Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2011; 7(3):286-296. doi:10.7150/ijbs.7.286 This issue Cite
Research Paper
Isolation and Identification of Insect Intestinal Mucin Haiim86 - The New Target for Helicoverpa Armigera Biocontrol
1. Biological Control Centre of Plant Pathogens and Plant Pests of Hebei Province, College of Life Sciences, Agricultural University of Hebei, Baoding 071001, China.
2. Biological Institute, Hebei Academy of Science, Shijiazhuang 050051, China
Received 2010-10-21; Accepted 2010-12-18; Published 2011-3-19
Abstract
There are many more glycoproteins in Helicoverpa armigera peritrophic membrane than midgut separated by SDS-PAGE analysis after Periodic acid-Schiff (PAS) and coomassie staining. The peritrophic membrane (PM) of H. armigera larvae contains about forty associated proteins. A cDNA library was constructed from H. armigera midgut mRNA to study the new target for pest biocontrol. An antiserum against Spodoptera exigua integral/total PM proteins cross reacted with several H. armigera PM proteins and was used to isolate a complete cDNA encoding an insect intestinal mucin (HaIIM86). The deduced protein sequence of the cDNA contained one potentially glycosylated, mucin-like domain, five cysteine-rich chitin-binding domains (CBDs) and two D-G rich regions. Mucin domain was lined between the first and second CBDs; the two additional D-G rich regions were proposed to internal reside at the amino terminus of the protein flanked by three cysteine-rich CBDs. HaIIM86 contains two D-G-rich tandem repeat domains flanked by cysteine-rich sequences in peritrophic membrane proteins which is not present in all the currently known PM proteins. Howerer the functions of D-G rich domains require further investigation. HaIIM86 was shown as 200kDa protein by SDS-PAGE analysis and appeared to be associated with the PM. HaIIM86 has chitin-binding activity and can be degraded into 90 and 70 kDa by HaGV Enhancin in vivo. The finding has shown that HaIIM86 is the target substrate for enhancin and the potential target for pest control.
Keywords: Peritrophic matrix, Helicoverpa armigera, cDNA expression library, insect intestinal mucin, biocontrol
INTRODUCTION
The digestive tract in insects is commonly lined with an invertebrate-unique structure, peritrophic membrane (PM), which have multiple physiological functions and serve as the first line of defense in the midgut[1, 2]. In the last 10 years, significant progress toward understanding the PM molecular structure and mechanism for PM formation has been made. It is known that proteins and chitin are major components of PMs, which play very important roles in the protection of insects from microbial infection [3]. Many factors, for example, calcoflour, Trichoplusia ni granulosis virus (TnGV) enhancin, chitinase and lectin can disrupt the PM formation and enhance pathogen infection in insect [4]. So, as a natural barrier to pathogenic microorganisms, the PM can be a potential target for pest biocontrol.
The first invertebrate intestinal mucin from larvae of Trichoplusia ni (TnIIM) was identified, which is similar to vertebrate mucus in biochemical characteristics [3, 5]. Identification of TnIIM provided the basis to developmental strategies of insect control by target PM the intestinal mucinous barrier. Recently, over 19 PM proteins have been identified from several insect species, including Mamestra configurata, Plutella xylostella, Aedes aegypti, and other agriculturally important insects [6-12]. To understand the function of PMs, it is necessary to study the characteristics and functions of PM IIMs.
Helicoverpa armigera is a world-wide pest. To understand the midgut physiology and digestive biochemistry, and the role of the PM in protection of the midgut epitheliums, we studied the proteins associated with the PM of H. armigera. Here we report the molecular cloning and sequencing of a cDNA encoding an intestinal mucin from H. armigera.
MATERIALS AND METHODS
Insect larvae
Larvae of H. armigera were obtained from a laboratory colony maintained in the entomology biochemistry and molecular pathology laboratory of Hebei Agricultural University (Baoding, Hebei). Larvae were reared on an artificial diet. Fifth instar larvae were used for dissection to isolate PMs and various tissues for analyses.
Proteins associated with larval PM and midguts
Midgut tissues were dissected from early to mid-fifth instar H. armigera larvae in cold Rinaldini's solution[5]. Food contents enclosed within the PM and other tissues attached (i.e. fat bodies, trachea, and malpighian tubules) were quickly removed from the midgut epithelium on ice. Isolated PMs were rinsed until clarity in ddH2O for several times. Both the PMs and midgut were collected in ddH2O and 2xSDS-PAGE loading sample buffer [13,14] was added in an equal volume. After 8% SDS-PAGE, the gel was stained by Periodic acid-Schiff (PAS) staining and followed by commassie staining.
