenna. A, Brightfield image of an antenna from an A. gambiae Kisumu adult female. The 13 flagellomeres are numbered from proximal (1) to distal (13). Scale bar 100 µm. B - D, WM-FISH with female antennae using an AgOR1-specific DIG-labelled antisense RNA probe. Receptor-expressing cells were visualized by red fluorescence. Pictures, which show more than one flagellomere were assembled from several images, each representing the projection of single images from a confocal image stack. The red fluorescence channel has been overlaid with the transmitted-light channel. Scale bars 20 µm. B, WM-FISH employing a probe for AgOR1 on an antenna of A. gambiae Kisumu. Segments 3 to 9 are shown. The AgOR1 probe labelled individual cells on each segment. C, AgOR1-expressing cells on segments 3 to 9 of an A. gambiae strain 16CSS antenna. D, Higher magnifications of segment 4 shown in C.
Topography of AgOR1-expressing cells on the female antenna. A, WM-FISH using a DIG-labelled AgOR1 antisense RNA probe and visualization by red fluorescence. The red fluorescence channel has been overlaid with the transmitted-light channel. Scale bars 10 µm. Left panel, AgOR1-expressing cells on segments 4, 5 and 6 of individual antennae from A. gambiae strain 16CSS (ant1) or Kisumu (ant2, ant3). Pictures were assembled from images of single segments. In corresponding segments the AgOR1 probe labelled a different number of cells, located at variable positions of the longitudinal axis of the antenna. Middle panel, projection (proj) and three single optical planes of the confocal image stack of segment 4 from antenna 1. Individual cells labelled by the AgOR1 riboprobe are detectable in different optical depths (numbers in µm), indicating different positions on the segment circumference. Right panel, projection of a confocal image stack and single optical planes showing labelled cells in a fourth antennal segment of an antenna, which was hybridized with a probe for AgOR7. B, Number of AgOR1-expressing cells on different antennal segments. The number of cells on each of the 13 flagellomeres was determined for 20 antennae. Bars of the same colour represent cells counted from one antenna.
Number and distribution of AgOR1- and AgOR2-expressing cells in the antennae of female and male A. gambiae strain Kisumu.
| flagellomere | AgOR1 | AgOR2 | ||
|---|---|---|---|---|
| ♀ (n=20) | ♂ (n=35) | ♀ (n=18) | ♂ (n=13) | |
| 1 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0.2 ±0.4 | 0 |
| 3 | 1.7 ±1.2 | 0 | 0.8 ±0.4 | 0 |
| 4 | 3.2 ±1.3 | 0 | 0.8 ±0.4 | 0 |
| 5 | 2.9 ±1.2 | 0 | 0.8 ±0.6 | 0 |
| 6 | 2.4 ±0.9 | 0 | 0.9 ±0.5 | 0 |
| 7 | 2.4 ±1.1 | 0 | 0.9 ±0.6 | 0 |
| 8 | 2.9 ±0.9 | 0 | 0.9 ±0.5 | 0 |
| 9 | 2.2 ±0.8 | 0 | 1.2 ±0.6 | 0 |
| 10 | 0.4 ±0.7 | 0 | 0.9 ±0.6 | 0 |
| 11 | 0.1 ±0.2 | 0 | 1 ±0.5 | 0 |
| 12 | 0.1 ±0.2 | 0 | 1.2 ±0.6 | 2.1 ±0.9 |
| 13 | 0.1 ±0.2 | 3.5 ±1 | 0.6 ±0.5 | 2.3 ±0.7 |
| Average number of cells/antenna | 18.1 ±3.5 | 3.5 ±1 | 10.5 ±1.5 | 4.4 ±1 |
Numbers represent the mean (±SD) number of AgOR1- and AgOR2-expressing cells in each of the 13 flagellomeres and in the complete antenna of female and male mosquitoes, respectively.
We further confirmed the consistency of WM-FISH labelling using a labelled antisense riboprobe specific to AgOR7, the A. gambiae orthologue of the Drosophila melanogaster olfactory co-receptor OR83b. A high number of labelled cells was found throughout each of the antennal segments 2 - 13. Consistent with an increase in the number of chemosensory sensilla from proximal to distal, more distal segments (6 - 13) contained considerably higher numbers of AgOR7 cells than proximal ones (2 - 5) (Fig. 2A, right panel). In control experiments employing a sense riboprobe for AgOR7 we never detected any stained cells (not shown). As illustrated by images taken from a fourth antennal segment (Figure 2A, right panel), cells labelled by the antisense riboprobe could be visualized below the antennal surface in different optical depths. This indicates AgOR7-expressing cells all around the circumference of the segment, resembling the arrangement of chemosensory sensilla on the antenna [4].
