Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2011; 7(6):823-836. doi:10.7150/ijbs.7.823 This issue Cite
Research Paper
Cloning and Characterization of a 2-Cys Peroxiredoxin in the Pine Wood Nematode, Bursaphelenchus xylophilus, a Putative Genetic Factor Facilitating the Infestation
1. Department of Entomology, China Agricultural University, Beijing 100193, China
2. Department of Entomology, University of Kentucky, Lexington, KY 40546-0091, USA
Received 2011-3-15; Accepted 2011-4-29; Published 2011-7-7
Abstract
The pine wood nematode, Bursaphelenchus xylophilus, is an invasive plant parasitic nematode and a worldwide quarantine pest. An indigenous species in North America and the causal agent of pine wilt disease, B. xylophilus has devastated pine production in Southeastern Asia including Japan, China, and Korea since its initial introduction in the early 1900s. The reactive oxygen species (ROS) is the first line of defense utilized by host plants against parasites, while nematodes, counteractively, employ antioxidants to facilitate their infection. Peroxiredoxins (Prxs) are a large class of antioxidants recently found in a wide variety of organisms. In this report, a gene encoding a novel 2-cysteine peroxiredoxin protein in B. xylophilus was cloned and characterized. The 2-cysteine peroxiredoxin in B. xylophilus (herein refers to as “BxPrx”) is highly conserved in comparison to 2-cysteine peroxiredoxins (Prx2s) in other nematodes, which have two conserved cysteine amino acids (Cp and Cr), a threonine-cysteine-arginine catalytic triad, and two signature motifs (GGLG and YF) sensitive to hydrogen peroxide. In silico assembly of BxPrx tertiary structure reveals the spatial configuration of these conserved domains and the simulated BxPrx 3-dimensional structure is congruent with its presumed redox functions. Although no signal peptide was identified, BxPrx was abundantly expressed and secreted under the B. xylophilus cuticle. Upon further analysis of this leader-less peptide, a single transmembrane α-helix composed of 23 consecutive hydrophobic amino acids was found in the primary structure of BxPrx. This transmembrane region and/or readily available ATP binding cassette transporters may facilitate the transport of non-classical BxPrx across the cell membrane. Recombinant BxPrx showed peroxidase activity in vitro reducing hydrogen peroxide using glutathione as the electron donor. The combined results from gene discovery, protein expression and distribution profiling (especially the “surprising” presence under the nematode cuticle), and recombinant antioxidant activity suggest that BxPrx plays a key role in combating the oxidative burst engineered by the ROS defense system in host plants during the infection process. In summary, BxPrx is a genetic factor potentially facilitating B. xylophilus infestation.
Keywords: Bursaphelenchus xylophilus, pine wilt disease, reactive oxygen species, Peroxiredoxin, hydrogen peroxide.
Introduction
The pinewood nematode, Bursaphelenchus xylophilus (Steiner and Buhrer) Nickle, is the pathogenic agent of pine wilt disease, which has caused serious damage to pine forests in Japan, Korea, and China [1]. Bursaphelenchus xylophilus is native to North America [2] and was introduced to Japan in the 1900s, South Korea in the 1980s, China in 1982 [3], and later to Portugal, Canada and 40 other countries [4, 5]. In China alone, the combined damage and management costs exceed 4 billion US dollars annually [6].
Biotic and abiotic factors affecting B. xylophilus infection
Bursaphelenchus xylophilus completes its life cycle within a temperature range of 25-30 °C; therefore, the population density of B. xylophilus normally peaks after summer, decreases from the beginning of the winter and reaches its lowest point the following spring [7]. Water status in pines also plays an important role in the pine-nematode relationship, and a reduced transpiration rate in pine can stimulate population growth of B. xylophilus [8]. In addition to abiotic factors, such as temperature and water stress, biotic factors also play a prominent role in the infection and distribution of this plant pathogenic nematode. A better understanding of these biotic factors will provide essential information to control the spread of this devastating disease in pine trees. The innate resistance of pine is one of the most important biotic factors limiting the occurrence of pine wilt disease. Bursaphelenchus xylophilus can rapidly reproduce and spread in susceptible pine trees, but its movement is restricted among resistant pine trees [9]. It has been reported that resistant pine species produce additional secondary metabolites to recognize and suppress the invasion of pathogens [10].
