Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2011; 7(5):505-516. doi:10.7150/ijbs.7.505 This issue Cite
Research Paper
Identification and Characterization of the Alternatively Spliced Nuclear Receptor Coactivator-6 Isoforms
1. Department of Molecular and Cellular Biology, Baylor College of Medicine, Houston, Texas 77030, USA.
2. Luzhou Medical College, Luzhou, Sichuan 646000, China
Received 2011-3-14; Accepted 2011-4-4; Published 2011-4-19
Abstract
The nuclear receptor coactivator-6 (NCOA6, AIB3, PRIP, ASC-2, TRBP, RAP250 or NRC) is a co-activator for nuclear hormone receptors and certain other transcription factors. NCOA6 plays an important role in embryonic development, adipocyte differentiation, metabolism and breast carcinogenesis. The human and mouse NCOA6 genes had 15 and 14 previously identified exons, respectively. This study further identified an alternatively spliced exon 11b (E11b) in human or E10b in mouse, which codes a short polypeptide and a Stop codon, resulting in splicing variants lacking the last four exon-coded polypeptide. Analyses of mouse testis NCOA6 mRNAs identified four alternatively spliced variants, NCOA6-α (without E10b), -β (without E10a and E10b), -γ (with E10a and E10b) and -δ (without E10a but with E10b). These isoforms were detected in multiple mouse tissues and in MDA-MB-435 human cells. NCOA6-α and -β are mainly located in the nucleus; NCOA6-γ is located in both cytoplasm and nucleus; and NCOA6-δ is mainly located in mitochondria. The C-terminus coded by the last four exons was responsible for locating NCOA6-α and -β into the nucleus. The human E11a or mouse E10a-coded region is responsible for distributing NCOA6-γ in both cytoplasm and nucleus, while the region coded by E8-E9 in human or E7-E8 in mouse is responsible for directing NCOA6-δ to mitochondria. Our assays also demonstrated that NCOA6-α and -β could significantly enhance estrogen receptor α-mediated transcription, but NCOA6-γ and -δ were unable to do so. These results suggest that the diverse physiological function of NCOA6 may be mediated by multiple isoforms expressed in different tissues and localized in different subcellular compartments.
Keywords: nuclear receptor coactivator-6, NCOA6
Introduction
The nuclear receptors (NRs) are hormone-inducible transcription factors that play important roles in various physiological processes by regulating gene expression. Hormone-activated NRs regulate their target genes mainly through recruiting coactivators or corepressors to influence chromatin remodeling and basal transcription factor assembly (1, 2). Among many identified NR coregulators, the nuclear receptor coactivator-6 (NCOA6) has many unique biological functions. NCOA6 is an essential coactivator for embryonic development, as knockout of the NCOA6 gene in mice results in embryonic lethality accompanied with defective development of placenta, heart and liver (3-6). Disruption of one NCOA6 allele in mice accelerates polyoma middle-T antigen-induced mammary tumorigenesis, decreases insulin secretion and reduces postnatal growth, suggesting a functional haplodeficiency of the NCOA6 gene (6-8). Furthermore, specific deletion of the NCOA6 gene in mouse mammary epithelial cells decreases mammary ductal growth regulated by estrogen and partially impairs milk synthesis (9). Moreover, the NCOA6 gene is amplified and overexpressed in subsets of the breast, colon and lung cancers, suggesting its potential role in carcinogenesis (10-16).
The NCOA6 gene is broadly expressed in many tissues (17). The human NCOA6 gene (NCBI Gene Database, ID 23054) contains 15 exons and codes for 2063 a.a. residues (18). The largest Exon 11 is about 3 kb in length. The start and stop codons are located in exons 3 and 15, respectively. The mouse NCOA6 gene (NCBI Gene Database, ID 56406) contains 14 exons and codes for 2069 a.a. residues. The start and stop codons are in exons 2 and 14, respectively. The 10th exon of the mouse NCOA6 gene is corresponding to the largest 11th exon of the human NCOA6 gene. The NCOA6 protein contains two LXXLL (L, leucine; X, any a.a.) motifs that serve as nuclear interaction motifs. Ectopic expression of the dominant negative mutant containing the N-terminal LXXLL motif in transgenic mice, which blocks NCOA6 interaction with multiple NRs, causes severe microphthalmia and posterior lenticonus with cataract as well as a variety of pathophysiological phenotypes in many other organs (19). Interestingly, loss-of-functional mutation of the C-terminal LXXLL motif in exon 10 of the mouse NCOA6 gene does not affect embryonic development and postnatal growth as well as the estrogen receptor α (ERα) function. Instead, it specifically disturbs the liver X receptor α (LXRα)-regulated lipogenesis and cholesterol/bile acid homeostasis in the liver (20). At the molecular level, NCOA6 may enhance NR-dependent transcription through its interaction and recruitment of multiple coactivator complexes, such as ASC-2 (NCOA6) complex, SRC-1/CBP (steroid receptor coactivator-1/cAMP response element-binding protein binding protein) complex, TRAP (thyroid receptor associated protein) or DRIP (vitamin D receptor-interacting protein) complex, and COAA (coactivator-associated coactivator) (11-16, 21, 22).
Although there are only about 30,000 protein-coding genes in the human genome, alternative splicing events of mRNAs and posttranslational modifications markedly increase the total number of protein molecules as well as their functional diversities. Since the NCOA6 gene has a complex structure and its protein plays important and diverse biological functions during development, organ function and carcinogenesis, it should be important to explore possible splicing variants of NCOA6 and their respective molecular and functional features. However, although the existence of a NCOA6 splicing variant has been suggested at the mRNA level, NCOA6 isoform proteins generated from alternative splicing are largely unknown. In this study, we have identified four NCOA6 splicing isoforms, examined their in vivo expression patterns and characterized their subcellular localizations and transcriptional activities. These findings should provide molecular structural bases for understanding the diverse biological function of the NCOA6 gene.