Construction of a cDNA expression library from the midgut of Helicoverpa armigera
Isolated midgut tissues were rinsed with cold Rinaldini's solution, quickly frozen in liquid nitrogen, and stored at -70 °C prior to use. H. armigera midgut mRNA was prepared using the RNeasy total RNA isolation kit and the Oligotex mRNA isolation kit (Qiagen Inc., Chatsworth, CA), according to the manufacturer's instructions. cDNA was synthesized from H. armigera midgut mRNA using the cDNA Synthesis Kit (Stratagene, La Jolla, CA), following the manufacturer's instructions.
The cDNAs prepared above were unidirectionally ligated into the Uni-ZAP XR vector (Stratagene, La Jolla, CA) between the EcoRI and XhoI sites by T4 DNA ligase. Packaged following the instructions provided by the manufacturer (Stratagene, La Jolla, CA), finally the constructed library was amplified.
cDNA Library screening and cloning of a cDNA coding for the PM protein HaIIM86
Screening of the cDNA expression library for IIM cDNA clones was conducted using a polyclonal antiserum specified Spodoptera exigua PM proteins in conjunction with the pico BlueTM Immunoscreening Kit (Stratagene, La Jolla, CA), according to the manufacturer's specifications. The first round of screening was performed at a high density (i.e. 50,000 plaques/15-cm plate). Positive plaques were selected and further purified by screening at a low plating density (i.e. 30-100 plaques/9-cm plate). From purified positive phages, the pBluescript® SK(-) phagemid was excised in vivo following the ZAP-cDNA Gigapack cloning kit protocol(Stratagene, La Jolla, CA).
Plasmid DNA was prepared using the CTAB method[13] (Sambrook, 1989) from positive cDNA clones and subjected to sequencing. Both strands of the cDNA were sequenced using T3 and T7 primers, complementary to the pBluescript® SK(-) sequences flanking the cDNA inserts. And primers within cDNA insert to cover the entire cDNA length. Selected positive cDNA clones were subjected to sequencing after restriction enzyme digestion analysis to exclude identical clones. DNA sequence analysis and a data base search were conducted using the DNAMAN software package and BLAST data base search programs. Domain analysis and protein glycosylation sites predicted see to Shi (2004) [9].
Preparation of antibodies reacting to HaIIM86
Antibodies reacting to the H.armigera PM protein, HaIIM86, were prepared from an antiserum made against the whole collection of H. armigera PM proteins as described by Guo[8].
Briefly, E. coli strain XL1-Blue carry pBluescript with the full-length cDNA for HaIIM86 was cultured in 12 ml LB medium and the expression of HaIIM86 was induced by addition of 100 ul of 0.5M isopropyl thiogalactopyranoside (IPTG). The E. coli cells were harvested after induced for 4 h by centrifugation and lysed by boiling in 0.5 ml of lysis buffer (2% SDS, 5% β-mercaptoethanol and 50mM Tris-HCl, pH 8.0) for 5 min. The cell lysate was clarified by centrifugation at 16,000g for 10 min, and the supernatant was collected and diluted with 10 ml of deionized water. The solubilized proteins were immobilized onto a piece of supported nitrocellulose membrane by incubation of the membrane in the lysate at room temperature for 1 h, followed by extensive washing with PBS 5 times and incubation in 3% bovine serum albumin (BSA) for 3 h to block nonspecific binding sites. The nitrocellulose membrane was then incubated with a 100-fold dilution of the antiserum with 3% BSA at room temperature for 3 h or at 4℃ overnight to allow the anti-HaIIM86 antibodies in the antiserum to bind to the blotted membrane. The membrane was then thoroughly washed five times with PBS and finally the bound antibodies specific to the HaIIM86 were eluted from the membrane by incubation in 5ml of 0.1M glycine buffer (pH 2.5) at room temperature for 10 min, followed by addition of 0.5 ml of 1M Tris-HCl buffer (pH 8.0) to neutralize the pH of the antibody preparation.
Recombinant HaIIM86 expression in E.coli
The cDNA for haiim86 was double digested from pHaIIM86 (plasmid from positive library clone) with BamHI and KpnI and ligated into pQE30 vector. The recombined plasmid, named of pQE-haiim86, was transferred into E.coli strain M15(pQEP4). Expression of the recombinant protein HaIIM86 was confirmed by SDS-PAGE and Western blot analysis.
