Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2011; 7(1):133-137. doi:10.7150/ijbs.7.133 This issue Cite
Short Research Communication
MicroRNA-137 Targets Carboxyl-terminal Binding Protein 1 in Melanoma Cell Lines
1. Laboratory of Signal Transduction & Molecular Targeted Therapy, State Key Laboratory of Biotherapy/ Department of Medical Oncology, West China Hospital, Sichuan University, Chengdu, Sichuan Province, 610041, China;
2. Department of Dermatology, The Sixth People's Hospital of Shanghai, Shanghai Jiaotong University, Yishan Road 600, Shanghai, 200211, China;
3. Department of Pathology, University of Colorado, Denver, Aurora, CO 80045, USA;
4. Department of Dermatology, University of Colorado, Denver, Aurora, CO 80045, USA;
5. Department of Medicine, University of Colorado, Denver, Aurora, CO 80045, USA.
# Co-first author
Received 2011-1-14; Accepted 2011-1-25; Published 2011-1-27
Abstract
Carboxyl-terminal binding protein 1 (CtBP1) is a transcriptional co-repressor that represses expression of various tumor suppressor genes. In the present study, we identified miR-137 as a potential regulator of CtBP1 expression in melanoma cells. Expression of miR-137 in melanoma cell lines was found to inversely correlate with CtBP1 levels. Target Scan predicted a putative site for miR-137 within the CtBP1 3′ untranslated region (3′UTR) at nt 710-716, which is highly conserved across species. To explore the mechanism of miR-137 targeting CtBP1, we performed an Argonaute 2 (Ago2)-pull down assay, and miR-137 was identified in complex with CtBP1 mRNA. miR-137 suppressed CtBP1 3' UTR luciferase-reporter activity, and this effect was lost with deletion of the putative 3' UTR target-site. Consistent with the results of the reporter assay, ectopic expression of miR-137 reduced expression levels of CtBP1. Furthermore, expression of miR-137 increased the immediate downstream effectors of CtBP1, such as E-cadherin and Bax. The human miR-137 gene is located at chromosome 1p22, which has previously been determined to be a susceptive region for melanoma. This study suggests miR-137 may act as a tumor suppressor by directly targeting CtBP1 to inhibit epithelial-mesenchymal transition (EMT) and inducing apoptosis of melanoma cells, thus illustrating a functional link between miR-137 and CtBP1 in melanoma development.
Keywords: CtBP1, miR-137, transcription, tumor suppressor, melanoma
Introduction
MicroRNAs (miRNAs) are highly conserved, small, non-coding RNAs that play a vital role in development, cell proliferation, differentiation, apoptosis, and metabolism. They function by binding to the 3′UTR of multiple target mRNAs. In mammals, miRNAs guide proteins of the Argonaute family to partially-complementary sequences typically located in the 3′UTR of specific target mRNAs, leading to translational repression or mRNA degradation [1] [2] [3].
The identification of miRNA as the causative force for chronic lymphocytic leukemia opened a new avenue in the field of oncogenesis. A variety of cancers, including colon, central nervous system, melanoma, prostate and breast were all found to be influenced by the dysregulation of miRNAs. miR-137 is located at the region on chromosome 1p22 where loss is associated with increased melanoma risk [4]. With the identification of miRNAs as important regulators of gene expression, we hypothesized miR-137 may regulate target expression associated with melanoma risk.
Previously miR-137 was shown to target the 3′UTR of the key regulator of melanogenesis and melanoma development, micropthalmia-associated transcription factor (MITF) [5]. CtBP1 is another predicted target of miR-137. CtBP1 was identified as a transcriptional co-repressor of several tumor suppressors [6]. CtBP1 has been shown to suppress the transcription of tumor suppressors such as E-cadherin in multiple cell lines [6]. Conversely, tumor suppressors, such as HIPK2, Ink4a/Arf, and APC, target CtBP1 degradation to induce apoptosis [6]. Therefore, we studied the regulation of CtBP1 and its down-stream targets by miR-137 in melanoma cell lines.