Materials and Methods
Plasmid construction. The pcDNA3-TRBP (NCOA6) expression plasmid containing human NCOA6-α cDNA with a 5' Flag tag was described previously (13). The other NCOA6 isoform cDNAs and cDNA fragments were amplified by PCR and subcloned into the same plasmid by replacing the NCOA6-α cDNA. To express GFP-NCOA6 fusion proteins, NCOA6 cDNA fragments were amplified by PCR and ligated to the 3' GFP coding sequence in the pEGFP-C3 plasmid (BD Biosciences Clontech, Mountain View, CA). All newly constructed plasmids were verified by DNA sequencing.
RT-PCR. Total RNA was extracted from cells and tissues using the TRIzol reagent (Invitrogen, Carlsbad, CA). The purified RNA was resuspended in RNase-free water. RNA quality was estimated by running 1% agarose gel. RNA samples were treated with RNase-free DNase I to remove residual genomic DNA. Two μg of total RNA was reversely transcribed into cDNA using the SuperScript II Reverse Transcriptase kit (Invitrogen, Carlsbad, CA). The PCR primers used for detecting mouse NCOA6 splicing variants include P16 (CAGAGTGACATTTCTGCAGG), P13 (ACCCCAGATAATCGTCCAAC), 65F (CCTCAACAGGACCCATCTTAC), P23 (CCTTGCAGTGCTGTGGC) and 3R (TCAAGTCATCTTCCAGCTCGC) (Fig. 1A). The GAPDH-F (CTGGTCACCAGGGCTGC) and GAPDH-R (GTGAGTTGTCATATTTCTCGTGG) primers were used for detecting GAPDH cDNA, which served as a control. The PCR primers used for detecting human NCOA6 splicing variants include hNCOA6-P1 (CAGAGTGACATATCTGCAGG) in exon 9, hNCOA6-P1R (CTCCAGCTCTGGTCTGGC) in exon 12, hNCOA6-P2F (GCAATCATGGTGGCATAGC) in exon 11a and hNCOA6-P2R (GCACTCGAGCTTGTTTACTTGGATTTTC) in exon 15.
Immunocytofluorescence. Cells cultured on slide cover slips were washed twice with PBS and fixed in freshly made 4% paraformaldehyde (pH7.4) for 10 minutes on ice. Fixed cells were permeabilized in PBS containing 0.1% Triton X-100 for 5 minutes, blocked with 1% BSA in PBS at room temperature for one hour, and then washed three times with PBS. The prepared cells were incubated overnight at 4oC with anti-Flag monoclonal antibody at 1:5000 dilution in PBS with 1% BSA (Sigma, St. Louis, MO). After washed 5 times in PBS, the cells were incubated in 1:200 dilution of the FITC-labeled goat-anti-mouse IgG (Zymed laboratories, South San Francisco, CA), washed twice in PBS and mounted in Vectashield mounting medium with DAPI (Vector, Burlingame, CA). The mitochondrial staining was performed using the MitoTracker and MitoFluor Mitochondriaon-Selective Probes kit (Molecular Probes, Eugene, OR) by following the manufacturer's instruction.
Immunoblotting. Whole cell lysate was extracted using RIPA buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS and 1% NP-40) containing proteinase inhibitor cocktail. Cytosolic and nuclear extracts were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Fisher Scientific, Rockford, IL). Mitochondrial extract was prepared using the Mitochondria Isolation Kit for Cultured Cells (Thermo Fisher Scientific, Rockford, IL). Cellular and subcellular extracts were separated in SDS-polyacrylamide gels and blotted onto nitrocellulose membranes. The blots were probed with individual primary antibodies followed by appropriate secondary antibodies conjugated with horseradish peroxidase (HRP) (BD Biosciences, San Jose, CA). The blots were developed using the chemiluminescent horseradish peroxidase substrate solution (Millipore Co., Billerica, MA) for about 5 min and then exposed to X-ray films. The primary antibodies included polyclonal antibodies against the mouse exons 3-4 (corresponding to human exons 4-5) coded region of NCOA6 (7) and monoclonal antibody against the human exons 13-15 (corresponding to mouse exons 12-14) coded region of NCOA6 (provided by Dr. Lee). Other primary antibodies included monoclonal antibodies against heat shock protein 90 (Hsp90) (Sigma, St. Louis, MO) and voltage-dependent anion-selective channel protein 1 (VDAC) (Calbiochem, Gibbstown, NJ) and polyclonal antibodies against poly-(ADP-ribose) polymerase (PARP) (Upstate Biotechnology, Lake Placid, NY).
Cell Culture, Transient Transfection and Luciferase Assay. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). In subcellular localization experiments, Hela cells and NCOA6 null mouse embryonic fibroblasts were transiently transfected with plasmids for expressing Flag-tagged NCOA6 isoforms and mutants or with plasmids for expressing GFP-NCOA6 fusion proteins. Cells expressing Flag-tagged proteins were analyzed by immunocytofluorescence, while live cells expressing GFP fusion proteins were analyzed by green fluorescent microscopy. To measure the coactivator activities of NCOA6 isoforms, Hela cells were cultured in 24-well plates in phenol red-free medium containing 5% of charcoal-stripped FBS and transfected with plasmid DNAs using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). Specifically, cells were co-transfected with RSV-ERα (0.2 μg) plasmids for ERα expression, ERE-tk-Luc plasmids (0.2 μg) for reporting ERα-dependent transcription, RSV-β-galacrosidase expression plasmids (0.1 μg) for normalizing transfectional efficiency and one of the pcDNA3-NCOA6 expression plasmids (0.3 μg of pcDNA-NCOA6-α, β, γ or δ). Transfected cells were treated with 10-8 M of 17β-estradiol for 24 hours, and then harvested and extracted with the cell lysis buffer in a Luciferase Assay Kit (Promega, Madison, WI). Luciferase activity in the cell lysates was assayed using the same kit. The β-galactosidase activity was assayed using the Mammalian β-Galactosidase Assay Kit (Thermo Fisher Scientific, Rochford, IL). The luciferase activity was normalized to the β-galactosidase activity in the same sample to represent the relative ERα transcriptional activity enhanced by individual NCOA6 isoforms.