Recombinant HaIIM86 expression and chitin binding analysis
To over express HaIIM86 in insect cells, a recombinant baculovirus was constructed using the Bac-to-Bac system from Invitrogen (Carlsbad, CA, USA). The cDNA for the HaIIM86 was excised from pHat46 by digestion with BamHI and KpnI and cloned into the vector pFASTBac1. The recombinant baculovirus with the cDNA for HaIIM86 was generated by following the protocols provided by the manufacturer.
Recombinant HaIIM86 was produced as a secreted protein by infecting BTI-Tn-5B1-4 (HighFive) cells with the recombinant baculovirus and the infected culture was maintained in TNM-FH medium supplemented with 10% fetal bovine serum (FBS). The cell culture medium containing the secreted recombinant HaIIM86 was collected at 72 h post-infection. The chitin-binding activity of HaIIM86 was analyzed using a chitin-binding assay described by Wang et al. (2004) [10]. Recombinant HaIIM86 was isolated by incubation of 1ml of HaIIM86 containing cell culture medium with 100 mg (wet weight) regenerated chitin to allow the HaIIM86 protein to bind to chitin at 4 ℃ for 1 h in the presence of a cocktail of protease inhibitors (0.5 mg/ml leupeptin, 1 mg/ml pepstatin, 1mM EDTA and 1mM phenylmethylsulfonyl fluoride). The regenerated chitin bound with HaIIM86 was washed thoroughly with PBS followed by centrifugations. Aliquot of the resulting chitin bound with HaIIM86 was incubated with PBS, 2% SDS, 6M Urea, 0.2% Calcofluor, 0.5M NaCl, or 20mM acetic acid respectively. After 15 min incubation at room temperature, the supernatants containing the HaIIM86 protein released from the chitin were collected after centrifugation and analyzed by SDS-PAGE.
Function and localization of HaIIM86 in H. armigera larvae
Recombined HaIIM86 expressed in insect cells and HaGV enhancin (50ng/mL) were mixed and incubated on 37 ℃ for 4 hrs or overnight. Products were examined by western blot analysis with HaIIM antiserum.
To identify and localize the HaIIM86 in H.armigera larvae, different instar larvae were dissected to isolate various tissues/structures, including hemolymph, tissue from the midgut, integument and the PM. Exuviae from the larvae molting from fourth instar to fifth instar and fecal pellets from fifth instar larvae were also collected. Midgut digestive fluid was collected from mid-fifth instar larvae by stimulating the larval mouth parts in a test tube to induce regurgitation of the midgut fluid. The samples were analyzed for the presence of HaIIM86 by Western blot analysis using the antibodies reacting to HaIIM86. Briefly, proteins from the samples were solubilized by boiling in SDS-PAGE sample buffer. Similar amounts of proteins from each tissue sample, which were estimated visually by SDS-PAGE analysis of the samples, and proteins from 1 PM, 1 ml midgut fluid and 1 exuvia were resolved by SDS-PAGE analysis, followed by transfer of the proteins onto nitrocellulose membrane. Then, the membrane was probed with antibodies reacting to HaIIM86 after treatment of the membrane with 1% BSA in PBS to block non-specific bindings. The positive antibody reaction was detected with goat anti-rabbit IgG antibodies conjugated with alkaline phosphatase and visualized by the colorimetric reaction with bromochloroindolyl phosphate/nitro blue tetrazolium as the substrate.
RESULTS
Proteins associated with larval PM and midguts
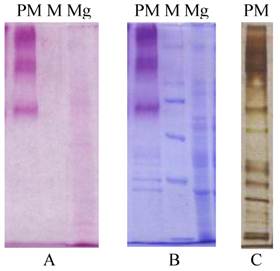
The peritrophic membrane (PM) of Helicoverpa armigera larvae was found to contain about forty associated proteins by SDS-PAGE analysis (Figure 1). And for the midgut there probably be tens. There are many more glycoproteins in Helicoverpa armigera peritrophic membrane than midgut separated by SDS-PAGE analysis after Periodic acid-Schiff (PAS) and coomassie staining (Fig.1).