Expression of AgOR1 in the male antenna
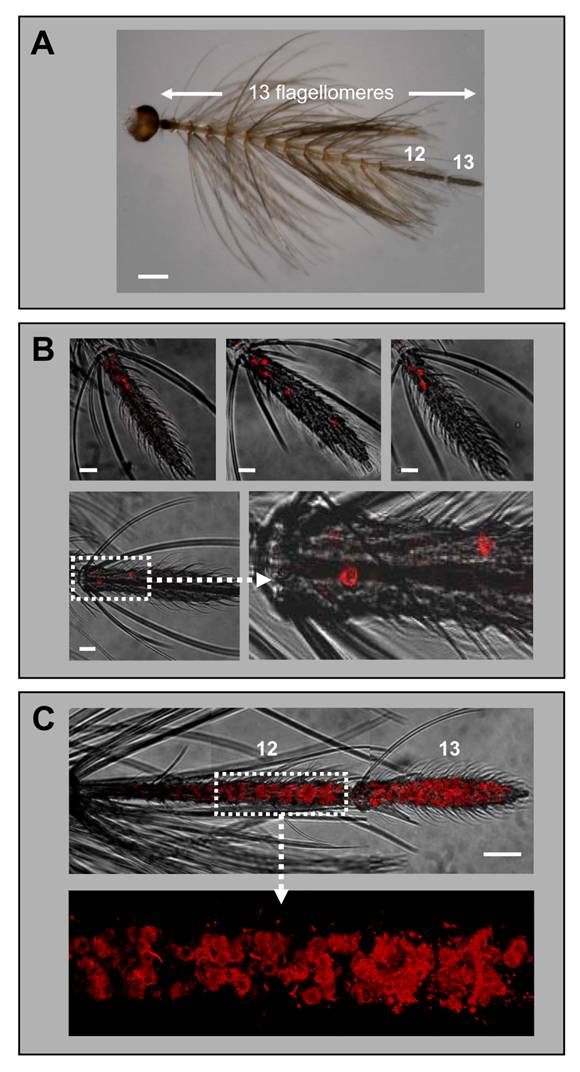
While earlier RT-PCR studies reported a female-specific expression of the AgOR1 gene [13], a more recent report has indicated a low level of AgOR1 transcripts also in male antennae [18]. Applying WM-FISH to complete male antennae of A. gambiae strain Kisumu, we routinely found 3 or 4 AgOR1-expressing cells, which were located only on the most distal flagellomere (Fig. 3B, upper panel). Less frequently 2, 5 or 6 cells were detected. The same result was obtained using antennae of A. gambiae strain 16CSS (Fig. 3B, lower panel). The average number of AgOR1-expressing cells on the male antenna was 3.5 (±1.0, n=35) (Table 1). Within the thirteenth flagellomere, AgOR1-expressing cells showed no stereotypic topography, but were most often located closer to its proximal end. To show that the applied WM-FISH method would allow labelling of receptor expressing cells all along segments 12 and 13, we employed the AgOR7-specific riboprobe (Fig. 2C). In agreement with the expression of AgOR7 in most ORNs we visualized a very high number of AgOR7-expressing cells along the entire longitudinal axis and the circumference of the last two distal flagellomeres. No labelled cells were found in the segments 1 - 11 (not shown).
Expression of AgOR2 in female and male antennae
Next we applied WM-FISH to determine the number and distribution of AgOR2-expressing ORNs in the antenna of A. gambiae strain Kisumu. In female antennae a riboprobe specific to AgOR2 routinely labelled one or two cells in segments 3 - 13 (Fig. 4A and 4B). Occasionally a segment within this antennal stretch carried no AgOR2 cell. Two AgOR2 cells were irregularly detected in segments 5 - 12. AgOR2-expressing cells were never found in segment 1 and rarely present in segment 2. The average number of AgOR2-expressing cells per antenna was 10.5 (±1.5, n=18), thus compared to AgOR1 a lower number of ORNs express AgOR2 (Table 1).