After millions of years of co-evolution with parasites, host plants have evolved various resistant mechanisms against the parasite infection. In many plants, the first line of defense involves an oxidative burst which generates toxic reactive oxygen species (ROS), including superoxide anion (O2-·), hydrogen peroxide (H2O2) and the hydroxyl radical (·OH) [11]. ROS production in plants is particularly important in the defense and recognition of novel pathogens [12], because it can affect many key events in plant-pathogen interactions, including signal transduction, antimicrobial effect, membrane lipoxidation, cell wall modification, induced resistance, and hypersensitive cell death [11]. Counteractively, parasites produce antioxidants to protect them from the host plant's ROS defenses [13]. Therefore, removing ROS in host plants is critical for the survival of parasites. Given the fact that B. xylophilus can successfully infest and spread in pine trees, antioxidant proteins likely play a key role in the host-parasite interaction.
Antioxidants and peroxiredoxins
The major antioxidants in eukaryotic organisms include superoxide dismutase which detoxifies the superoxide anion, catalase, glutathione peroxidase which is involved in the breakdown of cellular peroxides, and peroxiredoxin which functions to inactivate hydrogen peroxide [13, 14 and 15]. Based on the catalytic mechanisms and the presence of either one or two highly conserved cysteine residues, peroxiredoxins (Prxs) are classified into 1-Cys, typical 2-Cys, and atypical 2-Cys Prxs. Catalytic reactions of the typical 2-Cys, atypical 2-Cys, and 1-Cys Prx are through the formation of intermolecular disulfide, inner-molecular disulfide [16], and the participation of small chemical thiols as reduced agents, respectively [17]. Prxs are multifunctional proteins, for example they function as antioxidants, by reducing alkyl peroxide and hydrogen peroxide to alcohol or water, respectively [18, 19]. They can also protect against phospholipid peroxidation [20] and serve as a peroxynitrite reductase for the detoxification of the reactive nitrogen species (RNS) [21] to protect cells from oxidant-induced membrane damage and prevent cell death [22]. In addition, Prxs are also considered to be signal transmitters in phosphorylation and transcriptional regulation, presumably through regulation of hydrogen peroxide [23]. Differing from other antioxidants, Prxs have no cofactors, such as metals or prosthetic groups for its antioxidant activity. The highly conserved cysteine residues are their active sites [16]. The cysteine residue near the N-terminus is referred to as Cp, which exists in all Prxs; whereas the cysteine residue near the C-terminus is referred to as Cr, and it only exists in the 2-Cys Prxs [16].
Despite recent progresses in the genomic [24] and epigenomic [25] understanding of this world-wide quarantine pest, there is virtually no information available regarding the Prx gene in plant pathogenic B. xylophilus and its potential role in engineering the antioxidative counterattack against pine hosts. In this study, we pursue the following objectives: (1) Clone and sequence peroxiredoxin in the plant pathogenic B. xylophilus (BxPrx) using RACE-PCR; (2) Gain a better understanding of the transmembrane pathway of leader-less BxPrx based on the in silico structural analysis of BxPrx; and (3) Express recombinant BxPrx heterogeneously in a bacterial system and investigate the tissue localization of BxPrx by immunohistochemistry using a polyclonal antibody raised against recombinant BxPrx.
