Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2011; 7(1):9-17. doi:10.7150/ijbs.7.9 This issue Cite
Research Paper
Expression, Identification and Purification of Dictyostelium Acetoacetyl-CoA Thiolase Expressed in Escherichia coli
Department of Biochemistry and Molecular Biology, Faculty of Agriculture and Life Science, Hirosaki University, 3 Bunkyo-cho, Hirosaki, 036-8561, Japan
* Present address: Faculty of Life Sciences, University of Manchester, Manchester, M13 9PT, UK
Received 2010-11-18; Accepted 2010-12-22; Published 2010-12-30
Abstract
Acetoacetyl-CoA thiolase (AT) is an enzyme that catalyses the CoA-dependent thiolytic cleavage of acetoacetyl-CoA to yield 2 molecules of acetyl-CoA, or the reverse condensation reaction. A full-length cDNA clone pBSGT-3, which has homology to known thiolases, was isolated from Dictyostelium cDNA library. Expression of the protein encoded in pBSGT-3 in Escherichia coli, its thiolase enzyme activity, and the amino acid sequence homology search revealed that pBSGT-3 encodes an AT. The recombinant AT (r-thiolase) was expressed in an active form in an E. coli expression system, and purified to homogeneity by selective ammonium sulfate fractionation and two steps of column chromatography. The purified enzyme exhibited a specific activity of 4.70 mU/mg protein. Its N-terminal sequence was (NH2)-Arg-Met-Tyr-Thr-Thr-Ala-Lys-Asn-Leu-Glu-, which corresponds to the sequence from positions 15 to 24 of the amino acid sequence deduced from pBSGT-3 clone. The r-thiolase in the inclusion body expressed highly in E. coli was the precursor form, which is slightly larger than the purified r-thiolase. When incubated with the cell-free extract of Dictyostelium cells, the precursor was converted to the same size to the purified r-thiolase, suggesting that the presequence at the N-terminus is removed by a Dictyostelium processing peptidase.
Keywords: acetoacety-CoA thiolase, Dictyostelium discoideum, cloning, protein purification, protein processing.
Introduction
Thiolases are known to present as functionally various forms in prokaryotes and eukaryotes. They are divided into two different groups, β-ketoacyl-CoA thiolase (KT, degradative) and acetoacetyl-CoA thiolase (AT, biosynthetic). The former plays a key role in the β-oxidative degradation of fatty acids, whereas the latter catalyses the condensation of 2 molecules of acetyl-CoA to form acetoacetyl-CoA, or the CoA-dependent thiolysis of acetoacetyl-CoA. In eukaryotes, there are five types of thiolases, which are distinguished by their functions and subcellular localization; mitochondrial KT and AT, peroxisomal KT and AT, and cytosolic AT. Mitochondrial AT regulates the levels of acetoacetyl-CoA and acetyl-CoA and also is involved in the ketone body metabolism in mitochondria. Cytosolic AT catalyses the first reaction of the mevalonate biosynthesis pathway to yield acetoacetyl-CoA, and the formed acetoacetyl-CoA enters steroid biosynthesis pathway in peroxisomes. It is considered that peroxisomal AT found in yeast [1] and rat liver [2] catalyses the first reaction of peroxisomal cholesterol and dolichol synthesis [2]. Like this, ATs play very important functions in cells. In human, it is known that deficiency of mitochondrial AT, known as β-ketothiolase deficiency, causes a disease having the error of isoleucine and ketone body metabolism. Patients with this disorder have ketoacidotic attacks, and some patients die or have neurological sequelae. AT is an important one of the enzymes which are implicated in human diseases.
The cellular slime mold Dictyostelium discoideum is a simple eukaryote which is used as a model organism to study the macromolecular events coincident with and necessary for development. In the life cycle, there are two distinct stages of growth and differentiation. The amoebae grow vegetatively as single cells undergoing cell division under the food supply. Development can be synchronized and separated from vegetative growth, and upon starvation, vegetative growing amoeba cells undergo differentiation to form a fruiting body containing a stalk and spores. Previously, we isolated and identified several developmentally regulated genes during spore germination [3-6] or vegetative growth [7, 8]. During the course of our study on their genes, a unique partial cDNA clone was isolated, which has homology to known thiolases in Protein Database. So, we carried out the screening of its full-length cDNA clone. In this paper, we report cDNA cloning and the purification of the recombinant protein expressed in E. coli, and that the protein encoded in the cDNA clone is Dictyostelium AT. Also, we describe the processing of Dictyostelium AT precursor protein.
Materials and methods
Bacterial strains, media, and vectors
D. discoideum strain AX-3 was grown axenically in HL5 medium [9] supplemented with 100 μg/mL of streptomycin at 22°C on a reciprocal shaker (150 rpm). E. coli strains, DH-5α and XL-1 blue, were used for subcloning and were grown in Luria Bertani (LB) medium at 37°C. Plasmids pBluescript SKII(-) (Stratagene) and pTrc99a (Pharmacia Biotech) were used for subcloning and as an expression vector in E. coli, respectively. E. coli JM105 was used for expression of the recombinant thiolase (r-thiolase), according to the manufacturer's protocols.
