Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2011; 7(3):279-285. doi:10.7150/ijbs.7.279 This issue Cite
Research Paper
Identification of Triploid Individuals and Clonal Lines in Carassius Auratus Complex Using Microsatellites
1. Molecular Population Genetics Group, Temasek Life Sciences Laboratory, 1 Research Link, National University of Singapore, 117604 Singapore
2. Key Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Shanghai Ocean University, Ministry of Education, Shanghai 201306, China
Received 2011-1-24; Accepted 2011-3-11; Published 2011-3-18
Abstract
The Carassius auratus complex in natural populations includes diploid triploid and polyploidy individuals. Diploid individuals belong to the species Carassius auratus whereas triploid and polyploidy individuals are from the subspecies Carassius auratus gibelio. Triploid individuals are all female and reproduce clonally by gynogenesis. Therefore the Carassius auratus complex is an ideal system for studying evolution of unisexual reproduction. Identification of triploid individuals and clonal lines is the first step towards understanding of the evolution of unisexual clonal lines. We examined the ability of 10 microsatellites in identifying triploid individuals in 94 individuals from Japan and China. In 40 confirmed triploid individuals and eight confirmed diploid individuals, all triploid and diploid individuals can be identified by genotyping 10 microsatellite, and four triploid clonal lines were identified. Using the 10 microsatellites we genotyped 46 adult individuals (40 females and six males) from a natural population in China and found that all six males were diploid whereas the majority of females (36 of 40) were triploid and three triploid clonal lines were detected. In 18 diploid individuals from China, all individuals showed different genotypes, suggesting there is no diploid clonal line in diploid crucian carp. A phylogenetic analysis of 94 individuals from China and Japan showed that triploid individuals and clonal lines have originated recurrently.
Keywords: Triploid, identification, microsatellite, fish, evolution
Introduction
The Carassius auratus complex includes diploid triploid and polyploidy individuals from the family Cyprinidae and is distributed mainly in China, Japan and Russia [1, 2]. Diploid individuals belong to the species Carassius auratus whereas triploid and polyploidy individuals are from the subspecies Carassius auratus gibelio [3, 4]. Now they are also widely spread throughout Europe as an invasive species [5, 6]. Two different reproductive modes were revealed: allogynogenetic reproduction and gonochoristic reproduction [7-9]. In natural populations, the population structure is complex and there are diploid, triploid and polyploid individuals with the majority of individuals being triploid [5, 10]. The chromosome number of individuals from distinct natural populations are different, ranging from 100 to 200 [11]. The diploid crucian carp in China has 100 chromosomes, whereas the triploid gibel carp (or silver crucian carp) has 150 chromosomes or more [10, 12, 13]. The white crucian carp (C. auratus cuvieri) in Lake Biwa of Japan had 100 (diploid) or more chromosomes (triploid or tetraploid) [14]. Natural gynogenetic silver crucian carp is a unique triploid organism and reproduces clonally [15]. Therefore it could be an important model for studying the evolution of unisexual clonal lines.
Triploid organisms have been identified by various methods including analyzing the red blood cell size [7, 8], karyotyping [16, 17], flow cytometry [18] and isozyme analyses [19]. Although analyzing the red blood cell size and karyotyping are the most straightforward methods to assess ploidy level, they are time-consuming, labour-intensive and not able to differentiate different genotypes/clonal lines. Moreover, in some fish species karyotypes are extremely difficult to obtain due to the small and similar size of different chromosomes, their copious numbers, and/or the absence of suitable staining techniques [20]. Flow cytometry has been used to estimate ploidy level by assessing the total DNA content of cells in comparison with a known control. However, this method gives only an approximate estimation of the ploidy level [18] and measuring DNA content in cell is not only time-consuming, but also costly and labour-intensive.
Microsatellites are short (1-6 bp) repetitive DNA sequences [21], and are highly abundant and almost evenly distributed in genomes [22]. Microsatellites have been extensively applied for genome mapping, studying population genetics, ecology, evolution and conservation of endangered organisms and for identification of clonal lines [22-24]. A few microsatellites have been isolated from silver crucian carp [24-27]. Only four of these microsatellites have been used in identification of diploid and triploid individuals, as well as clonal lines [24].