Materials and Methods
Bursaphelenchus xylophilus culture and maintenance
The B. xylophilus isolate JSZJ1-6 [3] was kindly provided by Dr. Bingyan Xie (Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences). B. xylophilus was maintained at 25 °C for one week on a necrotrophic fungus, Botrytis cinerea, grown on a PDA (Potato Dextrose Agar) culture dish. Mixed stages of B. xylophilus were then harvested with a Baermann funnel, and washed with M9 buffer (KH2PO4 30 g/L, K2HPO4 60 g/L, NaCl 50 g/L) before subjecting the isolate to subsequent experiments [26].
BxPrx gene discovery and sequence analysis
A thorough search of available sequences at a web-based nematode genomics database, Nematode.Net v3.0 (http://www.nematode.net) showed that Prx2s are evolutionarily conserved in all extant nematode species. After alignment of the deduced protein sequences of Prx2 from EST libraries obtained at this website, the degenerate primers (Table. S1) were designed based on several highly conserved domains for the cloning of BxPrx. Total RNA was extracted from mixed stages of B. xylophilus using RNeasy Mini kit (Qiagen, Hamburg, Germany) according to the manufacture's protocols. The first strand cDNA used in 3′ RACE and 5′ RACE were synthesized with 3′ full RACE core set (Takara, Dalian, China) and Firstchoice® RLM RACE Kit (Ambion, Foster City, CA, USA), respectively. The 3′ end product of the BxPrx gene was obtained with oligodT-3 site adaptor primer (Takara) and the degenerate primer BxPrx3S. 5′ end sequence was obtained with 5′ RACE outer primer (Ambion) and BxPrx5R. The entire sequence was amplified by the primer set BxPrxSB and BxPrxRS (Table S1) which include restriction sites BamH I and Sal I to the 5′ and 3′ ends, respectively, and the resulting PCR products were validated by direct sequencing.
The deduced protein sequence, pI (Iso-electric Point) and MW (Molecular Weight) of BxPrx were predicted by DNAman v.5.2.2 (Lynnon Biosoft, Quebec, Canada). The catalytic triad of BxPrx was determined by a Conserved Domain search on the NCBI website (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) [27]. Protein tertiary structure was predicted by an online protein structure homology-modeling server SWISS-MODEL using the automated mode and modified by PyMOL-v1.3r1 software. All published EST sequences of Prx2 in nematodes were obtained from Nematode.Net (http://www.nematode.net). The signal peptide of BxPrx was determined using SignalP (http://www.cbs.dtu.dk/services/SignalP/) [28], and the transmenbrane region of BxPrx was predicted by TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/). The hydrophobicity profile of BxPrx was analyzed by DNAman v.5.2.2. Secretome 2.0 Server (http://www.cbs.dtu.dk/services/SecretomeP/) was used to characterize the non-classical secreted proteins.
Phylogenetic relationship of BxPrx with other Prx2s
All Prx2 sequences in animals which have the full open reading frames (ORFs) were extracted from GenBank, loaded in MEGA 4, and aligned. The phylogenetic relationship of BxPrx with other Prx2s was revealed using two different methods: (1) the neighbor-joining (NJ) analysis with an Amino: Poisson correction model; (2) the maximum parsimony (MP) analysis. The sites containing missing data or alignment gaps were removed in a pair-wise fashion. A total of 1,000 bootstrap replications were used to test topology. Bootstrap values (>50) were reported with each branch and the taxonomic origins of the sequences were highlighted.
Nuclear localization of BxPrx
Intracellular localization of BxPrx was investigated through transient expression of the Cam35S::GFP-BxPrx fusion construct which was bombarded into onion epidermal cells [29] using Model PDS-1000/He Biolistic Particle Delivery System (Baumbusch, LO.). Plasmid Cam35S::GFP -BxPrx was constructed by inserting the entire coding sequence of BxPrx into the Cam35S-GFP plasmid behind the Cam35S promoter (Cauliflower Mosaic virus 35S promoter) and GFP (Green Fluorescent Protein) reporter gene after digestion with BamH I and Sal I (Fig. S1A). The resulting in-frame fusion was confirmed by restriction mapping and DNA sequencing. Expression was detected against a blank Cam35S-GFP control with a Bx60 Olympus microscope using a blue wide band cube for fluorescence observation.