Results
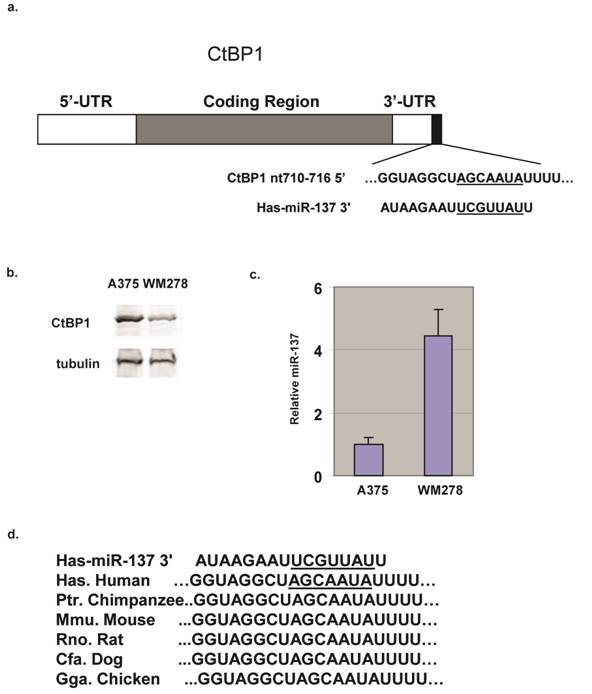
miRNAs have emerged as post-transcriptional regulators critically involved in the pathogenesis of human cancers. Previously, miR-137 has been shown to target MITF [5,7]. CtBP1 is another predicted target of miR-137 (Fig. 1a) and we found CtBP1 expression in melanoma cell lines was inversely correlated with miR-137 levels; CtBP1 expression in WM278 cells was significantly lower than in A375 cells (Fig. 1b) and its miR-137 is 4 fold higher (Fig. 1c). Target Scan revealed the 3'UTR of CtBP1 contains a miR-137 site at nt 710-716 (Fig. 1a), and the miR-137 binding site in CtBP1 3'UTR is conserved from human to chicken (Fig. 1d). Predictions of miRNA-mRNA interactions typically rely on bioinformatic algorithms, but these algorithms only suggest the possibility of miRNA binding and may falsely predict interactions.
CtBP1 is a predicted target for miR-137. (a) Diagram of CtBP1 mRNA depicting the predicted miR-137 site in the 3'UTR region. (b) CtBP1 protein levels in A375 and WM278 cells. Tubulin was used as a loading control. (c) miR-137 levels of the A375 and WM278 cells. miR-137 level was determined by the Taqman assay. Data were normalized to the 18s mRNA. Samples were assayed in triplicate for each experiment and at least two independent experiments were performed. Data are presented as mean ± SEM from a representative experiment. (d) Conservation of the miR-137 site in CtBP1 in different species.
We expressed miR-137 in HEK293 cells and analyzed the mRNAs associated with Ago2 using a biochemical approach based on the functional co-complex of Ago2 with miRNA and its target mRNA [8] to gain insight into the predicted miR-137 targeting of CtBP1. qRT-PCR shows less binding of a non-target mRNA in the presence or absence of miR-137, but CtBP1 mRNA binding to Ago2 in the presence of miR-137 was increased 3 fold compared to a control oligo (Fig. 2a).
miR-137 targets CtBP1 in melanoma cells. (a) CtBP1 mRNA binding to the Ago2 complex in the presence of miR-137. HEK293 cells were transfected with Ago2 in the presence (Ago2+miR137) and absence (Ago2) of miR-137, and the Ago2-complex was pulled down. Primer sets for CtBP1 and 18S (Non-target) were used to qRT-PCR-amplify the pull down samples. (b) miR-137 decreases CtBP1 3'UTR-reporter. A375 cell were transfected with luciferase constructs fused to CtBP1 3'UTR, either the wild type (WT) or the deletion of the putative miR-137 targeting site, along with control oligo (SC) or miR-137 by Lipofecatmine 2000 in suspension and were immediately transferred to 0.5 ml DMEM with FBS and incubated at 37°C for 48 h before luciferase assay. Relative luciferase expression levels were determined by normalizing with internal controls, values were expressed as ratios to the control sample. Samples were assayed in triplicate for each experiment and at least two independent experiments were performed. Data are presented as mean ± SEM from a representative experiment. (c) miR-137 decreases CtBP1 expression. A375 cell were transfected with the control oligo (SC) or miR-137 by Lipofecatmine 2000 in suspension and were immediately transferred to 0.5 ml DMEM with FBS and incubated at 37°C for 48 h before western blotting. Tubulin was used as a loading control. (d) miR-137 down-regulates E-cadherin and Bax genes. A375 cell were transfected with the control oligo or miR-137 and incubated at 37°C for 48 h before qRT-PCR for the mRNA levels of E-cadherin and Bax.