Expression of AgOR1 and AgOR7 in the antennae of males. A, Antenna of an A. gambiae Kisumu adult male. Of the 13 flagellomeres, only the last two distal segments (12 and 13) carry olfactory sensilla. Brightfield image. Scale bar 200 µm. B, Expression of AgOR1 in the male antenna. WM-FISH using antennae of A. gambiae strain Kisumu (upper three pictures) and strain 16CSS (image below with higher magnification of the boxed area). Pictures represent projections of confocal image stacks. Several labelled cells are visible only on segment 13. Scale bars 20 µm. C, WM-FISH on an A. gambiae strain 16CSS antenna employing a probe for AgOR7. A very large number of cells expressing AgOR7 are visualized by red fluorescence on segments 12 and 13. The picture was assembled from several images representing projections of confocal image stacks. The transmitted-light and red fluorescence channels have been overlaid. The boxed area is shown at higher magnification with only the red fluorescence channel displayed. Scale bar 50 µm.
Expression of AgOR2 in the antenna of female and male mosquitoes. DIG-labelled AgOR2 antisense RNA was used in WM-FISH to probe antennae of A. gambiae strain Kisumu. AgOR2-expressing cells were visualized by red fluorescence. Pictures of the antennae were assembled from single images representing projections of confocal stacks. The red fluorescence and the transmitted-light channel have been overlaid. A, WM-FISH employing the antenna of a female. One or two AgOR2-expressing cells are found on each of the segments shown. Scale bar 20 µm. B, Number of AgOR2-expressing cells on the different segments of female antennae. The number of cells on each of the 13 flagellomeres was determined for 18 antennae. Bars of the same colour represent the same antenna. C, Expression of AgOR2 in the antenna of a male. Two red-labelled AgOR2 cells are visible on each of the two distal segments. Scale bar 50 µm.
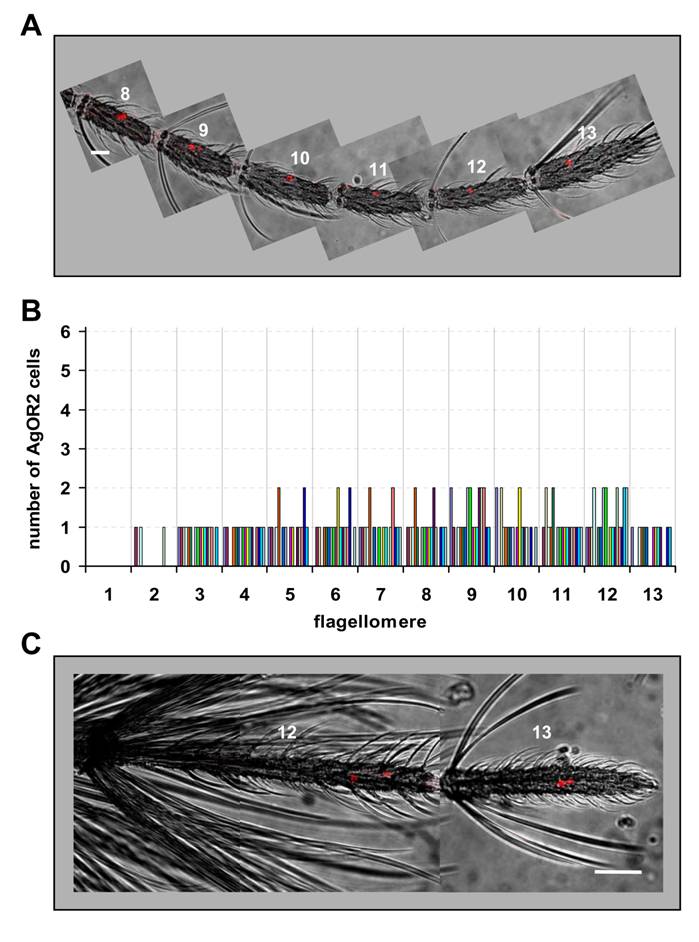
In contrast to AgOR1, cells expressing AgOR2 were found in both distal flagellomeres of the male antenna (Fig. 4C). One to three AgOR2 positive cells were regularly visualized in each of the segments 12 and 13. On average, 4.4 (±1.2, n=13) cells per male antenna were counted (Table 1). A similar result was obtained when antennae of A. gambiae strain 16CSS were used (not shown).