The purpose of this study is to establish a molecular method to identify triploid individuals and clonal lines in the Carassius auratus complex for studying the evolution of unisexual triploid clonal lines.
Materials and methods
Fish and DNA
DNA samples of 20 confirmed triploid silver crucian carp individuals from a wild population in Japan were obtained from Dr. Kenichi Ohara (Japan). The polyploidy levels of these individuals were determined by analyzing the red blood cell size using an existing method [7, 8]. Tissue and blood samples of 20 individuals were collected from two fish farms located in Jiangsu (14 individuals) and Zhejiang provinces (6 individuals), China. All twenty individuals from two farms were confirmed to be triploid by analyzing the red blood cell size. In addition, four confirmed diploid males and four confirmed diploid females collected from the Dayun River in Zhejiang province (China) were also used in this study. DNA was isolated from each fish using a method that we developed previously [28].
To understand the genetic composition of the Carassius auratus complex in the wild, we randomly collected 46 adult individuals from the Changshan River located in Zhejiang province, China. Sex of each fish was determined by checking gonads. Fin clips of all 46 individuals were collected and stored in 75% ethanol until DNA extraction. DNA was extracted using a existing method [28] and arrayed on 96-well plates for PCR.
Microsatellites and genotyping
Ten polymorphic dinucleotide microsatellites (J01, J04, J09, J12, J56, J58, J60, J62, J68 and J69, Table 1) developed previously [25] were genotyped using fluorescently labelled primers (FAM or Hex) and a DNA sequencer ABI 377 (Applied Biosystems).
Polymorphisms of 10 microsatellites in 94 individuals of silver crucian carp *
| Locus | A | HDI | HTRI | HT | HOM |
|---|---|---|---|---|---|
| J01 | 25 | 0.319 | 0.404 | 0.723 | 0.277 |
| J04 | 25 | 0.287 | 0.617 | 0.904 | 0.096 |
| J09 | 16 | 0.723 | 0.255 | 0.978 | 0.022 |
| J12 | 16 | 0.649 | 0.298 | 0.947 | 0.053 |
| J56 | 20 | 0.755 | 0.223 | 0.978 | 0.022 |
| J58 | 21 | 0.500 | 0.330 | 0.830 | 0.170 |
| J60 | 21 | 0.532 | 0.426 | 0.958 | 0.042 |
| J62 | 17 | 0.543 | 0.309 | 0.852 | 0.148 |
| J68 | 6 | 0.628 | 0.064 | 0.692 | 0.308 |
| J69 | 12 | 0.670 | 0.159 | 0.829 | 0.171 |
*: All 10 microsatellites were selected from our previous publication (see reference 25). A: Number of alleles; HDI: Proportion of diploid heterozygotes; HTRI: Proportion of triploid heterozygotes; HT: Proportion of all diploid and triploid heterozygotes and HOM: Proportion of homozygotes.
Briefly, amplification of DNA samples was performed in 25 μL volumes, each containing 1× polymerase chain reaction buffer (Finnzymes), 100 nM of each primer, 1.5 mM MgCl2, 50 μM of each dNTP, 0.5 unit of Taq polymerase (Finnzymes) and 10 ng of genomic DNA. PCR cycling conditions were as follows: an initial denaturation at 94°C for 2 minutes followed by 34 cycles of 30 s at 94°C, 30 s at 55°C, 30 s at 72°C, then a final extension step of 10 min at 72°C. Amplified fragments were separated on denaturing 8% polyacrylamide gels using the ABI 377 sequencer (Applied Biosystems). The fragment lengths of alleles were determined against the internal size standard GS-250-Rox (Applied Biosystems) using software GENESCAN and GENOTYPER (Applied Biosystems). Individuals with the same genotypes at all 10 loci were identified manually. The number of alleles, allele frequency, and expected and observed heterozygosity were calculated by direct counting. Genetic relatedness among individuals was estimated using the allele sharing index [29], where alleles were scored as either present (1) or absent (0). Phylogenetic trees based on NJ, ME and UPGMA were constructed using the default setting with software MEGA 4.0 [30].