Protein composition by SDS-PAGE analysis for H. armigera PM & midgut. A&B: Gels stained by Periodic acid-Schiff (PAS) before and after coomassie staining; C: Silver staining; PM, peritrophic membrane; Mg, midgut; M, protein Marker, 220, 170, 116, 76, 49kDa
Cloning and characterization of IIM cDNA
The original cDNA expression library contained 4.82×106 recombinants and the recombination rate was 99.79%. A positive cDNA clone for PM, named as HaIIM86, was identified and sequenced. The size of HaIIM86 is 3,147bp in length, and the longest open reading frame codes for 839 amino acids and is similar to Plutella xylostella intestinal mucin in structure (Fig.2). It contained five chitin binding domains and one threonine-rich mucin domain. Six tandem repeats of TTTQAPTTT are present in the mucin domain. The T residues in the repeating unit are predicted to be the potential O-glycosylation sites. In an aspartic acid-rich region, a repeating unit, DDDKPGCNGNCPE repeats 12 times, distinguishes the HaIIM from known IIMs[5-7, 9, 11, 15, 16]. The GenBank accession number for HaIIM86 was EU047712.
Characterization of HaIIM86 mucin-like domains
The former study reports that mucins are large glycoproteins generally with a high proline, threonine and serine content. They are normally associated with mucus layers at the interface between epithelia and their respective environments, particularly the lumens of hollow organs of vertebrate respiratory, digestive and urogenital tracts[15]. The mucin domain of HaIIM86 was similar in structure to 'mucin' domains. It exhibited a high proline (13.74%) and threonine (67.18%) content, accounting for 22.77% of the total amino acid residues; however, the serine content was not elevated. The mucin domains were also rich in alanine (10.69%) and glutamine (6.11%) (Table 1). Groups of 4 or 12 amino acids (VAPTAAPTTTTT and TAAP) were repeated 5 or 9 times within the individual mucin regions (Table 1). The repetitive regions within the mucin domains of T. ni[3] and M. configurata IIM[9] tended to be short, four to eight amino acids in length, and simple in structure with some elements being repeated as many as twenty-five times. In contrast, the tandem repeats of Px IIM were larger, up to 27 amino acids long, relatively complex and less frequent (Table 1).
Repeat sequences in mucin domains from known IIMs
| Domain/Protein | Repeat sequence | Number of repeats |
|---|---|---|
| HaIIM86 | VAPTAAPTTTTT TAAP /TTTTQAPTTTT | 5 9 8 |
| PxIIM Muc1 | PEV(V/T)T | 11 |
| PTT(L)E | 6 | |
| TTL(A/E)PETDA | 7 | |
| PxIIM Muc2 | SEAETVEVVTA(I/V)PTT(TE) | 9 |
| NAPETEATTVAETETPEVVTAVPTTTE | 4 | |
| PxIIM Muc3 | EEP(I/V)(A/V)TVTA(P/T) | 16 |
| EEPVATVTAT | 3 | |
| VVTVTAT | 2 | |
| EVVT | 2 | |
| M. configurata IIM | TAAP | 24 |
| TPVAVVT | 2 | |
| PTVAPETTA | 4 | |
| VTNA | 4 | |
| T. ni IIM | TTQAPT | 15 |
| AATTP | 6 | |
| TAAP | 25 | |
| TTVT | 5 |
Nucleotide sequence of the cDNA for Helicoverpa armigera intestinal mucin and its deduced amino acid sequence. Signal peptide domains (grey background), threonine-rich domains (Muc1, wavy underlined), cysteine-rich regions (CBD1-5, underlined), and the translation stop codon (in box) are indicated. Additional aspartic acid-rich regions are marked with _ _ _ _ _


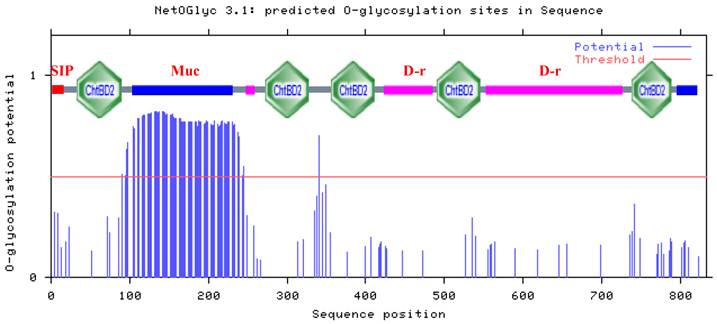
Of the 90 threonine residues in HaIIM86, 74 (82.2%) were predicted to be O-glycosylated (Fig. 3). Three potential amino N-glycosylation sites were identified at Asn 415, 423 and 562. The high degree of glycosylation imparts a viscousquality to mucus secretions, which is fundamental to their role in protecting epithelial cells from infection, dehydration, physical and chemical damage, as well as from the proteolytic and caustic or acidic environment of the gut[15-17]. Half the total mass of mature T. ni IIM was composed of carbohydrate moieties, with 86% of threonine and 22% of serine residues predicted to be O-glycosylated.The protein was highly resistant to proteolytic attack, a characteristic that was attributed to the high degree of O-glycosylation [3]. A similar proportion of the threonine residues within HaIIM was predicted to be glycosylated and thus comparable physical and biochemical properties may be inferred. Proline residues are often found interspersed among the glycosylated amino acids within IIMs.