Expression of AgOBP1 in the female and male antenna
Employing a probe specific to AgOBP1 in WM-FISH experiments with female antennae, we visualized AgOBP1 expression in the flagellomeres 3 - 13 (Fig. 5).
AgOBP1 expression in female and male antennae. WM-FISH using a probe for AgOBP1 on antennae of A. gambiae strain Kisumu. A, female antenna. Many AgOBP1-expressing cells are found on each of the flagellomeres 8 - 13. B, AgOBP1-positive cells on segments 3, 5 and 13 of a female antenna. The boxed areas are shown at higher magnification with only the red fluorescence channel displayed. C, Expression of AgOBP1 in the antenna of a male. Many red-labelled AgOBP1 cells are visible in segment 13, whereas only few positive cells were found on segment 12. Pictures in A and C were assembled from single images representing projections of selected images from confocal stacks. The red fluorescence and the transmitted-light channel have been overlaid. Scale bars 20 µm in A and C, 10 µm in B.
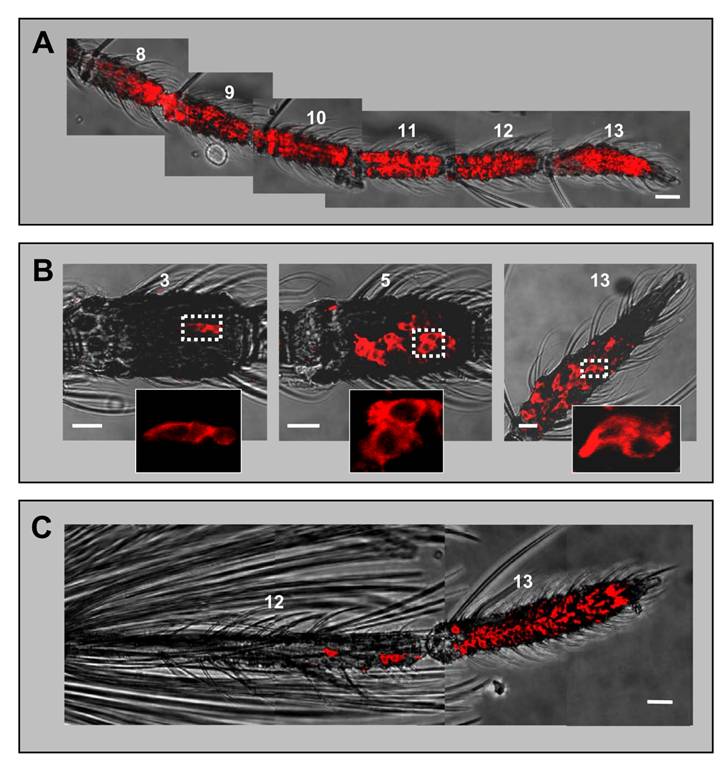
The shape of the FISH signal (flat and extended) was typical for labelling of support cells with binding protein-specific probes [23, 33] and was different from the labelling of ORNs generated by receptor-specific probes (round shape, Figs. 1 and 3). The highest numbers of AgOBP1-expressing cells were detected on more distal segments (Fig. 5A), while proximal segments (3 - 6) comprised reduced cell numbers with only a few labelled cells found in segment 3 (Fig. 5B). Consistent with the expression of binding proteins in only two of the three support cells of a chemosensory sensillum [7], the AgOBP1 riboprobe regularly visualized pairs of red-labelled cells (Fig. 5B). In male antenna large numbers of AgOBP1-expressing cells were found in the terminal segment, whereas segment 12 contained a rather low number (Fig. 5C). AgOBP1 cells were never detected in segments 1 - 11. Due to the high number and density of labelled cells in most fragments of the female and male antenna as well as the extended shape of the cellular labelling and overlap of the staining, a reliable determination of the total number of AgOBP1-expressing cells in the female and male antennae was not possible.