Results
Microsatellite polymorphism
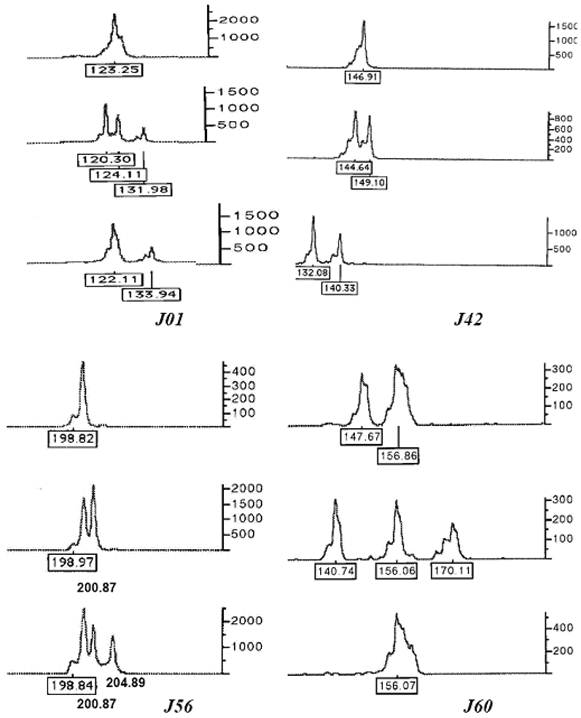
All ten microsatellites were polymorphic in 94 individuals (Table 1). A total of 179 alleles were detected. The loci J01 and J04 were most polymorphic (25 alleles), while the locus J68 was least polymorphic (6 alleles). The observed heterozygosity (including di-allelic and tri-allelic heterozygosity) ranged from 0.692 to 0.978 with an average of 0.870 ± 0.03. At some loci, triploid individuals clearly showed three alleles (see examples in Figure 1).
Some electrophoretograms of genotypes at microsatellite loci J01, J42, J56 and J60 showing one, two and three alleles at single locus. The numbers at the base of peaks are the sizes of alleles in bp, while the scale bar at right side shows height of alleles indicating the intensity of fluorescent signals.
Ability of microsatellites in identifying and distinguishing polyploidy individuals and triploid clonal lines
Using the genotypes of the 10 microsatellites, all 20 confirmed triploid silver crucian carp individuals from Japan were classified as triploids, and the 8 confirmed diploid individuals from China were designated as diploids. Furthermore, 20 confirmed triploid silver crucian carp collected from two farms in China were classified as triploids using microsatellite genotypes. Therefore, the accuracy of 10 microsatellites for identification of triploid and diploid individuals was 100% (Table 2). Using all 10 microsatellites, in the 20 triploid individuals from Japan, two clonal lines were identified. One clonal line contained three individuals and another line consisted of two individuals. The 20 triploid individuals from two farms in China contained two clonal lines, each from one farm. One line included 14 individuals, and another had six individuals. In the eight diploid individuals, there were no individuals showing the same genotypes at all 10 loci.
Characterization of a wild population in China
Forty-six individuals randomly collected from a wild population of the Carassius auratus complex in China were analyzed using 10 microsatellites. Among the 46 individuals, only six were males and were classified as diploids (Table 2). Among the 40 females, 36 (90%) were diagnosed as triploids and the remaining four were classified as diploids. In the 36 triploid individuals, three clonal lines were identified; one clonal line with four individuals, two lines each with two individuals. The overall heterozygosity was 0.87 ± 0.05 in 46 individuals (Table 2). The observed heterozygosity in diploid and triploid individuals was 0.81 ± 0.05 (n = 10) and 0.90 ± 0.04 (n = 36) respectively.