Prediction of O-glycosylation sites and structure in H. armigera intestinal mucin HaIIM86. Residues with an O-glycosylation potential exceeding the threshold level (red line) are expected to be glycosylated. SIP, signal peptide sequence; Muc, mucin-like domain; ChtBD2, chitin-binding domain; D-r, Glycine-Aspartic acid-rich domains. Asn-Xaa-Ser/Thr sequons in the sequence output below are highlighted in blue.
Characterization of HaIIM86 chitin-binding domains
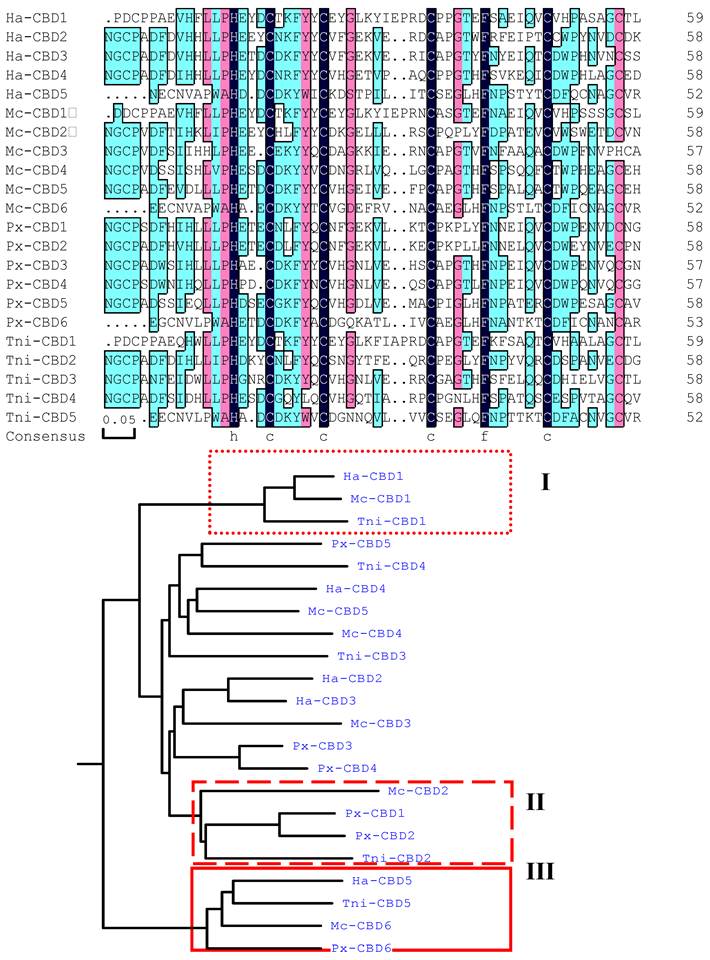
Six cysteine-rich regions were identified within HaIIM86, each of which possessed a register of six, spatially conserved, cysteine residues that form a putative CBD. These regions will hereafter be referred to as CBD (Fig. 4A)[10]. The consensus sequence of the HaIIM86 CBDs was CX13−15CX5CX9CX12CX7C, the same as that reported for PxIIM, T. ni[3] and M. configurata[9] IIMs. Cysteine-rich regions are also common to mucins[16]. The cysteine residues in vertebrate mucins are thought to facilitate mucin multimerization through the formation of intermolecular disulphide bonds[16, 18, 19]. However, the CBD of invertebrate mucins are believed to be involved in attachment to chitin and contribute to the PM structure[20]. The rigid structure of the mucin domains may ensure proper spacing of the CBDs associated with the chitin microfibrils such that the highly ordered, sieve-like structure of the PM is maintained.