Results
Alternative Splicing of the NCOA6 RNA Creates Four Isoforms
Several species of NCOA6 mRNAs have been detected in multiple human and mouse tissues by Northern blot in previous studies, suggesting the existence of multiple alternative splicing variants of NCOA6 in vivo (14, 15). Indeed, in addition to the full-length NCOA6-α, an alternative splicing variant, NCOA6-β, has been identified at its mRNA level from human testis (14). In comparison with human NCOA6-α with 2063 amino acids, human NCOA6-β isoform only contains 1070 amino acids (1-971 and 1965-2063), which is resulted from skipping over the largest 11th exon of the human NCOA6 gene (14). The mouse NCOA6 gene contains 14 exons, and its largest 10th exon is about 3 kb in length (20). To examine whether the NCOA6-β isoform was also present in mouse, we isolated RNA from the mouse testis and performed RT-PCR using several pairs of primers spanning exons 8-14 (Fig. 1A). As predicted, the NCOA6-α isoform was detected by primer pairs 65F/P23 and 65F/3R, exhibiting as a 248-bp and a 520-bp bands, respectively (Fig. 1B, the lower bands in the indicated lanes). The NCOA6-β isoform without exon 10 was detected by primer pairs P16/P23 (298 bp), P13/P23 (172 bp), P16/3R (570 bp) and P13/3R (444 bp), respectively (Fig. 1B, the lower bands in the indicated lanes). Surprisingly, each of the above RT-PCR reactions detected a higher band in addition to the expected NCOA6-α or β band. Sequence analysis of these higher bands identified a 110-bp insertion sequence in all of these longer RT-PCR products, and sequence alignment analysis matched this sequence to a region of “intron 10” (Fig. 1C). Similar assays using RNA isolated from the MDA-MB-435 human cells also detected a 75-bp insertion sequence corresponding to the similar “intron region” in the human NCOA6 gene (Fig. 1C). These results indicate that the initially designated “intron 10 or 11” of the mouse or human NCOA6 gene actually contains a small exon, which now can be designated as exon 10b (E10b) in mouse or exon 11b (E11b) in human. Accordingly, the formerly named mouse E10 or human E11 can be re-named as E10a or E11a (Fig. 1A and D).
Identification of NCOA6 splicing isoforms. A. A part of the mouse NCOA6 gene structure from exon 8 to exon 14. The boxes represent exons. The arrows indicate the locations and directions of the PCR primers. B. Predicted lengths of PCR product from the indicated PCR primer pairs (left panel) and the RT-PCR products amplified from the NCOA6 mRNA of mouse testis using the indicated PCR primer pairs. The alternative splicing isoforms identified from the upper and lower bands in each lane are labeled below the gel image. C. The DNA sequence of mouse exon 10b and human exon 11b in capital letters and its 5' and 3' intron sequences in small letters at the splicing junctions. Stop codons are italicized. n.t., nucleotide sequences; a.a., amino acid sequences coded by exon 10a in mouse and 11a in human. D. Diagram of the four NCOA6 isoforms derived from alternate splicing of human exons 11a and 11b (left panel). The lengths of coding nucleotides and a.a. residues as well as calculated molecular masses of individual isoforms are summarized in the right panel. GenBank accession numbers: JF707634 - JF707639.
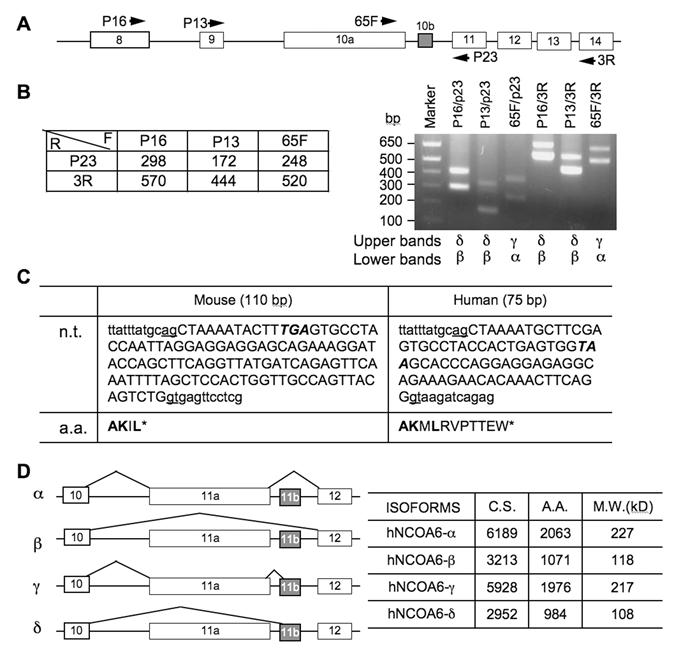
Both mouse E10b and human E11b are flanked with consensus RNA splicing sites and contain respective TGA and TAA stop codons (Fig. 1C). Before these stop codons, the mouse E10b or human E11b encodes for 4 or 11 a.a. residues (Fig. 1C). Therefore, the splicing variants with mouse E10b or human E11b should not have the polypeptide coded by the last four exons in both mouse and human NCOA6 genes (Fig. 1D). As detected by RT-PCR, either inclusion or exclusion of human E11a and/or E11b during RNA splicing generated four NCOA6 isoforms, designated as NCOA6-α (without E11b), β (without E11a and E11b), γ (with E11a and E11b) and δ (without E11a, but with E11b) (Fig. 1D). The predicted lengths of mRNA coding regions and amino acids as well as calculated molecular masses according to the amino acid compositions of human NCOA6 are listed in Figure 1D.