Phylogenetic relationships among individuals and clonal lines in the Carassius auratus complex
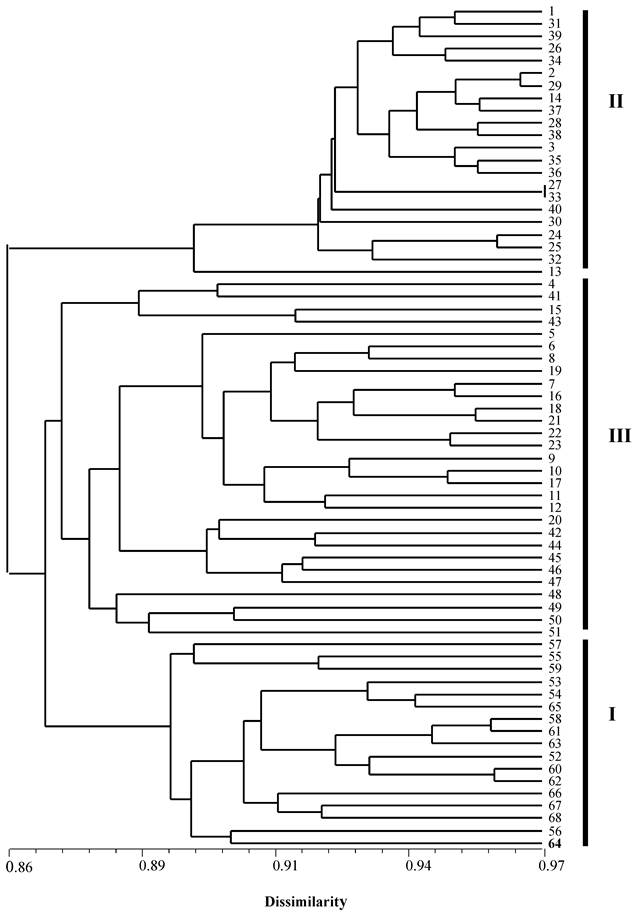
Among the 94 individuals used in this study, seven clonal lines each containing two or more genetically identical individuals, included a total of 33 triploid individuals. Therefore, in the phylogenetic analysis, only 68 individuals were used. All triploid individuals from Japan were located in cluster I while all individuals from China formed two clusters II and III, of which cluster III grouped with the Japanese cluster formed a paraphyletic cluster (Figure 2). The ten diploid males (fish: 5-10 and 17-20) and eight diploid females (fish: 1-4 and 16-19) were located in the cluster II and III. The five clonal lines (11, 12, 13, 14 and 15) and triploid individuals from China were also distributed in clusters II and III whereas the two Japanese clonal lines (58 and 61) and triploid individuals (53-68) were located in the cluster I.
Genetic characterization of the crucian carp from China and Japan with 10 microsatellites
| Japan | China | ||||
|---|---|---|---|---|---|
| Wild population | Farmed | Diploid | Wild population | ||
| N | 20 | 20 | 8 | 46 | |
| M | 0 | 0 | 4 | 6 | |
| F | 20 | 20 | 4 | 40 | |
| Diploid | 0 | 0 | 8 | 10 | |
| Triploid | 20 | 20 | 0 | 36 | |
| HDI | 0.35±0.14 | 0.66±0.13 | 0.72±0.07 | 0.57±0.08 | |
| HTRI | 0.65±0.14 | 0.18±0.10 | 0 | 0.30±0.06 | |
| HT | 1.00±0.00 | 0.84±0.10 | 0.72±0.07 | 0.87±0.05 | |
| No clonal lines | 2 | 2 | 0 | 3 | |
N: Number of individuals; M: Number of male individuals, F: Number of female individuals; Diploid: Number of diploid individuals; Triploid: Number of triploid individuals; HDI: Proportion of diploid heterozygotes; HTRI: Proportion of triploid heterozygotes; HT: Proportion of all diploid and triploid heterozygotes.
An NJ-phylogenetic tree showing the relationships among diploid, triploid individuals and clonal lines of crucian carp from China and Japan based on genetic dissimilarity at 10 microsatellite loci. 1-51: individuals from China, whereas 52-68: individuals from Japan. 1-4: diploid female individuals; 5-10 and 17-20: diploid male individuals; and branch 11, 12, 13, 14 and 15 are clonal lines containing 14, 6, 4, 2 and 2 individuals from China, while branch 58 and 61 are clonal lines containing three individuals and two individuals from a wild population in Japan.