Like that of T. ni, Px and M. configurata IIM, the sequence of the HaIIM86 carboxy-terminal CBD is distinct from those upstream (Fig. 4A). In fact, the carboxy-terminal domains from the four IIMs were more closely related to each other than to CBDs within the same protein (Fig. 4B). In addition to the six-cysteine register, the terminal CBDs of IIM possessed two additional conserved cysteine residues located four residues downstream from Cys1 and four residues upstream from Cys6 (Fig. 4A). Such conserved cysteine residues have not been described for any other CBD. It is not known whether these residues contribute an additional intramolecular disulphide bridge within the CBD or if they participate in intermolecular interactions leading to mucin multimerization as in vertebrate mucins[16, 18].
In this study, amino terminal CBD were classified into one group also (I and II regions in Fig.4B). Amino terminal CBD of PxIIM, the second CBD of M. configurata, T. ni and H. armigera, were grouped together. It is notable that PxIIM lacks amino acid sequence of 5' end referred by Sarauer[6]. So it is probably that amino terminal CBD in IIMs is conserved.
In addition to the conserved cysteine residues, aromatic residues were also present in conserved positions between Cys2 and Cys3 and between Cys4 and Cys5. These aromatic residues were invariably phenylalanine in HaIIM and PxIIM, but tyrosine was also observed in several of the Cys2-Cys3 locations in T. ni and M. configurata CBDs.
Alignment of insect intestinal mucin chitin-binding domains (CBD) from Mamestra configurata intestinal mucin, (Mc, genbank accession number AY057052), Plutella xylostella intestinal mucin (Px) (Genbank accession number AF545582), Trichoplusia ni intestinal mucin IIM14 (Tni) (Genbank accession number AF000605), and Helicoverpa armigera intestinal mucin (Ha) (Genbank accession number EU047712). A: The alignment of CBDs from known IIMs. B: Dendrogram shows the relationship among CBDs from known IIMs. The carboxyl-terminal CBD from each IIM formed a single group (in box).
Structural organization of insect intestinal mucin
Like Spodoptera exigua mucin SeIIM-8[11], HaIIM86 possess one type of region distinct from other known insect intestinal mucin: aspartic acid-rich (D-G rich) region. These two D-G rich regions were proposed to internal reside at the amino terminus of the protein flanked by three cysteine-rich CBDs. The first D-G rich region is shorter and the glycine and aspartic acid content is 32.71% and 26.55% separately. Sequence CDGSCPDNGGNDGN repeated for 2 times. Glycine and aspartic acid content is 41.24% and 28.25% in the second region, meanwhile repetitive sequence NDGG occurred 20 times. Compared with other IIMs, it was the first found for the two D-G-rich tandem repeat domains flanked by cysteine-rich sequences in peritrophic membranes proteins and also the first features in known proteins by now. But the function of this structure is not known now.
Recombinant HaIIM86 expression and chitin binding analysis
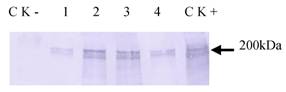
Haiim86 was cloned into pQE30 vector and about 200 kDa protein was detected in E. coli M15 by Western Blot analysis (data not shown). It was also expressed successfully in insect cells (Tn-5B1-4) with the recombinant baculovirus (Fig. 5). The recombinant protein, which was secreted into the cell culture medium, exhibited its activity to bind chitin (Fig. 6). The chitin-bound HaIIM86 could only be released from the chitin by the competitive chitin binding reagent Calcofluor (Sigma, St Louis, MO, USA). Treatments of the HaIIM86/chitin complex with PBS, 0.5M NaCl, 2% SDS, 20mM acetic acid and 0.1M sodium carbonate buffer (pH 10.5) did not result in detectable dissociation of HaIIM86 from chitin.
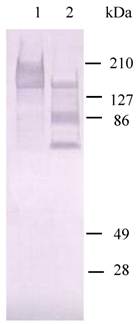
Western Blot analysis of expression of HaIIM86 in insect cells. 1-4, supernatant from cell culture infected by recombinant baculovirus for 24, 72, 96, 48hs; CK+, cells infected by recombinant baculovirus; CK-, Sf9 cells.
Characteristics and localization of HaIIM86 in H. armigera larvae
The function study on HaIIM86 demonstrated that it could be degraded into two protein bands ( 70 kDa and 90 kDa) by HaGV Enhancin (Fig.7). The phosphate-buffered saline buffer was used as negative controls. The former study showed that Enhancin has ability to attack the PM of H. armigera in vivo, but the destruction pattern and target are not clear. Here, IIM degradation assays in vitro showed that IIM of H. armigera is target for enhancin action.