Isolation of cDNA clone
Dictyostelium λzap cDNA library (kindly provided by Dr. Herbert L. Ennis, Columbia University) was screened using digoxigenin-labeled CT-7 cDNA (see Results) as a probe. Two large positive clones isolated, pBSGT-3 and pBSGT-23, were analyzed. The nucleotide sequences were determined by the dideoxy chain termination method [10] using Sequenase II kit (US Biochemical, USA). The DNA sequence data were analyzed using MacDNASIS (Hitachi Software Engineering, Japan).
Construction of plasmid pTrc-thio
To amplify the open reading frame (ORF) of pBSGT-3, two oligonucleotide primers, 5'-thiof (5'-CGCGCCATGGTTTCGGGCCTTTCAAAAG-3') and 3'-thior (5'-CGCGGGATCCTTATAATTTTTCTAAAAC-3') were designated. These primers contained NcoI and BamHI sites (underlined), respectively, and the bold-type and italic letters indicate the initiation and stop codons, respectively. PCR was performed on pBSGT-3 DNA as a template using these primers, as described previously [11]. The amplified PCR product was double-digested with NcoI and BamHI, and inserted into the corresponding sites downstream of trc promoter in pTrc99A to yield plasmid pTrc-thio. E. coli JM105 was transformed with pTrc-thio to obtain a transformant (pTrc-thio).
Purification of SGT3 protein (r-thiolase)
E. coli transformant (pTrc-thio) was cultured in 500 mL of LB medium (+50 μg/mL ampicillin) containing 0.1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) at 37°C for 10 h. Cells harvested by centrifugation (5,000 × g for 10 min) were suspended in Buffer A (10 mM potassium phosphate buffer (pH7.5), 5 mM β-mercaptoethanol, 1 mM EDTA, 5 μg/mL phenylmethanesulfonyl fluoride (PMSF) and 10% glycerol), and disrupted by sonication with an Ultrasonic disruptor (Model UD-201, TOMY, Japan). The homogenate was centrifuged at 12,000 × g for 15 min to remove cell debris and the resulting supernatant (crude extract) was used for purification of the r-thiolase. All procedures were done at 0-4°C. (1) Ammonium sulfate fractionation: The crude extract was brought to 30% saturation with powdered ammonium sulfate and stored on ice for 30 min. After the precipitate was removed by centrifugation (12,000 × g, for 30 min), the resulting supernatant was brought to 70% saturation. After standing on ice for 30 min, the precipitate was collected by centrifugation, and then dissolved in Buffer A, followed by dialysis against Buffer A overnight. (2) DEAE-cellulofine A-500 column chromatography: The 30-70% ammonium sulfate fraction was applied on a DEAE-cellulofine A-500 column (2 × 22 cm, Seikagaku-Kogyo, Japan) equilibrated previously with Buffer A. Proteins were eluted with a linear gradient from 0 to 0.4 M NaCl in Buffer A (200 × 200 mL). The thiolase activity was detected in the flowthrough fraction, and the enzyme fractions were pooled. (3) Hydroxyapatite column chromatography: The pooled enzyme fraction was applied on a Bio-Gel HTP column (2 × 10 cm, Bio-Rad) equilibrated previously with Buffer A. Proteins were eluted by step-wise elution of 50, 100, 150, and 200 mM potassium phosphate buffer (pH7.5) (each 100 mL). The enzyme fractions eluted with 150 mM potassium phosphate buffer were pooled, and concentrated by ammonium sulfate precipitation (80% saturation).
Thiolase activity assay
Thiolase activity was determined by measuring the decrease in absorbance at 303 nm at 30°C according to the method of Davis et al.[12]. The reaction mixture (a total volume of 1 mL) contained 0.1 M Tris-HCl (pH 8.3), 25 mM MgCl2, 50 mM KCl, 20 nM acetoacetyl-CoA and 20 nM CoA. The molar absorption coefficient of 16.9 mM-1 for acetoacetyl-CoA was used under the assay conditions. One unit of the thiolase activity was defined as the amount of enzyme that catalyses the cleavage of 1 μmol of acetoacetyl-CoA per min.