Discussion
Ability of 10 microsatellites in identifying and distinguishing triploid individuals and clonal lines
Microsatellites have been used to identify clonal lines in a lung fluke Paragonimus westermani [23] and the silver crucian carp [24]. In a previous study [24], Japanese scientists used four microsatellites for identifying clonal lines in sliver crucian carp and suggested that using more markers, more clonal lines could have been identified. In this study, we examined the efficiency of 10 microsatellites in identifying polyploidy individuals and clonal lines. Using 10 microsatellites all 48 individuals whose polyploidy levels were known, could be identified as diploid or triploid individuals with 100% accuracy and clonal line could be differentiated, indicating the high ability of the 10 microsatellites in identifying triploid individuals and clonal lines. Therefore, the set of 10 microsatellites could be used for identifying and differentiating triploid individuals and clonal lines of silver crucian carp. Ideally, all 10 markers could be multiplexed and electrophorezed in one run, which could reduce the cost and improve the accuracy of microsatellite genotyping [31, 32], thus ensuring the precision of indentifying and differentiating clonal lines. However, employment of diverse electrophoresis techniques across laboratories has resulted in inconsistent allele sizes, leading to difficulties in comparing genotyping results from different laboratories [33, 34]. Recent studies showed that genotyping results generated by capillary electrophoresis techniques are more reproducible than those generated by polyacrylamide gel electrophoresis [34]. Hence, the capillary electrophoresis techniques are recommended. In addition, individuals with known genotypes should be used as a control to allow cross-lab comparisons.
Genetic composition in the Carassius auratus complex
In several previous studies, triploid clonal lines have been identified in wild populations in Japan [24] and cultured populations in China [9]. In this study, we analyzed genetic composition of a Japanese wild population including 20 triploid individuals, two cultured populations from two farms in China and a wild population in China. In the Japanese population, using 10 microsatellites, we confirmed that all individuals were triploids. This result is in agreement with previous studies showing that all individuals of the white crucian carp, Carassius langsdorfii in Japan to be triploid [24, 35, 36]. However in this study we found only two clonal lines in 20 triploid individuals from Japan, whereas in the previous studies conducted in Japan, a lot of clonal lines were detected [24]. This difference may be due to the difference of sample size. Using more individuals, more clonal line may be found. The triploid fish from two farms in China consisted of only two clonal lines, each farm containing one line. In the wild population from China, 36 of 46 individuals were triploids, indicating that in the wild population the majority of individuals were triploids. This result is in agreement with other reports based on karyotyping [37, 38]. In this study, among the 36 triploid female individuals, only two clonal lines were detected. Among the individuals from China and Japan, none of the individuals and clonal lines shared the same genotypes at all 10 loci, suggesting origin of the triploid individuals and clonal lines happened after the separation of the Chinese and Japanese populations. In the 18 diploid individuals from China, all individuals showed different genotypes, suggesting there is no diploid clonal line in diploid crucian carp. Although it is reported that there are tetraploidy individuals of silver crucian carp in natural population [10], in this study we did not detect any tetraploidy individuals. This may be due to the fact that we have only collected samples from one population and the sample number is only 48. In the future it is necessary to analyze the genotypes of more individuals from various populations located in different regions.
Evolution of the triploid silver crucian carps
Phylogenetic analysis using genetic similarity of alleles at 10 microsatellites showed that the individuals from Japan formed one group while the individuals from China formed two clusters. One cluster of individuals from China was grouped with the cluster of Japanese individuals to form a paraphyletic cluster. The seven triploid clonal lines and the 18 diploid individuals were distributed in different clusters, suggesting recurrent origin of triploid clonal lines and triploid individuals. Future study should focus on genotyping more individuals from different natural populations to analyze the evolution of unisexual clonal lines in detail.