Discussion
In this study, we have explored the topography of cells expressing the olfactory receptor types AgOR1, AgOR2 and the odorant binding protein AgOBP1 in the antenna of A. gambiae. In the female antenna, OBP- and OR-expressing cells are located in about 750 chemosensory sensilla of four different types [4]. Each sensillum houses two to three ORNs summing up to a total estimated number of about 1500 - 1600 ORNs per antenna [34]. Of the 79 AgOR genes identified, 59 AgORs are expressed in the female antenna [18]. Assuming that an individual ORN expresses only one type of OR and that each receptor is expressed in an equal number of neurons, a defined AgOR would be expressed in about 25 ORNs. However, RT-PCR studies have detected differences in the level of antennal transcripts for the various AgORs [18], suggesting differences in the number of ORNs expressing individual receptor types. In support of this notion we have identified ~20 cells that express AgOR1 and ~10 cells that express AgOR2 per female antenna. Moreover, the observed labelling patterns indicate that distinct AgORs are expressed in only one of the 2-3 ORNs of an individual sensillum. Thus, the AgOR expression patterns in A. gambiae appear to be similar to Drosophila, where the repertoire of ORs is expressed in subsets of 2 to 50 ORNs per antenna and distinctive ORNs within a sensillum [35-37].
Only female mosquitoes take a blood meal and their antennae are populated with about three times as many olfactory sensilla as male antenna [5]. Therefore, female antennae are supposed to harbour significantly more sensilla that are tuned to detect human odors than males. With respect to this notion, the proposed role of AgOR1, AgOR2 and AgOBP1 in the detection of human host odorants [15, 16, 22] coincides with the higher number of cells expressing these proteins in the antenna of female mosquitoes compared to males. It remains to be seen whether a lower number of AgOR- and AgOBP-expressing cells in males affects the detection thresholds for odorants. Likewise, it is possible that in male mosquitoes a reduced number of odorant detection units on the antenna is compensated for more centrally in the antennal lobe or higher brain centers. However, it seems puzzling why males, which are solely nectar-feeders, should have the capacity to detect human host odorants. A possible explanation may be that odorants, which attract females may also attract males and thus lure them to the same location, a scenario, which may promote reproductive success. The mating biology of Anophelines is poorly understood [2, 38] and involvement of semiochemicals remains to be shown. Alternatively, ORs and OBPs, which participate in recognition of host odorants by the females may be used by males in a different context, for example to detect indole present in nectar sources [21].
Concerning the total number of AgOR1- and AgOR2-expressing cells, we found little variation between A. gambiae individuals. However, for both receptor types the number of AgOR-expressing cells in a given antennal segment was variable. In addition, the cells were not located on defined positions around the circumference and the longitudinal axis of a segment (Fig. 2). Consequently, our results for AgOR1 and AgOR2 indicate that chemosensory sensilla of distinct functional identity appear to be randomly distributed on the surface of certain flagellomeres. This notion is in agreement with recent electrophysiological recordings from olfactory sensilla on the antennae of female A. gambiae. Based on the response to odorants present in human emanations, the recordings identified different types of chemosensory sensilla, but did not find any evidence for a defined distribution pattern of chemospecific sensilla across an antennal segment [34].
The principles and mechanisms determining the number and the topography of receptor-specific ORNs on the mosquito antenna are unknown. In Drosophila, various transcription factors and regulatory elements in OR genes appear to control OR expression in subsets of ORNs located in certain sensillum types, specific areas of the antenna and distinct olfactory appendages [39-41]. It can be assumed that similar factors and mechanisms underlie the complex OR expression patterns in the mosquito antenna and dictate the expression of AgOR1 and AgOR2 in subpopulations of ORNs and circumscribed segments. However, the final positioning of ORNs expressing a distinct receptor on the circumference of the segments seems to have a stochastic component. In this respect, the antenna of A. gambiae is reminiscent of the olfactory epithelium of mammals, in which a given OR is expressed in a subset of ORNs localized in a broad but circumscribed zone. Within a zone, however, the receptor-expressing ORNs appear randomly dispersed [42-45].