Processing assay of r-thiolase precursor
E. coli transformant (pTrc-thio) was cultured in 100 mL of LB medium (+50 μg/mL Amp) at 37°C overnight in the presence of 1 mM IPTG. Cell pellet (0.9 g wet weight) collected by centrifugation was suspended in 8 mL of Buffer A and disrupted by sonication. The homogenate was centrifuged at 10,000 × g for 15 min and the pellet (inclusion body) was obtained. The inclusion body was washed with 10 ml of 1% Triton X-100/10 mM EDTA, and then dissolved in 1.5 mL of Buffer A containing 8 M urea, followed by dialysis against Buffer A containing 4M urea at 4°C for 12 h. The solution was again dialyzed against Buffer A (- urea) for 10 h, three times. Finally, the soluble r-thiolase precursor protein was obtained. D. discoideum strain AX-3 was grown axenically in HL5 medium up to a density of ~ 6. 0 × 106 cells/mL at 22°C on a reciprocal shaker (150 rpm). Cell-free extract from Dictyostelium cells was prepared as described previously [11]. For analysis of r-thiolase protein processing, the cell-free extract (15 μg protein) was added to the reaction mixture containing r-thiolase precursor (2.5 μg protein) in 50 mM potassium phosphate buffer (pH 6.7) and incubated at 37°C. The processing protein products were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) according to the method of Laemmli [13] and visualized by immunostaining with anti-thiolase antibody, as described for Western blot analysis.
Preparation of antibody against r-thiolase
The anti-thiolase antibody was prepared by Hokkaido System Science Co. (Sapporo, Japan). One female rabbit was immunized with the purified r-thiolase. The antiserum obtained was tested by ELISA and Western blot analysis. Antibody was purified from the antiserum as described [14].
Western blot analysis
To compare the molecular masses of the r-thiolase in the inclusion body expressed in E. coli and the purified r-thiolase, and to confirm processing of the r-thiolase precursor, samples were separated by 12.5% SDS-PAGE, electrotransferred onto a polyvinylidene difluoride (PVDF) membrane. Western blot analysis was performed using anti-thiolase antibody and anti-rabbit IgG-alkaline phosphatase (AP) (Sigma, USA) as primary and secondary antibodies, respectively, according to the method described previously [15].
Analytical methods
The protein concentration was measured by the method of Lowry et al. [16] using bovine serum albumin as a standard. The molecular mass of the expressed or the purified r-thiolase was estimated by SDS-PAGE. An electrophoresis calibration kit (Pharmacia Biotec) was used for the estimation of the molecular mass as standard proteins. Protein sequencing of the purified r-thiolase was carried out on a protein sequencer (Model PPSQ-10, SHIMADZU, Japan) according to the method described previously [17].
Nucleotide sequence accession number
The nucleotide sequence data reported in this paper are available in the DDBJ/EMBL/GenBank databases under the accession number AB212872.
Results
Cloning of the full-length cDNA
In a previous study of the Dictyostelium ubiquitin gene [7], we isolated a unique partial cDNA clone CT-7, by chance, which was not related to the ubiquitin clones. However, the deduced amino acid sequence of CT-7 clone was similar to known thiolases. So, we carried out cloning of cDNA from λzap cDNA library using this CT-7 cDNA as a probe, and several cDNA clones were isolated. Out of them, two large cDNA clones, pBSGT-3 and pBSGT-23, were analyzed. pΒSGT-3 clone was a full-length one of 1,353 bp long containing an ORF of 1,245 bp, whereas pBSGT-23 had an insert of 1,746 bp long containing an intron-like AT-rich sequence. When PCR amplification was performed with Dictyostelium genomic DNA as a template using 5'-thiof and 3'-thior as primers, the PCR product was 1.6 kbp long, which was larger by about 300 bp than the ORF of pBSGT-3. The nucleotide sequence of the PCR product was identical to that of pBSGT-23, indicating that pΒSGT-23 is an artificial clone containing a single intron of 271 bp between the codon (AAT) of Asn12 and the codon (GTA) of Val13 in the ORF. Genomic Southern analysis suggested that the SGT3 gene is a single copy in the Dictyostelium genome (data not shown).
Comparison of amino acid sequence of SGT3 protein with those of known thiolases
The deduced amino acid sequence of SGT3 predicted a protein of 414 amino acids with a molecular mass of 43,445. Its isoelectric point (pI) was 8.43. The amino acid sequence of SGT3 protein showed a significant degree of homology to other known thiolases. We compared the deduced amino acid sequence of SGT3 with those of thiolases from other organisms. The SGT3 protein showed 54% (73%), 52% (72%), and 42% (67%) sequence identity (similarity) to ATs of Saccharomyces uvarum [18], rat mitochondoria [19], and Zoogloea ramigera [20], respectively. Also, it showed 38% - 47% (59% - 65%) identity (similarity) to KTs of Alcaligenes eutorophus [21], rat mitochondria [22] and peroxisome [23], human peroxisome [24], and E. coli [25], respectively. The comparison results revealed that SGT3 protein is more closely identical to AT than KT, suggesting that pBSGT-3 clone encodes a Dictyostelium AT. So, we refer to SGT3 protein as Dictyostelium AT (Ddthiolase).