Acknowledgements
This study is supported by the internal fund of the Temasek Life Sciences Laboratory, Singapore. The authors would like thank Dr. Kenichi Ohara from Lake Biwa Museum, Japan for kindly supplying us tissue samples of 20 confirmed triploid individuals of Japanese sliver crucian carp. The authors thank Dr Mamta Chauhan for editing English of this paper.
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Nelson JS. Fishes of the World 4th edn. New York: John Wiley and Sons, Inc. 2006
2. Li FB, Gui JF. Clonal diversity and genealogical relationships of gibel carp in four hatcheries. Anim Genet. 2008;39(1):28-33
3. Gui JF, Zhou L. Genetic basis and breeding application of clonal diversity and dual reproduction modes in polyploid Carassius auratus gibelio. Sci China-C. 2010;53(4):409-415
4. Peng JX, Xie JL, Zhou L, Hong YH, Gui JF. Evolutionary conservation of Dazl genomic organization and its continuous and dynamic distribution throughout germline development in gynogenetic gibel carp. J Eexp Zool Part B. 2009;312(8):855-871
5. Vetešník L, Halačka K, Lusková V, Lusk S. Erythrocyte profile of diploid and triploid silver crucian carp (Carassius auratus). Acta Vet Brun. 2006;75:203-207
6. Balık I, Çubuk H, Çınar S. Spatial and seasonal variations in catch of silver crucian carp, Carassius gibelio (Bloch, 1782) in Lake Eğirdir, Turkey. Turk J Fish Aqua Sci. 2008;8:347-353
7. Sezaki K, Kobayasi H, Nakamura M. Size of erythrocytes in the diploid and triploid specimens of Carassius auratus langsdorfii. Japan J Ichthyol. 1977;24:135-140
8. Onozato H, Torisawa M, Kusama M. Distribution of the gyno-genetic polyploid crucian carp, Carassius auratus langsdorfii in Hokkaido, Japan. Japan J Ichthyol. 1983;30:184-190
9. Zhou L, Wang Y, Gui JF. Genetic evidence for gonochoristic reproduction in gynogenetic silver crucian carp (Carassius auratus gibelio Bloch) as revealed by RAPD assays. J Mol Evol. 2000;51:498-506
10. Zhou L, Gui JF. Karyotypic diversity in polyploid gibel carp, Carassius Auratus Gibelio Bloch. Genetica. 2002;115:223-232
11. Abramenko MI, Kravchenko OV. Evolutionary cytogenetics. Cytogenet Cell Genet. 1998;81:123-135
12. Fan Z, Shen J. Studies on the evolution of bisexual reproduction in crucian carp (Carassius auratus gibelio Bloch). Aqua Int. 1990;84:235-244
13. Zhu HP, Ma DM, Gui JF. Triploid origin of the gibel carp as revealed by 5S rDNA localization and chromosome painting. Chrom Res. 2006;14:767-776
14. Yamashita M, Onozato H, Nakanishi T, Nagahama Y. Breakdown of the sperm nuclear envelope is a prerequisite for male pronucleus formation: direct evidence from the gynogenetic crucian carp Carassius auratus langsdorfii. Dev Bio. 1990;137:155-160
15. Gui JF. A unique study system: gynogenetic fish Carassius auratus gibelio. Sci Found China. 1996;4:44-46
16. Kim DS, Nam YK, Park IS. Survival and karyological analysis of reciprocal diploid and triploid hybrids between mud loach (Misgurnus mizolepis) and cyprinid loach (Misgurnus anguillicaudatus). Aquaculture. 2005;135:257-265
17. Kunitake H, Nakashima T, Mori K, Tanaka M. Somaclonal and chromosomal effects of genotype, ploidy and culture duration in Asparagus officinalis L. Euphytica. 1998;102(3):309-316
18. Normark BB. The evolution of parthenogenesis in the Aramigus tessellatus species complex (Coleoptera: Curculionidae): Evidence from mitochondrial DNA sequences. Evolution. 1996;50:734-745
19. Arai K, Mukaino M. Electrophoretic analysis of the diploid progenies from triploid × diploid crosses in the loach Misgurnus anguillicaudatus (Pisces: Cobitidae). J Eexp Zool Part A. 1998;280:368-374