In Vivo Expression of the NCOA6 mRNAs and Protein Isoforms
To examine the expression patterns of NCOA6 splicing isoforms in vivo, we isolated RNA samples from different mouse organs and performed RT-PCR with isoform-specific primer pairs. NCOA6-α mRNA was detected in all organs examined by primers 65F in E10a and P23 in E11. Although NCOA6-β mRNA was also detected in all organs examined by primers P16 in E8 and P23, its expression levels were relatively high in the testis, ovary, kidney, lung and heart but low in the skeletal muscle and uterus (Fig. 2A). Only very low levels of NCOA6-γ mRNA was detected in the testis, cerebellum, liver and skeletal muscle by primers 65F and P23, while NCOA6-δ mRNA was only detected in the testis by primers P16 and P23 (Fig. 2A). These results suggest that NCOA6 isoforms are expressed in a tissue-specific manner.
Next, we analyzed the expression patterns of NCOA6 isoform mRNAs in the mouse mammary glands at different developmental stages. We found that the NCOA6-α mRNA was expressed in all stages including adult virgin, day 18 pregnant, day 15 lactation and day 3 involution mammary glands of female mice (Fig. 2B). The NCOA6-β mRNA was expressed at a low level in the virgin mouse mammary gland, but its level was markedly increased in the mammary glands of pregnant and lactation mice. At involution day 3, the NCOA6-β mRNA was barely detectable (Fig. 2B). However, only low levels of NCOA6-δ mRNA could be detected in the virgin and involution mouse mammary glands (Fig. 2B). These results suggest that NCOA6 isoforms are expressed in the mammary gland in a developmental stage-specific manner.
Expression of the NCOA6 isoforms in mouse tissues. A. Detection of NCOA6 mRNAs in different mouse organs by RT-PCR. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) RT-PCR products served as an endogenous control. B. Analysis of NCOA6 isoform mRNA levels by RT-PCR in the mouse mammary glands at different developmental stages. Vir, P18, L15 and In3, mammary glands from virgin, pregnant day 18, lactation day 15 and involution day 3 female mice, respectively. C. Western blot analysis of mouse testis tissue extracts using antibodies against NCOA6 peptide regions coded by mouse exons 3-4 and exons 12-14, respectively.
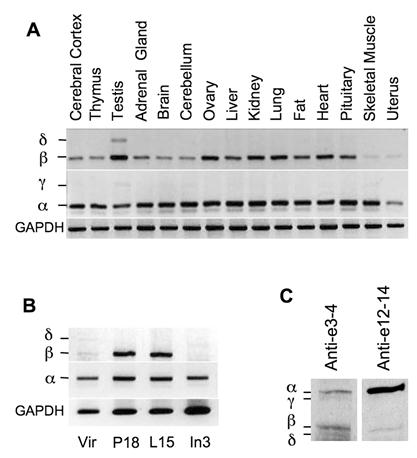
NCOA6 isoform proteins were analyzed by Western blot using polyclonal antibodies generated from recombinant polypeptides encoded by specific exons. All four isoforms were detected in the mouse testis tissue extract by Western blot using a polyclonal antibody against a common N-terminal region encoded by mouse exons 3 and 4. Among these isoforms, NCOA6-α and NCOA6-β protein levels were much higher than NCOA6-γ and δ protein levels (Fig. 2C, lane 1). On the other hand, NCOA6-α and β isoforms were detected in the same tissue extract by Western blot using a monoclonal antibody against a C-terminal region coded by mouse exons 12-14, a region in NCOA6-α and β but not in the other two isoforms (Fig. 2C, lane 2). It was noticed that the C-terminal antibody detected a strong NCOA6-α band but a weak NCOA6-β band, which was probably caused by a limited access of this antibody to the C-terminus of the NCOA6-β isoform since the NCOA6-α and NCOA6-β isoforms showed comparable band intensities as assayed by the N-terminal antibody (Fig. 2C, compare lanes 1 and 2). These results demonstrated that all four NCOA6 isoform proteins are expressed in vivo, while NCOA6-α and β are major isoforms expressed in the testis.