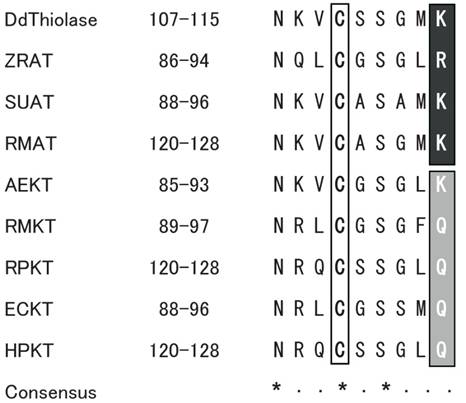
The amino acid sequence of Ddthiolase was aligned with those of several representative ATs from other organisms described above. Two cysteine and one histidine residues, which are important for the catalytic reaction and the enzymatic activity, are highly conserved among the thiolases of different species [26, 27]. The three residues correspond to Cys110, Cys400, and His370 in the Ddthiolase sequence. The sequence around Cys110 at positions 107 - 115 is in good agreement with the highly conserved active site in other thiolases, which was proposed as active sites [21] (Fig. 1). In Ddthiolase, Lys residue resides at the last position of the active site. A basic amino acid residue at this position is specific for AT, whereas KT has a Gln residue at this position [25], except for KT of Alcaligenes eutorophus [21]. These facts support that Ddthiolase is an AT.
Comparison of active site around the conserved cysteine residue. The putative active site of Ddthiolase is aligned with the homologous region of known thiolases. The numbers indicate the positions of amino acids shown in this alignment. The conserved cysteine residues are boxed. The conserved residues specific for ATs and KTs are highlighted on black and gray backgrounds, respectively. The consensus sequence is indicated underneath. Asterisks (*) and dots (·) indicate the identical and similar residues, respectively. Abbreviations used: DdThiolase, Dictyostelium AT (in this paper, DDBJ/EMBL/GenBank accession No. AB212872); ZRAT, Zoogloea ramigera AT (J02631) [20]; SUAT, Saccharomyces uvarum AT (X07976) [18]; RMAT, rat mitochondrial AT (NM017075) [19]; AEKT, Alcaligenes eutorophus KT (J04987) [21]; RMKT, rat mitochondorial KT (X05341) [22]; RPKT, rat peroxisomal KT (J02749) [23]; ECKT, Escherichia coli KT (J05498) [25]; HPKT, human peroxisomal KT (X12966) [24].
Expression of the SGT3 protein (r-thiolase)
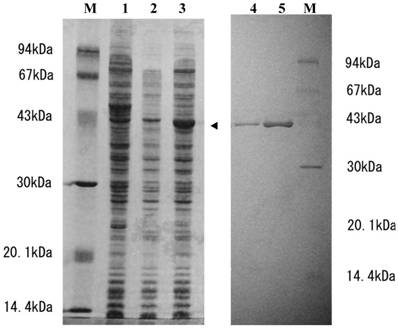
To determine whether or not SGT3 protein exhibits thiolase activity, we prepared E. coli transformant (pTrc-thio) under the control of a ptrc promoter. When the transformant was cultivated in the presence of 1 mM IPTG, a 43-kDa protein was strongly expressed (Fig. 2, lane 3), whose molecular mass was in a good agreement with that calculated from the ORF of pBSGT-3. But, no thiolase activity was detected in the supernatant or the precipitate (inclusion body) fractions prepared from the transformant. When the inclusion body was solubilized with 8 M urea and then activated according to the general method [28] as described in Materials and methods, the thiolase activity was expressed with a specific activity of 0.3 mU/mg protein. Endogeneous thiolase activity was not detectable under the conditions used. These results suggest that SGT3 protein is a thiolase.
SDS-PAGE analysis of SGT3 protein expressed in E. coli and the purified r-thiolase. Samples were applied on 12% SDS-PAGE gel and after electrophoresis, the gel was stained with coomassie brilliant blue (Quick-CBB, Wako, Japan). Lane 1, crude extract of E. coli (without plasmid); lane 2, crude extract of E. coli transformant (pTrc99a); lane 3, crude extract of E. coli transformant (pTrc-thio) with IPTG induction (1 mM IPTG, for 3 h); lanes 4 and 5, 1 and 3 μg of the purified r-thiolase, respectively; lane M, protein size markers. Protein expressed in E. coli transformant is indicated by an arrow head.