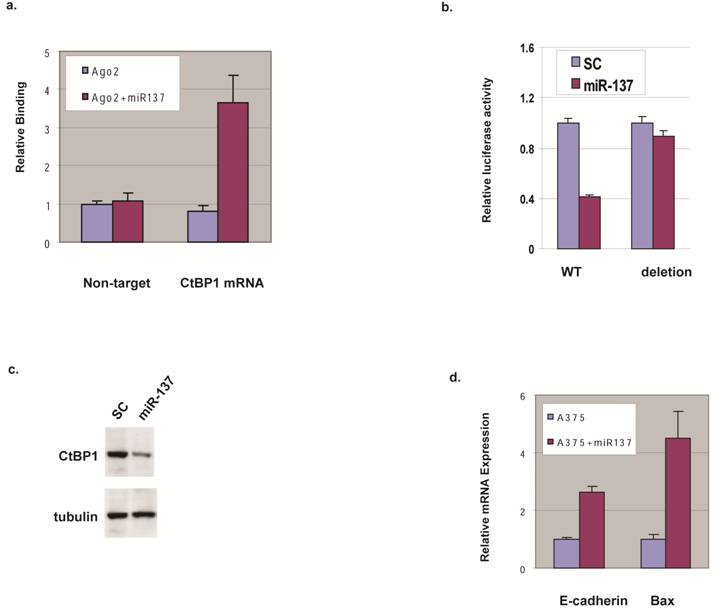
To assess the contribution of the predicted miR-137 binding site, we constructed luciferase-reporter genes with the CtBP1 3'UTR with and without deletion at the miR-137 binding region and tested their expression with miR-137 addition. Consistent with binding data, miR-137 transfection repressed luciferase expression when CtBP1 3'UTR was fused to the luciferase reporter (Fig. 2b). Supporting the role of the putative miR-137 targeting site on CtBP1, miR-137 transfection did not decrease luciferase expression when fused with CtBP1 3'UTR harboring the deletion (Fig 2b). These data suggest that miR-137 down-regulates CtBP1, probably through specific interaction with the putative site at CtBP1's 3'UTR, and CtBP1 is a direct target for miR-137.
To explore if miR-137 controls protein expression levels of CtBP1 in melanoma cells, we transfected melanoma cell line A375 that has relatively high CtBP1 expression with miR-137 and analyzed CtBP1 protein by western blotting. As shown in Fig. 2c, over-expression of miR-137 led to a significant decrease in CtBP1 protein compared with the control. Therefore, we conclude miR-137 plays a critical role in CtBP1 expression.
We studied the function of human miRNAs and to determine if the downstream targets of CtBP1 are affected by ectopic expression of miR-137 by performing qRT-PCR to assess mRNA levels of E-cadherin and Bax in A375 cells transfected with miR-137 or control oligo. As shown in Fig. 2d, mRNA levels of E-cadherin and Bax increased with miR-137 expression, mimicking the functional outcome of CtBP1 knockdown. This data suggests the function of miR-137 in melanoma is, at least in part, through its regulation of CtBP1.
Discussion
miRNAs are involved in many different cell processes including cell-cycle regulation, differentiation, proliferation and apoptosis [reviewed in [9]]. Improper expression of miRNA genes is seen in both benign and malignant cancers, and miRNA genes can act as tumor suppressors or oncogenes. miRNA expression profiles can be used to classify solid tumors [10] and previous study has shown that miRNA expression differs between melanoma cell lines [11]. The incidence of malignant melanoma is increasing at a greater rate than any other cancer type, thus it remains a significant public health threat. Molecular mechanisms involved in the development of melanoma are being intensely researched. Research by others found miR-137 is located at chromosome 1p22, a melanoma susceptibility hot spot. In this study, we identified CtBP1 as a novel target for miR-137. Given the critical role of CtBP1 in tumorigenesis, we hypothesize that miR-137 might function as a tumor suppressor in melanoma by targeting CtBP1. Our data showing regulation of CtBP1 down-stream genes such as E-cadherin and Bax by miR-137 further support this notion. Our future studies will assess if other CtBP1 targets, such as PTEN and p53, are controlled by miR-137, thus further linking the miR-137-CtBP1 axis with melanoma development.