Subcellular localizations of the NCOA6 isoforms
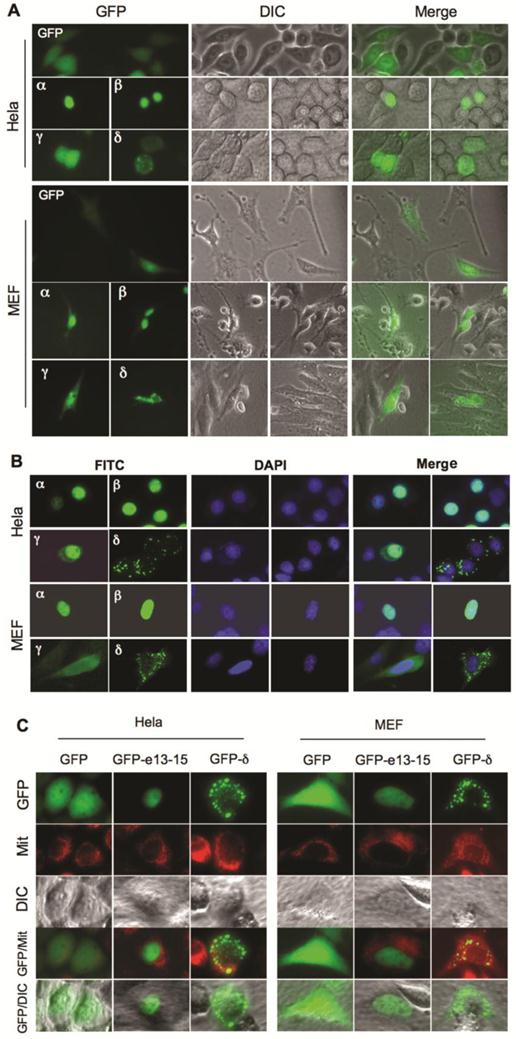
GFP fusion proteins were expressed in Hela and NCOA6 null MEF cells to investigate the subcellular localizations of the NCOA6 isoforms. Fluorescent microscopy revealed that GFP was distributed in the entire cells. GFP-NCOA6-α and -β fusion proteins were mainly located in the nuclei of both Hela and NCOA6 null MEF cells. However, GFP-NCOA6-γ was found in both cytosol and nucleus and GFP-NCOA6-δ was distributed in the cytoplasm as granules (Fig. 3A). To validate these observations, we also expressed Flag-tagged NCOA6 isoforms in Hela and NCOA6 null MEF cells and performed immunocytofluorescence staining. The immunostaining signals of Flag-tagged NCOA6-α and -β were located in the nucleus, the NCOA6-γ signal was in the entire cells, while the NCOA6-δ signal was scattered in the cytoplasm as granules (Fig. 3B). Since the cytoplastic distribution of NCOA6-δ showed a granular pattern, we expressed GFP-NCOA6-δ in Hela and NCOA6 null MEF cells and stained mitochondria with a mitochondrial tracker dye. We found that most of the GFP-NCOA6-δ signals were overlapping with the mitochondrial signals (Fig. 3C). Taken together, these results demonstrated that NCOA6-α and -β are mainly located in the nucleus, NCOA6-γ is located in both cytoplasm and nucleus, and NCOA6-δ is located in the cytoplasm with a large portion in the mitochondria.
Identification of the Sequences Responsible for the Different Subcellular Localizations of the NCOA6 Isoforms
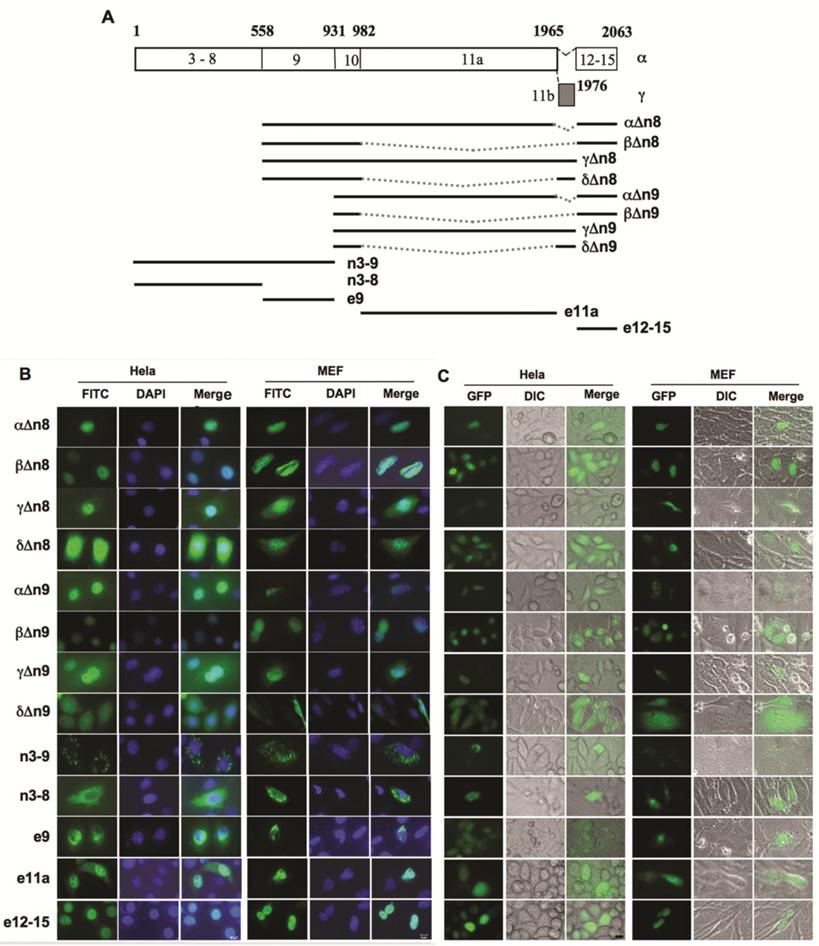
In order to map the specific regions of NCOA6 responsible for nuclear translocalization, we constructed a series of Flag-tagged deletion constructs of human NCOA6 isoforms and expressed these NCOA6 protein fragments in both Hela and NCOA6 null MEF cells (Fig. 4A). Immunostaining analysis revealed that the NCOA6-αΔn8, -βΔn8 and -γΔn8 deletion mutants lacking the N-terminal 558 a.a. (exons 3-8) and the NCOA6-αΔn9, -βΔn9 and -γΔn9 deletion mutants lacking the N-terminal 931 a.a. (exons 3-9) exhibited similar subcellular localizations as their corresponding full-length NCOA6-α, -β and -γ isoforms in Hela cells. Specifically, the α and β deletion mutants were located in the nucleus and γ deletion mutants were located in both cytoplasm and nucleus (Fig. 4B). These results indicate that the N-terminus of these three isoforms is not essential for determining their subcellular localizations.
However, the NCOA6-δΔn8 and -δΔn9 proteins were distributed in the entire cells, which was different from the mitochondrial localization of the full-length NCOA6-δ (Fig. 4B). Interestingly, NCOA6-n3-9 (exons 3-9) with the N-terminal 931 a.a. showed a mitochondrial localization similar to NCOA6-δ, while NCOA6-n3-8 (exons 3-8) with the N-terminal 558 a.a. had a larger portion in the entire cytoplasm versus the mitochondrial distribution of NCOA6-n3-9 (Fig. 4B). The e9 (exon 9)-coded polypeptide was mainly located in the perinuclear area and the nuclear membrane (Fig. 4B). These results suggest that the region of 558-931 a.a. polypeptide coded by exon 9 plays an important but insufficient role in direction of NCOA6-δ to the mitochondrium, and both E8 and E9-coded regions may be involved in efficient localization of NCOA6-δ to the mitochondrium.