Recombinant protein expression, purification and validation
A recombinant expression plasmid was constructed by ligating the entire open reading frame of BxPrx into a bacterial expression vector pET-28a (Fig. S1B). The resulting recombinant construct was validated by the direct sequencing of PCR products and then transformed into an Escherichia coli BL21 (DE3) competent cell. Escherichia coli was cultured at 37 oC for 16 hrs with shaking at 250 rpm. It was then diluted 1:50 (V:V) into liquid LB (Lysogeny Broth), and the liquid medium was shaken continuously until the OD600 value reached 0.4-0.6. The induction conditions, such as isopropyl β-D-1-thiogalactopyranoside (IPTG) concentration and incubation temperature, were optimized to maximize the yield of soluble recombinant BxPrx. A series of IPTG concentrations ranging from 0, 1, 2, and 5 mM was tested. The induction was then carried out at two different temperatures: 37 oC, 250 rpm shaking for three hours, and 20 oC, 150 rpm shaking for 16 hrs. 100 ml of bacterial culture from each treatment was precipitated by centrifugation at 4,000 g for 15 min. Then the pellets were dissolved in the lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM Imidazole, pH 7.0), and sonicated (250 W, 10 s × 10 times, 30 s interval, in ice) to break up the bacteria. After centrifugation at 4 oC, 12,000 rpm for 20 min, the supernatant of sonicated fraction was mixed with Ni-NTA agarose (Qiagen, Hamburg Germany). To purify recombinant BxPrx, the resulting protein was washed (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 7.0) and then eluted (50 mM NaH2PO4, 300 mM NaCl, 250 mM Imidazole, pH 7.0). A total of 10 elution fractions were collected with 500 µl in each fraction.
After SDS-PAGE and staining by coomassie blue, the band of the purified BxPrx protein was cut out from the gel. After being dipped in a 100 µl discoloration solution (H2O: methonal: acetic acid = 5: 4: 1) for 20 min, dehydrated with 100 µl acetonitrile for 10 min, and alkylated with 50 µl indoacetamide for 30 min in dark, the product was treated with 15 µl 0.01 µg/µl trypsin for 16 h, and then was extracted with 100 µl 5 % TFA (trifluoroacetic acid) at 40 oC for 1h. After drying under nitrogen and mixed with 5 µl 0.1 % TFA, the sample was loaded into an ABI-4800 MALDI-TOF-TOF for a confirmation test of the induced recombinant protein.
BxPrx activity assay
The activity of recombinant BxPrx on the reduction of hydrogen peroxide (H2O2) and cumene hydroperoxide (C9H12O2) was measured at OD340 according to Kwatia [30]. The reaction mixture contained 5 mM potassium phosphate (pH 7.0), 1 mM EDTA, 0.1 mM NADPH, 1mM L-glutathione, and 0.1 unit/ml glutathione reductase (Sigma). One unit of activity is defined as the amount of enzyme required to cause the oxidation of 1 nmol of NADPH per minute.
BxPrx antiserum production and immunoblot analysis
A polyclonal antiserum against recombinant BxPrx was obtained by consecutive 4-week injections of the purified protein into a rabbit. For the first injection, 1 mg of BxPrx protein was injected concurrently with 1 mg Freund's Complete Adjuvant (Sigma, St. Louis, MO). In the following 3 weeks, 1 mg of protein was injected mixed with 1 mg of Freund's Incomplete Adjuvant (Sigma, St. Louis, MO). The serum supernatant were obtained after centrifugation of collected blood samples subjected to immunoblot analysis at 4 °C, 12,000 rpm for 15 min.