Purification and characterization of the r-thiolase
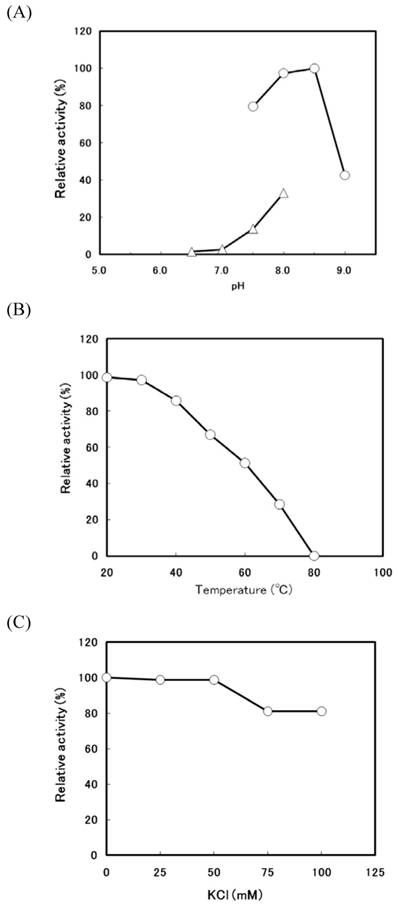
When the transformant (pTrc-thio) was cultivated at 37°C in the low concentration (0.1 mM) of IPTG, a large amount of thiolase activity was detected in the soluble fraction. The r-thiolase was purified from the soluble fraction (crude extract) by the 30-70% saturated ammonium sulfate fractionation followed by two steps of column chromatography using DEAE-cellulofine and Bio-Gel HTP. The results of the purification of the r-thiolase are shown in Table 1. The enzyme was purified 42.7-fold with a yield of 24.4%. The purified enzyme had a specific activity of 4.70 U/mg protein and showed a single band on a 12.5% SDS-PAGE gel (Fig. 2, lanes 4 and 5), indicating that it is homogeneous. For the characterization of the purified r-thiolase, the effects of pH, temperature, and salt on the enzyme activity were examined. The activity was highest at pH 8.0 - 8.5, and it decreased significantly under conditions below pH 7.0 and above pH 9.0 (Fig. 3A). The enzyme was stable up to 40 °C, and its activity decreased to about 50% at 60 °C (Fig. 3B). To examine whether the thiolase activity is activated by KCl or not, we measured the enzyme activity in the presence of KCl. Fity mM KCl did not activate the activity and also not inhibit. But, the activity was slightly inhibited (about 20% inhibition) by high concentration of KCl (Fig. 3C).
Purification of recombinant Dictyostelium acetoacetyl-CoA thiolase
| Fraction | Total protein | Total activity | Specific activity | Yield | Fold |
|---|---|---|---|---|---|
| (mg) | (U) | (U/mg protein) | (%) | (x) | |
| Crude extract | 455.5 | 52.0 | 0.11 | 100 | 1.0 |
| 30 - 70% (NH4)2SO4 | 292.8 | 32.5 | 0.11 | 62.5 | 1.0 |
| DEAE-Cellulofine | 21.3 | 19.4 | 0.91 | 37.3 | 8.3 |
| Hydroxyapatite | 2.7 | 12.7 | 4.70 | 24.4 | 42.7 |
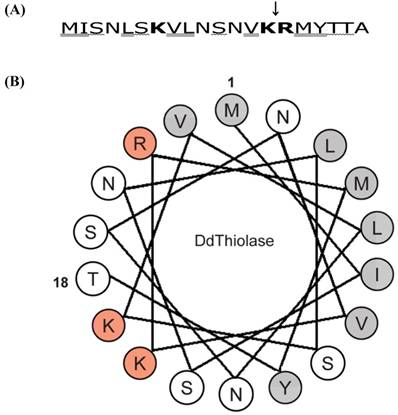
Next, we determined the N-terminal sequence of the purified r-thiolase. As a result, it was found that the N-terminal sequence is (NH2)-Arg-Met-Tyr-Thr-Thr-Ala-Lys-Asn-Leu-Glu-, which corresponds to the sequence from positions 15 to 24 of Ddthiolase. This indicates that the N-terminal 14 residues were removed by an E. coli protease. The cleavage site is between Lys at position 14 and Arg at position 15, as shown in Fig. 4A. These results suggest that the N-terminal presequence of r-thiolase might be removed by E. coli protease OmpT, which cleaves the peptide bond between consecutive basic amino acid residues [29, 30].
Effects of pH, temperature, and salt on the activity of the r-thiolase. (A) Optimum pH for the activity of the r-thiolase. The enzyme activity was measured under the standard enzyme assay conditions, except that the following buffers were used; 50 mM potassium phosphate buffer (pH 6.5 - 8.0, open triangles) and 50 mM Tris-HCl buffer (pH 7.5 - 9.0, open circles). The relative activity is expressed as the percentage of the maximum activity attained under the assay conditions used. (B) Thermostability of the r-thiolase. After the purified r-thiolase (0.52 μg, 2.45 mU) was incubated at the indicated temperatures for 10 min with 10 mM potassium phosphate buffer (pH 7.5), the remaining activity was measured under the standard enzyme assay conditions. The remaining activity is expressed as the percentage of the original enzyme activity. (C) Effect of KCl. Using the purified r-thiolase (0.52 μg, 2.45 mU), the activity was measured under the standard enzyme assay conditions at the KCl concentrations indicated. The relative activity is expressed as the percentage of the activity obtained in the absence of KCl.
Characteristics of the N-terminal sequence of Ddthiolase. (A) The first 20 amino acid sequence of Ddthiolase. Positively charged, hydroxylated, and hydrophobic residues are indicated by bold face, wavelined, and double-underlined, respectively. A downward arrow indicates cleavage site by E. coli protease OmpT, which has specificity for the paired basic residues. (B) Helical wheel analysis. Residues 1-18 of the N-terminal sequence of Ddthiolase are plotted circularly in an ideal α-helix, with 3.6 residues per helical turn. Positively charged and hydrophobic residues are represented by red and gray circles, respectively. The other residues are indicated open circles. The numbers 1 and 18 indicate the first Met and the 18th Thr, respectively.