NCOA6-γ contains 984 a.a. residues coded by the largest human E11a that does not exist in NCOA6-β and -δ, but does not have the 98 a.a. residues coded by E12-E15 that exists in NCOA6-α and -β (Fig. 4A). Based on the subcellular localizations of NCOA6-α and -β in the nucleus and -γ in both cytoplasm and nucleus, we hypothesized that the C-terminal 98 a.a. polypeptide coded by E12-E15 contains nuclear localization sequence and the E11a-coded polypeptide can partially translocate into the nucleus as NCOA6-γ does. Indeed, the E11a-coded 984 a.a. polypeptide was observed in both cytoplasm and nucleus and the C-terminal 98 a.a. polypeptide was observed in the nucleus in both Hela and MEF cells (Fig. 4B).
Furthermore, we made GFP fusion constructs for all of the deletion mutants of NCOA6 isoforms described above (Fig. 4A) and expressed these GFP fusion proteins in both Hela and NCOA6 null MEF cells. Fluorescent microscopy analysis of GFP signals in these live cells demonstrated identical subcellular distribution patterns of these GFP-fused deletion mutants of NCOA6 isoforms to those Flag-tagged isoforms described above (Fig. 4C).
Subcellular locations of NCOA6 isoforms. A. Subcellular localizations of GFP-fused hNCOA6 isoforms in transfected Hela and NCOA6 null MEF cells. Live cell images were taken by GFP fluorescent microscopy (left column) and diachronic in contrast image (DIC) microscopy (central column). The merged images are also shown (right column). B. Subcellular locations of Flag-tagged hNCOA6 isoforms in Hela and NCOA6 null MEF cells. FITC (green) signals indicate the cellular locations of the Flag-tagged hNCOA6 isoforms. DAPI staining indicates the cell nuclei. C. Hela cells and NCOA6 null MEF cells were transfected with GFP, GFP-hNCOA6-e13-15 or GFP-hNCOA6-δ expression plasmid. Cells were stained with mitochondrial tracker (red color) and examined by fluorescent and DIC microscopy. Merged images of GFP and mitochondrial (Mit) staining as well as GFP and DIC are shown.
Identification of NCOA6 isoform sequences responsible for their subcellular localizations. A. Diagram showing a.a. sequences of hNCOA6-α and -γ coded by specific exons, as well as all other constructed and expressed isoform fragments used in mapping the regions responsible for their subcellular localizations. B. Subcellular localizations of Flag-tagged hNCOA6 isoform fragments (green) in transfected Hela and NCOA6 null MEF cells. Cell nuclei were stained with DAPI. C. Subcellular localizations of GFP-fused hNCOA6 isoform fragments.
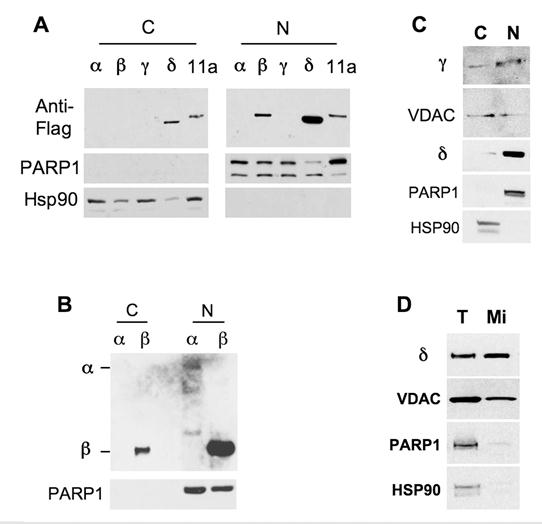
Moreover, we prepared cytoplasmic fractions and nuclear extracts from Hela cells transfected with Flag-tagged NCOA6 isoforms and performed Western blot analysis. The high quality of the preparations was confirmed by checking the PARP1 nuclear protein and the Hsp90 cytoplasmic protein (Fig. 5). In the first experiment, although the NCOA6-α and γ isoforms were undetectable due to insufficient DNA inputs in the transfection, NCOA6-β was solely detected in the nuclear extract (Fig. 5A). In the second experiment with enhanced expression of both NCOA6-α and -β in Hela cells, all of the NCOA6-α protein and most of the NCOA6-β protein were detected in the nucleus, while a small portion of NCOA6-β was seen in the cytoplasmic fraction, probably due to its over production (Fig. 5B). In the third experiment with enhanced NCOA6-γ expression, NCOA6-γ protein was detected in both cytoplasmic and nuclear fractions (Fig. 5C). In agreement with the subcellular localizations of NCOA6-γ, its fragment (a.a. 982-1965) coded by human E11a was also detected in both cytoplasmic and nuclear fractions (Fig. 5A). Interestingly, the NCOA6-δ protein was repeatedly detected in both the cytoplasmic (20%) and the nuclear (80%) fractions when samples were prepared using a cytoplasmic and nuclear extraction kit (Fig. 5A and 5C). However, examination of VDAC, a mitochondrial protein marker, revealed that these nuclear fractions were contaminated by mitochondrial proteins (Fig. 5C). Thereafter, we purified mitochondria and measured the amount of NCOA6-δ in the mitochondrial extract with minimal PARP1 nuclear and Hsp90 cytosol protein contaminations (Fig. 5D). Indeed, we found abundant NCOA6-δ protein in the purified mitochondrial extract (Fig. 5D). These results are consistent with the conclusions made from the fluorescent microscopic analyses.