To test the quality of resulting anti-BxPrx serum, 30 µg of total protein extract from B. xylophilus and B. mucronatus were separately loaded in 12 % SDS-PAGE for the immunoblot analysis. The protein bands were transferred from SDS-PAGE to a nitrocellulose (NC) membrane. Goat anti-rabbit IgG-alkaline phosphatase (Fermentas, MD) (diluted by 1: 10000) was used as the secondary antibody. Immunoreactions were visualized by adding 5-bromo-4-chloro-3-indolyl-phosphate and p-nitroblue tetrazolium chloride (Promega, WI).
Immunohistolocalization
Mixed stages of B. xylophilus were fixed in the solution of paraformaldehyde (4 %) and glutaraldehyde (2 %) in 0.1 M phosphate buffered saline (PBS) (pH 7.4), dehydrated in an ethanol series and embedded in LR (London Resin Company) White resin with polymerization at 60 °C. Ultrathin sections (60-80 nm) were cut with a diamond knife on a Reichert Ultracut Ultramicrotome (Reichert Company, Vienna, Austria). For immunohistochemistry, the grids were subsequently floated on droplets of the following solutions: PBS (containing 50 mM glycine), PBGT (PBS containing 0.2 % gelatine, 1 % bovine serum albumin and 0.02 % Tween-20), the primary antibody (Sigma) diluted with PBGT, and the secondary antibody diluted with PBGT. Optional silver intensification increased the size of the gold granules from 10 to about 40 nm, and 2 % of uranyl acetate increased the tissue contrast for observation in the transmission electron microscope (HITACHI H-7500, Hitachi, Tokyo, Japan). The primary antibody was diluted 1: 2000 and incubated overnight at 4 °C. The secondary antibody, an anti-mouse IgG which was coupled to 10 nm colloidal gold (AuroProbe EM, GAR G10; Amersham Biosciences, Little Chalfont, Buckinghamshire, UK), was diluted 20-fold and incubated at room temperature for 60-90 min.
Results
Cloning and sequence analysis of BxPrx
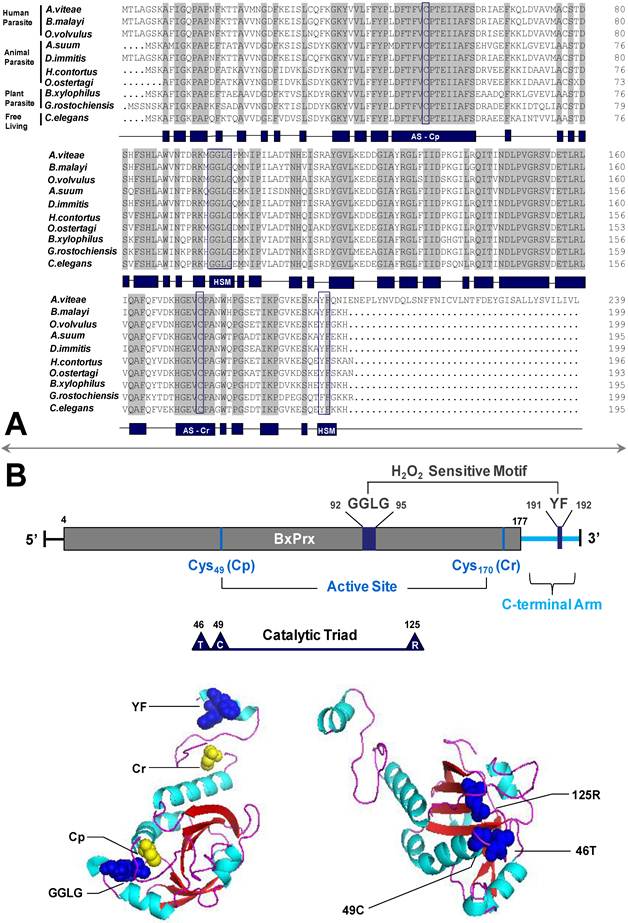
The full length cDNA of BxPrx was determined through RACE-PCR and was deposited in GenBank under accession number EU095848. The cDNA of BxPrx contains 588 bp ORF encoding 195 amino acids. The predicted isoelectric point (pI) and molecular weight (MW) are 6.54 and 21,854 Da, respectively. BxPrx contained two conserved functional cysteine regions flanked by valine and proline residues (VCP). The cysteines, located at the peptide positions 49 and 170, respectively, are generally referred to as Cp and Cr (Fig. 1A and 1B). BxPrx is, therefore, classified as a 2-cys peroxiredoxin [17]. Moreover, Cp is a part of the catalytic triad. Two conserved motifs GGLG and YF are also identified in BxPrx, which are critical as a signal transmitter through sensitive recognition of H2O2 changes. Prx2 have been found in human, animal, plant-parasitic, and free living nematodes, and BxPrx shares a high degree of similarity with Prx2 from other nematodes, such as Ascaris suum [159/195 (81 %)], Caenorhabditis elegans [157/195 (80 %)], Globodera rostochiensis [153/193 (79 %)].