Processing of the r-thiolase precursor
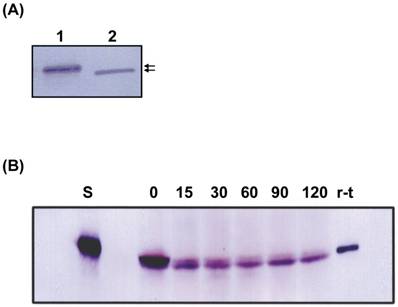
When the molecular size of the r-thiolase in the inclusion body was compared with that of the purified r-thiolase, the former was slightly larger than the latter (Fig. 5A). This result suggests that the r-thiolase might be a precursor form carrying still the N-terminal presequence. So, the r-thiolase isolated from the inclusion body was incubated at 37°C with the Dictyostelium cell-free extract. With increase of incubation times, the r-thiolase was converted to the smaller molecule, which size is the same to that of the purified r-thiolase (Fig. 5B). This result suggests that the r-thiolase in the inclusion body is a precursor with the N-terminal presequence, which is removed by a Dictyostelium processing peptidase.
Western blot analysis. (A) Samples were subjected to SDS-PAGE, and Western blot analysis was performed using anti-thiolase antibody. Lane 1, inclusion body (10 μg protein); lane 2, the purified r-thiolase (0.5 μg protein). The positions of thiolase precursor and r-thilase were indicated by arrows. (B) Time course of processing of the r-thiolase precursor. The r-thiolase precursor (2.5 μg protein) was mixed with the cell-free extract (15 μg protein) prepared from Dictyostelium cells and incubated at 37°C for the time indicated in figure. Processing peptides were subjected to SDS-PAGE followed by Western blot and immunostaining using anti-thiolase antibody and anti-rabbit IgG-AP. Numbers indicate incubation times (min) of the reaction. Lanes S and r-t contain substrate (r-thiolase precursor) alone and the purified r-thiolase, respectively.
Discussion
When we started the study on Ddthiolase, its structure and function remained unknown. During the course of this thiolase work, the sequence of AT gene was reported from the Dictyostelium Genome Project [31]. Searches of the Dictyostelium database (http://dictybase.org) revealed that three single genes of AT (Dicty gene ID DDB_G0271544), KT (DDB_G0274339), and a putative thiolase (DDB_G0269588) exist in the Dictyostelium genome. The nucleotide sequence of pBSGT-23 is identical to that of the AT gene in the Dictyostelium database. Dictyostelium KT and the putative thiolase remain to be characterized.
The majority of mitochondrial proteins are synthesized in the cytosol as a precursor protein with an N-terminal mitochondrial targeting signal (MTS) sequence, which is a signal for import of proteins to mitochondria [32-34]. Although MTS sequence is different in length and in sequence, it is rich in positively charged, hydroxylated, and hydrophobic amino acids to form an amphiphilic α-helix structure. As shown in Fig.4A, Ddthiolase has 3 positively charged, 5 hydroxylated and 8 hydrophobic amino acids in the N-terminal region. A helical wheel analysis of the first 18 residues of Ddthiolase shows that an amphiphilic α-helix structure would be formed, although its amphiphilicity is slightly weak (Fig.4B), suggesting that the N-terminal sequence may function as a MTS to direct Ddthiolase to mitochondria. As shown in (Fig. 5B), the r-thiolase precursor was processed to the smaller form by Dictyostelium processing peptidase. Since Ddthiolase possesses a MTS sequence at the N-terminus as described above, this processing peptidase appears to be a mitochondrial processing peptidase (MPP), which cleaves the MTS presequence to make a mature protein. The cleavage site of Ddthiolase presequence by MPP is considered to be between Tyr at position 17 and Thr at position 18 from the tendancy of the MPP cleavage site according to the “R-2/3 rule” [35, 36]. The correct cleavage site of Ddthiolase precursor remains to be identified.
Peroxisomal proteins have peroxisomal targeting signal (PTS) necessary for import of those proteins to peroxisomes. Two types of PTS are well known; PTS-1 is a tripeptide motif of Ser-Lys-Leu (SKL) sequence, which is located at the C-terminus [37], and PTS-2 is a nonapeptide motif of the consensus sequence (Arg/Lys)(Leu/Val/Ile)(Xxx)5(His/Gln)(Leu/Ala) [(R/K)(L/V/I)X5(H/Q)(L/A)] within the N-terminal region, where X indicates any amino acid [38]. Ddthiolase has Glu-Lys-Leu (EKL) similar to PTS-1 sequence at the C-terminus, suggesting that this thiolase may be localized to peroxisomes. Taken together, Ddthiolase appares to be a dual-localization enzyme containing MTS and PTS-1 sequences at the N- and C-terminus, respectively. The subcellular localization of Ddthiolase remains to be determined. It was reported that rat mitochondrial AT has both MTS and PTS-1 sequences at the N- and C-terminus, respectively [39]. Also, several proteins, such as 3-hydroxymethylglutaryl-CoA lyase [40], dienoyl-CoA isomerase [41], and malonyl-CoA decarboxylase [42], contain both N-terminal MTS and C-terminal PTS-1. It is of interest to understand the dual-localization mechanism of proteins to mitochondria and/or peroxisomes.