Numerous studies characterizing expression profiles of miRNAs in cancer have shown down-regulation of miRNA expression in cancer cell lines compared with normal tissues [12] [13]. Furthermore, the recent identification of altered miRNA expression in a variety of cancer sites has confirmed this is another class of molecules capable of functioning as tumor suppressors or oncogenes [14]. miRNAs binding to the target gene 3′UTR is usually imperfect and therefore multiple targets have been identified for a single miRNA. As mentioned above, miR-137 in melanoma cells also targets MITF, believed to play a significant role in melanoma-genesis [15,16]. This mode of regulation could be used as a therapeutic strategy by manipulating the miRNA targeting multiple gene pathways for a synergistic effect.
In oral squamous carcinoma miR-137 is silenced by hypermethylation [17]. In rectal cancer miR-137 is up-regulated after preoperative capecitabine chemoradiotherapy [18]. miR-137, along with miR-124, induce differentiation of glioblastoma multiforme-derived stem cells and induce glioblastoma multiforme cell cycle arrest, which was associated with decreased expression of cyclin-dependent kinase 6 (CDK6) and phosphorylated retinoblastoma proteins [19]. In addition, the ectopic transfection of miR-137 into an oral squamous cell carcinoma significantly reduces cell growth, by down-regulation of the translation of CDK6 or E2F6 transcription factors [17]. Recently, miR-137 was shown to target CDC42 in colorectal cells, inducing cell cycle G1 arrest and inhibiting invasion [20]. The identification of the multi-functional transcriptional co-repressor CtBP1 as a miR-137 target further highlights the importance of miR-137 loss/silence in various human tumors. Determining if miR-137 regulates CtBP1 and other targets in cancer patients awaits further investigation.
Acknowledgements
This work was supported by the NIH grant R01CA115468 (to Q.Z.).
Conflict of Interests
The authors declare no conflict of interests.
References
1. Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo J. 2002;21(17):4663-4670
2. Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8(2):93-103
3. Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17(3):118-126
4. Walker GJ, Indsto JO, Sood R. et al. Deletion mapping suggests that the 1p22 melanoma susceptibility gene is a tumor suppressor localized to a 9-Mb interval. Genes Chromosomes Cancer. 2004;41(1):56-64
5. Bemis LT, Chen R, Amato CM. et al. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008;68(5):1362-1368
6. Chinnadurai G. The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res. 2009;69(3):731-734
7. Haflidadottir BS, Bergsteinsdottir K, Praetorius C, Steingrimsson E. miR-148 regulates Mitf in melanoma cells. PLoS One. 2010;5(7):e11574
8. Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4(2):76-84
9. Mueller DW, Bosserhoff AK. Role of miRNAs in the progression of malignant melanoma. Br J Cancer. 2009;101(4):551-556
10. Lu J, Getz G, Miska EA. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834-838
11. Gaur A, Jewell DA, Liang Y. et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67(6):2456-2468
12. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857-866
13. Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12(12):580-587
14. Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1-12
15. Du J, Widlund HR, Horstmann MA. et al. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6(6):565-576
16. McGill GG, Horstmann M, Widlund HR. et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109(6):707-718
17. Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68(7):2094-2105
18. Svoboda M, Izakovicova Holla L, Sefr R. et al. Micro-RNAs miR125b and miR137 are frequently upregulated in response to capecitabine chemoradiotherapy of rectal cancer. Int J Oncol. 2008;33(3):541-547
19. Silber J, Lim DA, Petritsch C. et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14
20. Liu M, Lang N, Qiu M. et al. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest and inhibits invasion in colorectal cancer cells. Int J Cancer. 2010 [Epub ahead of print]
Author contact
![]() Corresponding author: Qinghong Zhang, PhD, Department of Dermatology, University of Colorado, Denver, Aurora, CO 80045. (303)-724-4051; Fax: (303)-724-4048; Email: Qinghong.Zhangedu
Corresponding author: Qinghong Zhang, PhD, Department of Dermatology, University of Colorado, Denver, Aurora, CO 80045. (303)-724-4051; Fax: (303)-724-4048; Email: Qinghong.Zhangedu

 Global reach, higher impact
Global reach, higher impact