20. Mair GC. Chromosome-set manipulation in tilapia - Techniques, problems and prospects. Aquaculture. 1993;111(1-4):227-244
21. Weber JL. Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics. 1990;7(4):524-530
22. Goldstein DB, Schlotterer C. Microsatellites: Evolution and Applications. Oxford: Oxford University Press. 1999
23. van Herwerden L, Blair D, Agatsuma T. Genetic diversity in parthenogenetic triploid Paragonimus westermani. Int J Parasi. 1999;29(9):1477-1482
24. Ohara K, Dong S, Taniguchi N. High proportion of heterozygotes in microsatellite DNA loci of wild clonal silver crucian carp, Carassius langsdorfii. Zool Sci. 1999;16(6):909-913
25. Yue GH, Orban L. Polymorphic microsatellites from silver crucian carp (Carassius auratus gibelio Bloch) and cross-amplification in common carp (Cyprinus carpio L.). Mol Ecol Notes. 2002;2(4):534-536
26. Yue GH, Ho MY, Orban L, Komen J. Microsatellites within genes and ESTs of common carp and their applicability in silver crucian carp. Aquaculture. 2004;234(1-4):85-98
27. Guo W, Gui JF. Microsatellite marker isolation and cultured strain identification in Carassius auratus gibelio. Aqua Int. 2008;1(16):497-510
28. Yue GH, Orban L. A simple and affordable method for high throughput DNA extraction from animal tissues for PCR. Electrophoresis. 2005;26:3081-3083
29. Nei M. Genetic distance between populations. Am Nat. 1972;106(949):283-292
30. Tamura K, Dudley J, Nei M, Kumar SP. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596-1599
31. Yue GH, Beeckmann P, Bartenschlager H, Moser G, Geldermann H. Rapid and precise genotyping of porcine microsatellites. Electrophoresis. 1999;20(17):3358-3363
32. Giardina D, Peconi C, Cascella R, Sinibaldi C, Cuzzola VF, Nardone AM, Bramanti P, Novelli G. A multiplex molecular assay for the detection of uniparental disomy for human chromosome 7. Electrophoresis. 2009;30:2008-2011
33. Zhu ZY, Wang CM, Lo LC, Lin G, Feng F, Tan J, Chou R, Lim HS, Orban L, Yue GH. A standard panel of microsatellites for Asian seabass (Lates calcarifer). Anim Genet. 2010;41:208-212
34. Vemireddy LR, Archak S, Nagaraju J. Capillary electrophoresis is essential for microsatellite marker based detection and quantification of adulteration of Basmati rice (Oryza sativa). J Agr Food Chem. 2007;55(20):8112-8117
35. Ohara K, Dong S, Taniguchi N. Identification and distribution of clonal lines detected by DNA polymorphism in silver crucian carp, Carassius langsdorfii collected from the Monobe and Niyodo rivers. Japan J Ichthyol. 1998;45:21-27
36. Ohara K, Taniguchi N. Preliminary study on genetic diversity evaluated by 11 microsatellite markers in Kuromejina Girella leonina and Mejina Girella punctata. Fish Sci. 2003;69(4):861-863
37. Chan SS, Jian YJ. Kyrotyping of silver crucian carp. Acta Hydro Sin. 1988;4:381-384
38. Gui JF, Liang SC, Zhu LF, Jiang YG. Discovery of multiple tetraploids in artificially propagated populations of allogynogenetic silver crucian carp and their breeding potentialities. Chin Sci Bull. 1993;38:327-331
Author contact
![]() Corresponding author: Dr G.H. Yue, Temasek Life Sciences Laboratory, 1 Research Link, National University of Singapore, 117604 Singapore. Tel: 65-68727405; Fax: 65-68727007; Email: genhuaorg.sg
Corresponding author: Dr G.H. Yue, Temasek Life Sciences Laboratory, 1 Research Link, National University of Singapore, 117604 Singapore. Tel: 65-68727405; Fax: 65-68727007; Email: genhuaorg.sg

 Global reach, higher impact
Global reach, higher impact