In summary, when Ddthiolase was expressed as a soluble enzyme in an E. coli expression system, it was an active enzyme, of which the N-terminal presequence was cleaved off by E. coli protease. On the other hand, expressed as an insoluble enzyme in the inclusion body under the high expression conditions, the r-thiolase was a precursor protein with the N-terminal presequence, which might be protected by aggregating from the cleavage by E. coli protease. In in vitro studies of protein processing, precursor proteins are indispensable as substrates for processing peptidases. Generally, precursor proteins are labeled with radioactively amino acids using an in vitro transcription/translation kit and used as a substrate for processing peptidase, and the detection is dependent to autoradiography. As shown in this paper, however, if precursor proteins could be prepared using an E. coli expression system, in vitro further studies of signal peptidase or processing peptidase would be carried out using them as substrates by SDS-PAGE/Western blot and immunostaining using their antibodies.
Acknowledgements
This work was supported in part by The Japan Association of Chemistry.
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Kurihara T, Ueda M, Kanayama N. et al. Peroxisomal acetoacetyl-CoA thiolase of an n-alkane-utilizing yeast, Candida tropicalis. Eur J Biochem. 1992;210:999-1005
2. Antonenkov VD, Croes K, Waelkens E. et al. Identification, purification and characterization of an acetoacetyl-CoA thiolase from rat liver peroxisomes. Eur J Biochem. 2000;267:2981-2990
3. Ennis HL, Giorda R, Ohmachi T. et al. Characterization of genes that are developmentally regulated during Dictyostelium discoideum spore germination. Develop Genet. 1988;9:303-313
4. Shaw DR, Richter H, Giorda R. et al. Nucleotide sequences of Dictyostelium discoideum developmentally regulated cDNAs rich in (AAC) imply proteins that contain clusters of asparagine, glutamine or threonine. Mol Gen Genet. 1989;218:453-459
5. Giorda R, Ohmachi T, Ennis HL. Organization of a gene family developmentally regulated during Dictyostelium discoideum spore germination. J Mol Biol. 1989;205:63-69
6. Giorda R, Ohmachi T, Shaw DR. et al. A shared internal threonine-glutamic acid-threonine-proline repeat defines a family of Dictyostelium discoideum spore germination specific proteins. Biochemistry. 1990;29:7264-7269
7. Ohmachi T, Giorda R, Shaw DR. et al. Molecular organization of developmentally regulated Dictyostelium discoideum ubiquitin cDNAs. Biochemistry. 1989;28:5226-5231
8. Ohmachi T, Fukuoka R, Kimura Y. et al. The characterization of two Dictyostelium discoideum genes encoding ribosomal proteins with sequence similarity to rat L27a and L37a. Biosci Biotechnol Biochem. 1998;62:2008-2015
9. Sussman R, Sussman M. Cultivation of Dictyostelium discoideum in axenic medium. Biochem Biophys Res Commun. 1967;29:53-55
10. Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463-5467
11. Nagayama K, Itono S, Yoshida T. et al. Antisense RNA inhibition of the β subunit of the Dictyostelium discoideum mitochondrial processing peptidase induces the expression of mitochondrial proteins. Biosci Biotechnol Biochem. 2008;72:1836-1846
12. Davis JT, Moore RN, Imperiali B. et al. Biosynthetic thiolase from Zoogloea ramigera 1. Preliminary characterization and analysis of proton transfer reaction. J Biol Chem. 1987;262:82-89
13. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London). 1970;227:680-685
14. Talian JC, Olmsted JB, Goldman RD. A rapid procedure for preparing fluoresein-labeled specific antibodies from whole antiserum: its use in analyzing cytoskeletal architecture. J Cell Biol. 1983;97:1277-1282
15. Nagayama K, Ohmachi T. Mitochondrial processing peptidase activity is controlled by the processing of α-MPP during development in Dictyostelium discoideum. Microbiology. 2010;156:978-989
16. Lowry OH, Rosebrough NJ, Farr AL. et al. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265-275
17. Ohmachi T, Nishino M, Kawata M. et al. Identification, cloning, and sequencing of the genes involved in the conversion of D,L-2-amino-△2-thiazoline-4-carboxylic acid to L-cysteine in Pseudomonas sp. strain ON-4a. Biosci Biotechnol Biochem. 2002;66:1097-1104