Previous studies on moths [46-48] and Drosophila [49, 50] have indicated that an interplay between suitable OBP-subtypes in the sensillum lymph and receptor subtypes in the dendritic membrane of sensory neurons is essential for a sensitive and specific reception of pheromones. It is presently unclear to which extent a similar interplay of distinct AgOBP/AgOR pairs may be necessary for a sensitive and selective detection of host odorants. AgOBP1 is expressed in numerous support cells in female and male antennae. This implies that the “indole-binding protein” AgOBP1 [22] plays a role in olfactory sensilla with different functions and that it may co-operate with various AgOR-types in the detection of indole and/or other odorants. In fact, in recordings from sensilla trichodea of the A. gambiae antenna, different classes of ORNs have been identified, which displayed distinct but overlapping odorant response spectra and also responded to indole [34]. Moreover, recent functional characterization of the AgOR repertoire in Xenopus oocytes [17] and in the “empty neuron” system of Drosophila [16] revealed several AgOR-types including AgOR2 with distinct but overlapping ligand spectra, including indole. Based on these findings, it may be possible that AgOBP1 binds indole as well as a spectrum of other odorants and transports them to AgOR2 and other receptor types. This would imply a minor contribution of the binding protein to the selectivity of the odorant detection system. However, AgOBP1 in functionally different sensilla may co-localize with other AgOBPs. In such sensilla, AgOBP1 may specifically transfer indole to a receptor, while other AgOBP-types deliver other odorants. In this respect, it is interesting that AgOBP1 shares significant similarity to the Drosophila OBPs OS-E and OS-F [8], which are co-expressed with an additional OBP-type in the same sensillum [51]. Moreover, in a first set of double WM-FISH experiments we found a partial overlap in the expression of AgOBP1 and AgOBP4 [52]. Additionally, it is possible that AgOBPs may act as dimers. In fact, recent studies have indicated that certain AgOBPs may form homodimers and heterodimers with various other AgOBPs. AgOBP1 and AgOBP4 were also capable of interacting with each other [53]. For insect as well as vertebrate OBPs [54, 55], dimerization has been proposed to create new binding sites at the interface between the two subunits. In this way, AgOBP1 located in functionally different sensilla may interact with different AgOBPs and thereby form dimers.
Here we could show that WM-FISH approaches in combination with laser scanning microscopy allow the visualization of AgOBP- and AgOR-expressing cells in the mosquito antenna. This procedure should now permit us to explore the expression pattern of the whole OBP- and OR-repertoire of A. gambiae. Furthermore, extensive application of double WM-FISH approaches, employing differentially labelled probes and fluorophore-detector ranges, may allow the identification of combinations of AgOBPs that are co-expressed in the support cells of individual olfactory sensilla. In the same way, the combination of AgOBP- and AgOR-specific probes will open the possibility to evaluate if cells expressing distinct protein subtypes are located in the same sensillum. Such a survey would provide a first clue for OBP and OR subtypes, which may interact in the process of detecting and discriminating distinct odorants.
Acknowledgements
We are grateful to Dr. Günther Nentwig, Manfred Arnold and Jörg Egger (Bayer CropScience, Monheim, Germany) and to Prof. Patrick M. Guerin (Institute of Biology, University of Neuchâtel, Switzerland) for collaboration and providing Anopheles gambiae eggs, larvae and adults. Drs. Dan Woods (Inscent, Inc., Irvine, CA) and Marika Walter (University of California, Irvine, CA) are acknowledged for providing us with an AgOBP1 cDNA clone and the antennal cDNA library used for the isolation of AgOR1, AgOR2 and AgOR7 sequences. We also thank Nelli Dick for excellent technical assistance.
This work was supported by grants from the European Community´s Seventh Framework Programme (FP7/2007-2013) to J.K. and K.I. under agreement FP7-222927.
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Smallegange RC, Qiu YT, van Loon JJ, Takken W. Synergism between ammonia, lactic acid and carboxylic acids as kairomones in the host-seeking behaviour of the malaria mosquito Anopheles gambiae sensu stricto (Diptera: Culicidae). Chem. Senses. 2005;30:145-52