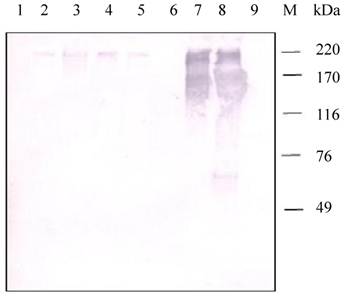
Western blot analysis of samples from different tissues of H. armigera larvae positively detected the presence of a HaIIM86-like protein (Fig.8). In contrast to the observation in H.armigera larvae, positive reactions were detected in the proteins from the PM, midgut, pupa, fecal pellets, but negative from fat bodies and hemolymph.
Analysis of chitin-binding activity of recombinant HaIIM86. M, protein marker, 220, 116, 96, 66, 43kDa; HaIIM86 treated with 0.2% Calcoflour (1) and 1% Calcoflour (2).
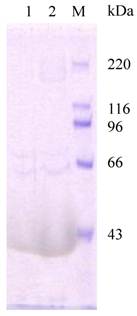
HaIIM86 is the target for enhancin action. The degradation of HaIIM86 was analyzed by Western blot analysis using anti-HaIIM86 antiserum. Lane 1, HaIIM86; lane 2, HaIIM86 treated by enhancin
Detection of HaIIM86 in H. armigera tissues. 1, fat bodies; 2, pupa; 3, fecal pellets; 4-5, PM from early and middle of fifth instar larvae; 6, hemolymph; 7-8, midgut from early and middle of fifth instar larvae;9, DH5a(pMal- SeCBP66); M, Protein Marker
DISCUSSION
Proteins are the primary components of the PM and the formation of PMs has been suggested to involve binding of PM proteins to chitin fibrils [2, 3, 14, 19, 20]. In this study, HaIIM86 is a full length PM protein while we isolated many partial chitin-binding proteins which posses different numbers of chitin-binding domains and IIM which posses both mucin domains and CBDs. The proteins are composed of numerous chitin binding domains, indicated a mechanism for these PM proteins to adapt and function in the proteinase rich gut environment. It was previously demonstrated that the PM proteins could be degraded before and/or after being assembled into the PM structure in T. ni [10]. Here the chitin binding domains in H.armigera PM is shorter than that in T.ni. And recombined HaIIM86 was expressed successfully in insect cells with the recombinant baculovirus, exhibited its activity to bind chitin. It is notable that recombined HaIIM86 could be degraded by enhancin. Wang reviewed the molecular structure of the PM and using PM as a target to conduct pests control in 2001. This strategy of pests control using PM as a target includes degradation of PM-associate proteins and interfering chitin fibrils binding with PM-associate proteins [3, 19]. So this study may contribute to the discovery of the biocontrol target on HaIIM86 for H.arnigera control in the coming day.
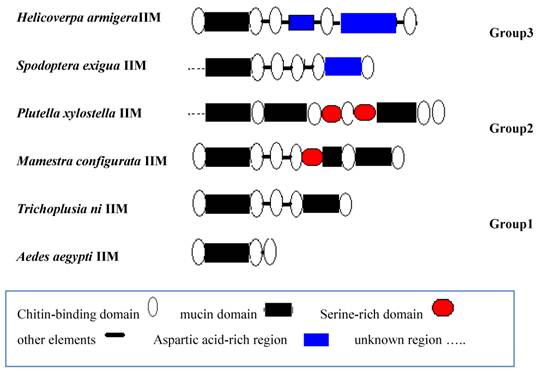
We analyzed the sequence structure of known IIMs from different insects and classified IIM into three groups (Fig.9). Group1: There are several CBDs and mucin domains in structure. For example, T.ni and Aedes aegypti IIM belongs to this group. Group 2: There are several CBDs, mucin domains and serine-rich domains in structure just as P.xylostella and M. configurata IIM. The third group contains not only CBDs, mucin domain, but also D-G rich regions.
Up to date, the molecular structure and the function of PM and how the PM work are not very clear. Therefore, there is much work we should do. In one hand, our description of HaIIM86 here expands knowledge of the insect PM. In another hand, as the target for enhancin, it may prove to be a novel strategy for insect control.