A catalytic triad consisting of threonine, cysteine, and arginine are found to be in close spatial proximity and forms a triangle shape in their 3-D tertiary structure to facilitate the redox catalytic reaction (Fig. 1B). The two motifs GGLG and YF are located at the opposite ends in the tertiary configuration, and Cp is closer to the ATP binding motif GGLG to initiate the catalytic reaction of BxPrx with the supply of ATP. Motif YF helix is located above the Cr in their spatial configuration, and this arrangement could potentially block the reduction reaction of BxPrx from the oxidized status and it is important for Prx2's function as a signal transmitter.
Phylogenetic analysis
The phylogenetic relationship of BxPrx with other Prx2s from the animal kingdom is depicted in Fig. 2. Phylogenies derived from both neighbor joining (NJ) and maximum parsimony (MP) methods were generally congruent, and the results of the two phylogenetic trees and associated bootstrap supports are displayed in Fig. 2. Prx2 sequences came from five clades of nematodes with different life histories (Table 1).
BxPrx homologues in nematodes
| Clade | Species | Host | Contig ID | RFα | H-Score§ | e-Value£ |
|---|---|---|---|---|---|---|
| Clade I | Xiphinema index | Plants | XI01176 | +2 | 806 | 6.0e-81 |
| Trichinella spiralis | Human | TS00204 | +3 | 573 | 4.7e-56 | |
| Clade III | Ascaris suum | Human | AS00117 | +1 | 849 | 5.1e-85 |
| Toxocara canis | Dogs, cats, and human | TX02017 | +2 | 786 | 2.2e-78 | |
| Dirofilaria immitis | Dogs and cats | DI00483 | +2 | 758 | 2.0e-75 | |
| Brugia malayi | Human | BM01405 | +2 | 438 | 2.1e-41 | |
| Clade IVa | Strongyloides stercoralis | Human | SS01309 | +2 | 805 | 5.5e-81 |
| Parastrongyloides trichosuri | Possums | PT00761 | +3 | 804 | 7.6e-81 | |
| Strongyloides ratti | Mice | SR00321 | +1 | 650 | 1.5e-64 | |
| Clade IVb | Zeldia punctata | Free living (bacterivore) | ZP00073 | +1 | 830 | 6.9e-83 |
| Heterodera glycines | Plant (soybean) | HG03313 | +2 | 817 | 1.7e-81 | |
| Meloidogyne hapla | Plants (root knot) | MH01325 | +2 | 810 | 1.1e-80 | |
| Meloidogyne paranaensis | Plant (coffee) | MP00535 | +2 | 807 | 2.0e-80 | |
| Meloidogyne incognita | Plants (root knot) | MI02462 | +3 | 807 | 2.1e-80 | |
| Pratylenchus penetrans | Plant (meadow) | PE00136 | +3 | 718 | 5.6e-71 | |
| Meloidogyne arenaria | Plants (root knot) | MA03144 | +1 | 680 | 6.1e-67 | |
| Meloidogyne javanica | Plants (root knot) | MJ00986 | +2 | 611 | 1.1e-59 | |
| Globodera pallida | Plant (potato cyst) | GP03375 | +2 | 560 | 2.7e-54 | |
| Ditylenchus africanus | Plant (peanut) | DA00204 | +3 | 450 | 1.3e-53 | |
| Globodera rostochiensi | Plant (potato cyst) | GR13953 | +2 | 282 | 4.8e-43 | |
| Meloidogyne chitwoodi | Plants (root knot) | MC03207 | +1 | 239 | 6.4e-20 | |
| Clade V | Caenorhabditis elegans | Free living (bacterivore) | F09E5.15 | +1 | 843 | 5.7e-84 |