2. Takken W, Knols BG. Odor-mediated behavior of Afrotropical malaria mosquitoes. Annu. Rev. Entomol. 1999;44:131-57
3. Foster WA. Mosquito sugar feeding and reproductive energetics. Annu. Rev. Entomol. 1995;40:443-74
4. Pitts RJ, Zwiebel LJ. Antennal sensilla of two female anopheline sibling species with differing host ranges. Malaria J. 2006;5:26
5. McIver SB. Sensilla of mosquitoes (Diptera: Culicidae). J. Med. Entomol. 1982;19:489-535
6. Steinbrecht RA, Gnatzy W. Pheromone receptors in Bombyx mori and Antheraea pernyi. I. Reconstruction of the cellular organization of the sensilla trichodea. Cell Tissue Res. 1984;235:25-34
7. Steinbrecht RA, Ozaki M, Ziegelberger G. Immunocytochemical localization of pheromone-binding protein in moth antennae. Cell Tissue Res. 1992;270:287-302
8. Xu PX, Zwiebel LJ, Smith DP. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol. Biol. 2003;12:549-60
9. Vogt RG. Odorant binding protein homologues of the malaria mosquito Anopheles gambiae; possible orthologues of the OS-E and OS-F OBPs of Drosophila melanogaster. J. Chem. Ecol. 2002;28:2371-6
10. Zhou JJ, Huang W, Zhang GA, Pickett JA, Field LM. "Plus-C" odorant-binding protein genes in two Drosophila species and the malaria mosquito Anopheles gambiae. Gene. 2004;327:117-29
11. Biessmann H, Walter MF, Dimitratos S, Woods D. Isolation of cDNA clones encoding putative odourant binding proteins from the antennae of the malaria-transmitting mosquito, Anopheles gambiae. Insect Mol Biol. 2002;11:123-32
12. Fox AN, Pitts RJ, Zwiebel LJ. A cluster of candidate odorant receptors from the malaria vector mosquito, Anopheles gambiae. Chem. Senses. 2002;27:453-9
13. Fox AN, Pitts RJ, Robertson HM, Carlson JR, Zwiebel LJ. Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc. Natl. Acad. Sci. USA. 2001;98:14693-7
14. Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, Cravchik A, Collins FH, Robertson HM, Zwiebel LJ. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176-8
15. Hallem EA, Nicole FA, Zwiebel LJ, Carlson JR. Olfaction: mosquito receptor for human-sweat odorant. Nature. 2004;427:212-3
16. Carey AF, Wang G, Su CY, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464:66-71
17. Wang G, Carey AF, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4418-23
18. Iatrou K, Biessmann H. Sex-biased expression of odorant receptors in antennae and palps of the African malaria vector Anopheles gambiae. Insect Biochem. Mol. Biol. 2008;38:268-74
19. Meijerink J, Braks MA, Brack AA, Adam W, Dekker T, Posthumus MA, van Beek TA, van Loon JJA. Identification of olfactory stimulants for Anopheles gambiae from human sweat samples. J. Chem. Ecol. 2000;26:1367-82
20. Blackwell A, Johnson SN. Electrophysiological investigation of larval water and potential oviposition chemo-attractants for Anopheles gambiae s.s. Ann. Trop. Med. Parasitol. 2000;94:389-98
21. Knudson JT, Eriksson R, Gershenzon J, Stahl B. Diversity and distribution of floral scent. Botanical Review. 2006;72:1-20
22. Biessmann H, Andronopoulou E, Biessmann MR, Douris V, Dimitratos SD, Eliopoulos E, Guerin PM, Iatrou K, Justice RW, Krober T, Marinotti O, Tsitoura P, Woods DF, Walter MF. The Anopheles gambiae Odorant Binding Protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS ONE. 2010;5:e9471
23. Krieger J, Grosse-Wilde E, Gohl T, Dewer YME, Raming K, Breer H. Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc. Natl. Acad. Sci. U. S. A. 2004;101:11845-50
24. Krieger J, Grosse-Wilde E, Gohl T, Breer H. Candidate pheromone receptors of the silkmoth Bombyx mori. Eur. J. Neurosci. 2005;21:2167-76
25. Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307:1638-42
26. Mitsuno H, Sakurai T, Murai M, Yasuda T, Kugimiya S, Ozawa R, Toyohara H, Takabayashi J, Miyoshi H, Nishioka T. Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur. J. Neurosci. 2008;28:893-902
27. Krieger J, Gondesen I, Forstner M, Gohl T, Dewer Y, Breer H. HR11 and HR13 receptor-expressing neurons are housed together in pheromone-responsive sensilla trichodea of male Heliothis virescens. Chem. Senses. 2009;34:469-77
28. Pitts RJ, Fox AN, Zwiebel LJ. A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5058-63