18. Dequin S, Gloecker R, Herbert CH. et al. Cloning, sequencing and analysis of the yeast S. uvarum ERG10 gene encoding acetoacetyl-CoA thiolase. Curr Genet. 1988;13:471-478
19. Fukao T, Kamijo K, Osumi T. et al. Molecular cloning and nucleotide sequence of cDNA encoding the entire precursor of rat mitochondrial acetoacetyl-CoA thiolase. J Biochem. 1989;106:197-204
20. Peoples OP, Masamure S, Walsh CT. et al. Biosynthetic thiolase from Zoogloea ramigera III. Isolation and characterization of the structural gene. J Biol Chem. 1987;262:97-102
21. Peoples OP, Sinskey AJ. Poly-β-hydroxybutyrate biosynthesis in Alcaligenes eutrophus H16. Characterization of the genes encoding β-ketothiolase and acetoacetyl-CoA reductase. J Biol Chem. 1989;264:15293-15297
22. Arakawa H, Takiguchi M, Amaya Y. et al. cDNA-derived amino acid sequence of rat mitochondrial 3-oxoacyl-CoA thiolase with no transient presequence: structural relationship with peroxisomal isozyme. EMBO J. 1987;6:1361-1366
23. Hijikata M, Ishii N, Kagamiyama H. et al. Structural analysis of cDNA for rat peroxisomal 3-ketoacyl-CoA thiolase. J Biol Chem. 1987;262:8151-8158
24. Bout A, Teunissen Y, Hashimoto T. et al. Nucleotide sequence of human peroxisomal 3-oxoacyl-CoA thiolase. Nucl Acids Res. 1988;16:10369
25. Yang S-Y, He Yang X-Y, Healy-Louie G. et al. Nucleotide sequence of the fadA gene. Primary structure of 3-ketoacyl-coenzyme A thiolase from Escherichia coli and the structural organization of the fadAB operon. J Biol Chem. 1990;265:10424-10429
26. Anderson VE, Bahnson BS, Wlassics ID. et al. The reaction of acetyldithio-CoA, a readily enolized analog of acetyl-CoA with thiolase from Zoogloea ramigera. J Biol Chem. 1990;265:6255-6261
27. Williams SE, Palmer MAJ, Peoples OP. et al. Biosynthetic thiolase from Zoogloea ramigera. Mutagenesis of the putative active-site base Cys-378 to Ser-378 changes the partitioning of the acetyl S-enzyme intermediate. J Biol Chem. 1992;267:16041-16043
28. Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual; 2nd ed. New York: Cold Spring Harbor Laboratory Press. 1989
29. Grodgerg J, Dunn JJ. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988;170:1245-1253
30. Sugimura K, Nishihara T. Purification, characterization, and primary structure of Escherichia coli protease VII with specificity for paired basic residues: identity of protease VII and OmpT. J Bacteriol. 1988;170:5625-5632
31. Eichinger L, Pachebat JA, Glockner G. et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43-57
32. von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986;5:1335-1342
33. Roise D, Schatz G. Mitochondrial presequences. J Biol Chem. 1988;263:4509-4511
34. Roise D, Theiler F, Horvath SJ. et al. Amphiphilicity is essential for mitochondrial presequence function. EMBO J. 1988;7:649-653
35. Gavel Y, von Heijne G. Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng. 1990;4:33-37
36. Gakh O, Cavadini P, Isaya G. Mitochondrial processing peptidases. Biochim Biophys Acta. 2002;1592:63-77
37. Elgersma Y, Vos A, van der Berg M. et al. Analysis of the carboxyl-terminal peroxisomal targeting signal 1 in a homologous context in Saccharomyces cerevisiae. J Biol Chem. 1996;271:26375-26382
38. Swinkles BW, Gould SJ, Bondar AG. et al. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991;10:3255-3262
39. Oliver LM. Kovacs W, Masuda K, et al. Identification of peroxisomal targeting signals in cholesterol biosynthetic enzymes: AA-CoA thiolase, HMG-CoA synthase, MPPD, and FPP synthase. J Lipid Res. 2000;41:1921-1935
40. Ashmarina LI, Pshezhetsky AV, Branda SS. et al. 3-Hydroxy-3-methylglutaryl coenzyme A lyase: targeting and processing in peroxisomes and mitochondria. J Lipid Res. 2000;40:70-75
41. Filppula SA, Yagi AI, Kilpeläinen SH. et al. △3,5-△2,4-Dienoyl-CoA isomerase from rat liver: molecular characterization. J Biol Chem. 1998;273:349-355
42. Voilley N, Roduit R, Vicaretti R. et al. Cloning and expression of rat pancreatic β-cell malonyl-CoA decarboxylase. Biochem J. 1999;340:213-217
Author contact
![]() Corresponding author: Tel; +81-172-39-3774, Fax; +81-172-39-3750, e-mail; tohmachihirosaki-u.ac.jp.
Corresponding author: Tel; +81-172-39-3774, Fax; +81-172-39-3750, e-mail; tohmachihirosaki-u.ac.jp.

 Global reach, higher impact
Global reach, higher impact