| Nippostrongylus brasiliensis | Rats | NB00963 | +2 | 835 | 3.3e-83 | |
| Ancylostoma ceylanicum | Human and hamster | AE00970 | +2 | 833 | 5.4e-83 | |
| Caenorhabditis remanei | Free living (bacterivore) | CR02463 | +1 | 831 | 8.1e-83 | |
| Pristionchus pacificus | Free living (bacterivore) | PP00536 | +3 | 831 | 8.4e-83 | |
| Haemonchus contortus | Sheep and goats | HC02224 | +2 | 792 | 1.2e-78 | |
| Ancylostoma caninum | Dogs | AC03236 | +2 | 761 | 2.5e-75 | |
| Ostertagia ostertagi | Cattles | OS00892 | +3 | 367 | 1.2e-67 |
“α”: Reading Frame. “§”: High Score, species of the first three highest score were highlighted. “£”: Smallest sum probability P (N)
Sequence and structural analysis of BxPrx. (A) Alignment of BxPrx with orthologues in other nematode species. Grey blocks represent conserved amino acids across all nematodes. The schematic drawing of conserved domains of BxPrx is shown in blue blocks. Two conserved functional active cysteine sites (AS-Cp and AS-Cr) and two H2O2 sensitive (HS) motifs (GGLG and YF) are highlighted in blue boxes. Accession numbers of Prx2s used in this study are Acanthocheilonema viteae [AAL91102], Brugia malayi [Q17172], Onchocerca volvulus [AAC32810], Ascaris suum [Q9NL98], Dirofilaria immitis [AAC38831], Haemonchus contortus [AAT28331], Ostertagia ostertagi [CAD20737], Bursaphelenchus xylophilus [ABW81468], Globodera rostochiensis [CAB48391], and Caenorhabditis elegans [NP_872052]. (B) Schematic drawing of BxPrx primary and tertiary structures. The grey bar shows the conserved typical 2-Cys Prx domain. The two conserved active sites (Cp and Cr), H2O2 sensitive motifs (GGLG and YF), and catalytic triad (Thr-Cys-Arg) are highlighted in BxPrx tertiary structure as colored spheres. One sphere represents one of the atoms in the corresponding amino acids.
Phylogenetic relationship of BxPrx with Prx2s from other organisms. (A) Neighbor-joining (NJ) consensus tree. (B) Maximum parsimony (MP) consensus tree. The branch length of the NJ and MP tree indicates evolutionary distance. Accession numbers of the Prx2s used in this study are Xenopus tropicalis [NP_989001], Ascaris suum [Q9NL98], Onchocerca volvulus [AAC32810], Onchocerca ochengi [AAC77922], Dirofilaria immitis [AAC38831], Brugia malayi [Q17172], Acanthocheilonema viteae [AAL91102], Litomosoides sigmodontis [AAG10102], Ostertagia ostertagi [CAD20737], Haemonchus contortus [AAT28331], Caenorhabditis elegans [NP_872052], Globodera rostochiensis [CAB48391], Bursaphelenchus xylophilus [ABW81468], Fenneropenaeus indicus [ACS91344], Bombus ignitus [ACP44066], Helicoverpa armigera [ABW96360], Bombyx mori [NP_001037083], Osmerus mordax [ACO09982], Thunnus maccoyii [ABW88997], Danio rerio [NP_001002468], Mus musculus [NP_035693], Rattus norvegicus [AAH58481], and Homo sapiens [NP_005800]. Calculated bootstrap value is attached to each branch and the value is a confidence level supporting this arrangement. BxPrx is highlighted in a box.