Distribution of NCOA6 isoform proteins in subcellular extracts. A. Transfected Hela cells expressing the indicated Flag-tagged hNCOA6 isoforms or human exon 11a-coded peptide (11a) were subjected to preparation of cytoplasmic (C) and nuclear (N) fractions, followed by Western blot analysis with antibodies against Flag-tagged isoforms, PARP1 (a nuclear protein control) and Hsp90 (a cytosolic protein control). B. Western blot analysis of another batch of cytosolic and nuclear fractions prepared from Hela cells transfected with more plasmid DNA versus panel A. C. Western blot analysis of cytosolic and nuclear fractions prepared from Hela cells expressing NCOA6-γ and -δ proteins. VDAC served as a mitochondrial protein control. D. Western blot analysis of total lysate (T) and purified mitochondrial (Mi) extract of Hela cells transfected with NCOA6-δ.
Different Activities of the NCOA6 Isoforms in Mediating Estrogen Receptor-dependent Transcriptional Activation
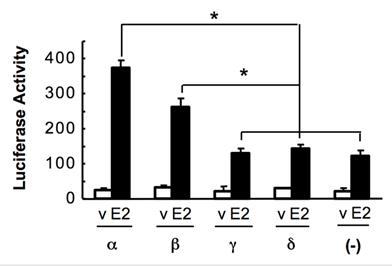
NCOA6-α is an established coactivator for ERα (12, 20). To measure the coactivator activities of the NCOA6 isoforms, we co-transfected Hela cells with ERα, ERE-tk-Luc reporter and one of the NCOA6 isoform expression plasmids. The cells were treated with or without 17β-estradiol. The reporter activity assays revealed that NCOA6-α and -β isoforms significantly enhanced ERα-mediated transcriptional activation, while NCOA6-γ and -δ isoforms were incapable to co-activate ERα-mediated transcription (Fig. 6).
Assessment of the coactivator activities of the NCOA6 isoforms. Hela cells were co-transfected with ERα, ERE-tk-Luc reporter and one of the NCOA6 isoform expression plasmids or a mock plasmid as indicated. A β-galactosidase expression vector was also co-transfected as an internal control. Cells were treated with either 17β-estradiol or its vehicle (ethanol) and subjected to luciferase and β-galactosidase activity assays. The luciferase activity was normalized to the β-galactosidase activity. *, P < 0.05 by unpaired t test.
Discussion
In this study, we found that both human and mouse NCOA6 genes contain two alternative exons, the 11a and 11b in human and the 10a and 10b in mouse. Alternative splicing events of these exons resulted in endogenously expressed four isoforms, NCOA6-α, -β, -γ and -δ. Because the human exon 11b or mouse 10b contains a stop codon, the NCOA6-δ and -γ isoforms with this exon lack the a.a. sequence coded by the last four exons in both human and mouse NCOA6 genes, while the NCOA6-α and -β isoforms without this exon contain the common sequence coded by the last four exons. Importantly, a previously unknown strong nuclear localization signal was mapped to the C-terminal sequence coded by the last four exons in both NCOA6-α and -β isoforms. This nuclear localization region plays a dominant role to direct NCOA6-α and -β to the nucleus. The lack of this region in NCOA6-γ and -δ provides an explanation for their different subcellular localizations from the other two isoforms. We found that NCOA6-γ is distributed in both cytoplasm and nucleus, which may be determined by a weak nuclear localization signal residing in the region coded by human E11a (a.a. 982-1965) or mouse E10a. NCOA6-δ is mainly located in the mitochondrium, and the responsible region for this localization is within the N-terminal 931 a.a. residues of the human protein. Most likely, this region is coded by E8 and E9 in human or E7 and E8 in mouse.
Since the mitochondrial localization sequence is common in all isoforms, an intriguing question is why other isoforms except NCOA6-δ do not go to the mitochondrium. Two possibilities might be speculated. First, the region coded by the largest E11a in human or E10a in mouse in NCOA6-α and -γ may inhibit the mitochondrial localization signal in these two isoforms. Second, the strong C-terminal nuclear localization sequence in NCOA6-α and -β plays a dominant role to direct these two isoforms to the nucleus. Another mystery is that preliminary analysis failed to locate any typical putative sequence of nuclear localization signal in the two mapped regions containing nuclear localization functions. Thus, the minimal nuclear localization domains of NCOA6 remain to be determined.
A transcriptional coactivator must locate in the nucleus to coactivate transcription factor-regulated nuclear gene transcription. In agreement with their nuclear location, NCOA6-α and -β isoforms can strongly potentiate the ERα-dependent transcriptional activation. In contrast, the other two isoforms failed to coactivate ERα-mediated gene transcription. Since NCOA6-δ is mainly located in the mitochondrium, its incapability to coactivate ERα should be expected. However, NCOA6-γ is located in both cytoplasm and nucleus, but it is also incapable to serve as an ERα coactivator in the nucleus. These data suggest that the C-terminal 98 a.a. residues in NCOA6-α and -β but not in NCOA6-γ plays important roles in both nuclear localization and transcriptional activation. Previous studies have found that the C-terminus of NCOA6-α serves as a part of the interactive domains for DRIP130, DNA-dependent protein kinase C, CBP and CoAA (reviewed in (16)). Thus, it is possible that the lack of this region in NCOA6-γ may make this isoform incapable to interact with some of these factors required for transcriptional activation.