29. Tsitoura P, Andronopoulou E, Tsikou D, Agalou A, Papakonstantinou MP, Kotzia GA, Labropoulou V, Swevers L, Georgoussi Z, Iatrou K. Expression and membrane topology of Anopheles gambiae odorant receptors in lepidopteran insect cells. PLoS ONE. 2010 in press
30. Timmermann SE, Briegel H. Water depth and larval density affect development and accumulation of reserves in laboratory populations of mosquitoes. Bull. Soc. Vector Ecol. 1993;18:174-87
31. Angerer LM, Angerer RC. In situ hybridization to cellular RNA with radiolabelled RNA probes. In: (ed.) Wilkinson DG. In situ hybridization. Oxford: IRL Press. 1992
32. Belton P. Attraction of male mosquitoes to sound. J. Am. Mosq. Control Assoc. 1994;10:297-301
33. Forstner M, Gohl T, Breer H, Krieger J. Candidate pheromone binding proteins of the silkmoth Bombyx mori. Invert. Neurosci. 2006;6:177-87
34. Qiu YT, van Loon JJ, Takken W, Meijerink J, Smid HM. Olfactory Coding in Antennal Neurons of the Malaria Mosquito, Anopheles gambiae. Chem. Senses. 2006;31:845-63
35. Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725-36
36. Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147-59
37. Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327-38
38. Howell PI, Knols BG. Male mating biology. Malar J. 2009;8(Suppl 2):S8
39. Ray A, van Naters WG, Shiraiwa T, Carlson JR. Mechanisms of odor receptor gene choice in Drosophila. Neuron. 2007;53:353-69
40. Clyne PJ, Certel SJ, de Bruyne M, Zaslavsky L, Johnson WA, Carlson JR. The odor specificities of a subset of olfactory receptor neurons are governed by Acj6, a POU-domain transcription factor. Neuron. 1999;22:339-47
41. Miller CJ, Carlson JR. Regulation of Odor Receptor Genes in Trichoid Sensilla of the Drosophila Antenna. Genetics. 2010;186:79-95
42. Miyamichi K, Serizawa S, Kimura HM, Sakano H. Continuous and Overlapping Expression Domains of Odorant Receptor Genes in the Olfactory Epithelium Determine the Dorsal/Ventral Positioning of Glomeruli in the Olfactory Bulb. J. Neurosci. 2005;25:3586-92
43. Iwema CL, Fang H, Kurtz DB, Youngentob SL, Schwob JE. Odorant receptor expression patterns are restored in lesion-recovered rat olfactory epithelium. J. Neurosci. 2004;24:356-69
44. Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597-609
45. Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mamalian olfactory epithelium. Cell. 1993;74:309-18
46. Forstner M, Breer H, Krieger J. A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth Antheraea polyphemus. Int. J. Biol. Sci. 2009;5:745-57
47. Grosse-Wilde E, Gohl T, Bouche E, Breer H, Krieger J. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur. J. Neurosci. 2007;25:2364-73
48. Grosse-Wilde E, Svatos A, Krieger J. A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem. Senses. 2006;31:547-55
49. Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133:1255-65
50. Xu PX, Atkinson R, Jones DNM, Smith DP. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45:193-200
51. Shanbhag SR, Smith DP, Steinbrecht RA. Three odorant-binding proteins are co-expressed in sensilla trichodea of Drosophila melanogaster. Arthropod Structure & Development. 2005;34:153-65
52. Qiao H, Xiaoli H, Schymura D, Ban L, Field L, Dani FR, Michelucci E, Caputo B, della Torre A, Iatrou K, Krieger J, Zhou JJ, Pelosi P. Coomparative interactions between odorant-binding proteins of Anophels gambiae. Cell. Mol. Life Sci. 2010 in press
53. Andronopoulou E, Labropoulou V, Douris V, Woods DF, Biessmann H, Iatrou K. Specific interactions among odorant-binding proteins of the African malaria vector Anopheles gambiae. Insect Mol. Biol. 2006;15:797-811
54. Tegoni M, Ramoni R, Bignetti E, Spinelli S, Cambillau C. Domain swapping creates a third putative combining site in bovine odorant binding protein dimer. Nature Struct. Biol. 1996;3:863-7
55. Bianchet MA, Bains G, Pelosi P, Pevsner J, Snyder SH, Monaco HL, Amzel LM. The three-dimensional structure of bovine odorant binding protein and its mechanism of odor recognition. Nature Struct. Biol. 1996;3:934-9
Author contact
![]() Corresponding author: Dr. Jürgen Krieger, University of Hohenheim, Institute of Physiology (230), Garbenstrasse 30, 70599 Stuttgart, Germany. E-mail: juergen.kriegerde
Corresponding author: Dr. Jürgen Krieger, University of Hohenheim, Institute of Physiology (230), Garbenstrasse 30, 70599 Stuttgart, Germany. E-mail: juergen.kriegerde

 Global reach, higher impact
Global reach, higher impact