Structure analysis of known insect intestinal mucin proteins
Acknowledgements
This work was supported by National Program on Key Basic Research Projects (“973” program, 2009CB118902), National Natural Science Foundation of China(30771447), supported by the earmarked fund for Modern Agro-industry Technology Research System and Hebei Natural Science Foundation (C200700540).
CONFLICT OF INTERESTS
The authors have declared that no conflict of interest exists.
References
1. Leanhe MJ. Peritrophic matrix structure and function. Annu Rev Entomol. 1997;42:525-50
2. Tellam RL, Wijffels G, Willadsen P. Peritrophic matrix proteins. Insect Biochem Mol Biol. 1999;29:87-1011
3. Wang P, Granados RR. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc Natl Acad Sci USA. 1997;94:6977-6982
4. Ji HH, Yuan ZM. Peritrophic membrane: a potential target for biocontrol of pest insects. Acta Entomol Sin. 2005;48(6):968-974
5. Wang P, Granados RR. Molecular Cloning and Sequencing of a Novel Invertebrate Intestinal Mucin cDNA. J Biol Chem. 1997;272(26):16663-16669
6. Sarauer BL, Gillott C, Hegedus D. Characterization of an intestinal mucin from the peritrophic matrix of the diamondback moth, Plutella xylostella. Insect Mol Biol. 2003;12(4):333-43
7. Devenport M, Alvarenga PH, Shao L. et al. Identification of the Aedes aegypti peritrophic matrix protein AeIMUCI as a heme-binding protein. Biochem. 2006;45(31):9540-9
8. Guo W, Li GX, Pang Y. et al. A novel chitin binding protein identified from the peritrophic membrane of the cabbage looper, Trichoplusia ni. Insect Biochem Mol Biol. 2005;35:1224-1234
9. Shi X, Chamankhah M, Visal-Shah S. et al. Modeling the structure of the type I peritrophic matrix: characterization of a Mamestra configurata intestinal mucin and a novel peritrophin containing 19 chitin binding domains. Insect Biochem Mol Biol. 2004;34(10):1101-15
10. Wang P, Li GX, Granados RR. Identification of two new peritrophic membrane proteins from larval Trichoplusia ni: structural characteristics and their functions in the protease rich insect gut. Insect Biochem Mol Biol. 2004;34:215-2271
11. Zhang X, LI GX, GUO W. Molecular Cloning and sequencing of an insect intestinal mucin cDNA from Spodoptera exigua. Scientia Agricultura Sinica. 2008;41(11):3898-3904
12. Campbell PM, Cao AT, Hines ER. et al. Proteomic analysis of the peritrophic matrix from the gut of the caterpillar, Helicoverpa armigera. Insect Biochem Mol Biol. 2008;38(10):950-8
13. Sambrook J, Fritisch EF, Maniatis T. Molecular cloning A laboratory manual, 2nd ed. Beijing: Science Press. 1989
14. Pauchet Y, Muck A, Svatos A. et al. Mapping the larval midgut lumen. proteome of Helicoverpa armigera, a generalist herbivorous insect. J Proteome Res. 2008;7(4):1629-39
15. Van Klinken BJ-W, Dekker J, Buller HA. et al. Mucin gene structure and expression: Protection vs. adhesion. Am J Physiol. 1995;269:G613-G627
16. Perez-Vilar J, Hill RL. The structure and assembly ofsecreted mucins. J Biol Chem. 1999;274:31751-31754
17. Van den Steen P, Rudd PM, Dwek RA. et al. Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol. 1998;33:151-208
18. Gum JR, Hicks JW, Toribara NW. et al. The human MUC2 intestinal mucin has cysteine-rich subdomains located both upstream and downstream of its central repetitive region. J Biol Chem. 1992;267:21375-21383
19. Wang P, Granados RR. Molecular structure of the peritrophic membrane (PM): identification of potential PM target sites for insect control. Insect Biochem Physiol. 2001;47:110-118
20. Shen Z, Jacobs-Lorena M. Evolution of chitin-binding proteins in invertebrates. J Mol Evol. 1999;48:341-347
Author contact
![]() Corresponding author: Wei Guo, Agricultural University of Hebei, Baoding, Hebei 071001, People's Republic of China. Tel: 86-0312-7528178. Email: guoweiedu.cn, guowei_hbaucom.cn
Corresponding author: Wei Guo, Agricultural University of Hebei, Baoding, Hebei 071001, People's Republic of China. Tel: 86-0312-7528178. Email: guoweiedu.cn, guowei_hbaucom.cn

 Global reach, higher impact
Global reach, higher impact