The presence of four NCOA6 isoforms should largely increase functional diversity and specificity of the NCOA6 gene products. For example, NCOA6-β does not contain the second LXXLL motif and should not interact with LXR in the liver since this LXXLL motif specifically interacts with LXR (23). Therefore, the previously observed phenotypes in mice with the mutated second LXXLL motif should be attributed to an impaired function of NCOA6-α for LXR (20). In addition, it would be very interesting to find out what NCOA6-γ and -δ isoforms do in the cytosol and mitochondrium in the future. Recent studies have demonstrated that nuclear receptor coregulators and their isoforms may also play important functions in the cytoplasm in addition to their nuclear function. For example, the steroid receptor coactivator-3 (SRC-3) and its splicing variant SRC-3Δ4 in the cytoplasm can suppress the translation of proinflammatory cytokine mRNAs and bridge epidermal growth factor receptor (EGFR)-mediated activation of focal adhesion kinase, respectively (24, 25).
The detection of all four NCOA6 isoforms in human cells and mouse tissues indicate that these isoforms may play important roles in vivo. In mice, we found that NCOA6-α and -β are broadly expressed at relatively high levels in different tissues, while NCOA6-γ and -δ are expressed at relatively low levels in selective tissues, such as testis and/or mammary gland. More interestingly, the expression of NCOA6-β mRNA is low in the mammary glands of virgin and involution day 3 female mice but high in the mammary glands of pregnant and lactation female mice. These findings suggest that the diverse physiological function of the NCOA6 gene is mediated by multiple isoforms that are dynamically expressed in different tissues at different developmental stages.
Acknowledgements
We thank Dr. Lan Ko for the NCOA6 expression plasmid and Dr. Jae Woon Lee for NCOA6 monoclonal antibody. This work was partially supported by National Institutes of Health grants CA119689, DK058242, CA112403 and DK59820.
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17:1681-92
2. Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615-30
3. Kuang SQ, Liao L, Zhang H. et al. Deletion of the cancer-amplified coactivator AIB3 results in defective placentation and embryonic lethality. J Biol Chem. 2002;277:45356-60
4. Antonson P, Schuster GU, Wang L. et al. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Mol Cell Biol. 2003;23:1260-8
5. Zhu YJ, Crawford SE, Stellmach V. et al. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. J Biol Chem. 2003;278:1986-90
6. Mahajan MA, Das S, Zhu H. et al. The nuclear hormone receptor coactivator NRC is a pleiotropic modulator affecting growth, development, apoptosis, reproduction, and wound repair. Mol Cell Biol. 2004;24:4994-5004
7. Zhang H, Kuang SQ, Liao L. et al. Haploid inactivation of the amplified-in-breast cancer 3 coactivator reduces the inhibitory effect of peroxisome proliferator-activated receptor gamma and retinoid X receptor on cell proliferation and accelerates polyoma middle-T antigen-induced mammary tumorigenesis in mice. Cancer Res. 2004;64:7169-77
8. Yeom SY, Kim GH, Kim CH. et al. Regulation of insulin secretion and beta-cell mass by activating signal cointegrator 2. Mol Cell Biol. 2006;26:4553-63
9. Qi C, Kashireddy P, Zhu YT. et al. Null mutation of peroxisome proliferator-activated receptor-interacting protein in mammary glands causes defective mammopoiesis. J Biol Chem. 2004;279:33696-701
10. Guan XY, Xu J, Anzick SL. et al. Hybrid selection of transcribed sequences from microdissected DNA: isolation of genes within amplified region at 20q11-q13.2 in breast cancer. Cancer Res. 1996;56:3446-50
11. Zhu Y, Kan L, Qi C. et al. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J. Biol. Chem. 2000;275:13510-6
12. Lee SK, Anzick SL, Choi JE. et al. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J Biol Chem. 1999;274:34283-93
13. Ko L, Cardona GR, Chin WW. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6212-7
14. Caira F, Antonson P, Pelto-Huikko M. et al. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J Biol Chem. 2000;275:5308-17
15. Mahajan MA, Samuels HH. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol Cell Biol. 2000;20:5048-63
16. Mahajan MA, Samuels HH. Nuclear hormone receptor coregulator: role in hormone action, metabolism, growth, and development. Endocr Rev. 2005;26:583-97
17. Zhang H, Liao L, Kuang SQ. et al. Spatial distribution of the messenger ribonucleic acid and protein of the nuclear receptor coactivator, amplified in breast cancer-3, in mice. Endocrinology. 2003;144:1435-43
18. Antonson P, Al-Beidh F, Matthews J. et al. The human RAP250 gene: genomic structure and promoter analysis. Gene. 2004;327:233-8
19. Kim SW, Cheong C, Sohn YC. et al. Multiple developmental defects derived from impaired recruitment of ASC-2 to nuclear receptors in mice: implication for posterior lenticonus with cataract. Mol Cell Biol. 2002;22:8409-14
20. Li Q, Chu MJ, Xu J. Tissue- and nuclear receptor-specific function of the C-terminal LXXLL motif of coactivator NCoA6/AIB3 in mice. Mol Cell Biol. 2007;27:8073-86
21. Iwasaki T, Chin WW, Ko L. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM). J Biol Chem. 2001;276:33375-83
22. Goo YH, Sohn YC, Kim DH. et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140-9
23. Kim SW, Park K, Kwak E. et al. Activating signal cointegrator 2 required for liver lipid metabolism mediated by liver X receptors in mice. Mol Cell Biol. 2003;23:3583-92
24. Yu C, York B, Wang S. et al. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol Cell. 2007;25:765-78
25. Long W, Yi P, Amazit L. et al. SRC-3Delta4 mediates the interaction of EGFR with FAK to promote cell migration. Mol Cell. 2010;37:321-32
Author contact
![]() Corresponding author: Jianming Xu, Ph.D., Department of Molecular and Cellular Biology, Baylor College of Medicine. One Baylor Plaza, Houston, TX 77030. E-mail: jxuedu.
Corresponding author: Jianming Xu, Ph.D., Department of Molecular and Cellular Biology, Baylor College of Medicine. One Baylor Plaza, Houston, TX 77030. E-mail: jxuedu.

 Global reach, higher impact
Global reach, higher impact