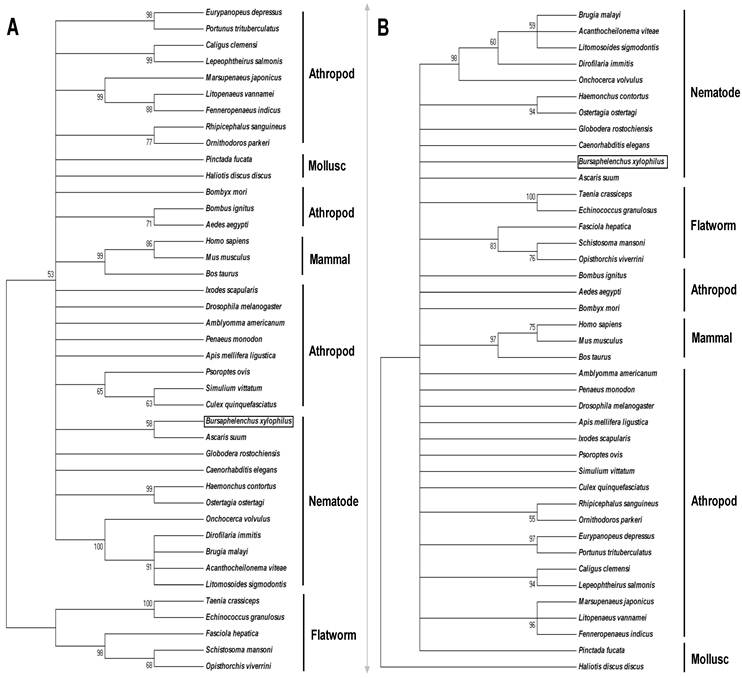
The deduced ORFs have the two conserved functional cysteine sites which suggest a similar antioxidative function through the same catalytic mechanism. Among all the sequences, Prx2 from A. suum has the highest identity with BxPrx which is consistent with the results of phylogenetic analysis, in which BxPrx has the closest evolutionary relationship with a Prx2 in A. suum. Additionally, two main clades are resolved from the NJ tree, flatworms and a cluster of nematodes, arthropods and mammals; In MP tree, all the Prx2s originated from a same evolutionary root, only with an isolated branch of Haliotis duscus discus from taxonomic level of mollusc.
Cellular distribution of BxPrx
Based on the primary structure analysis, there is no signal peptide at the N-terminal of BxPrx, which is consistent with other known Prx2s. However, the cellular distribution study using a GFP reporter system demonstrated that BxPrx is located within both the cytoplasm and nucleus of the onion epidermis cell (Fig. 3A and 3B). The green florescent signals emitted by the blank Cam35S-GFP construct were detected in all portions of the onion cell, including the nucleus, cytoplasm and vacuole (Fig. 3A); whereas signals from the Cam35S::GFP-BxPrx construct were predominately located in the nucleus and cytoplasm (Fig. 3B).
Intracellular localization of BxPrx. (A) 35S::GFP, GFP fluorescence signal was ubiquitously distributed in the whole cell. (B) 35S::GFP-BxPrx, GFP signal was strictly located in the cytoplasm and nucleus. The photo was created by overlaying the differential interference contrast light micrographs with the fluorescence micrographs. C & D presents two plausible transmembrane mechanisms for BxPrx to reach the cytoplasm. (C) Transmembrane region within BxPrx. A 23-amino acid transmembrane region was predicted by TMHMM. The sequence of the predicted transmembrane region is provided, and the transmembrane helix is highlighted in red. Hydrophobicity profile of BxPrx was analyzed by DNAman. The 23 consecutive hydrophobic amino acids coincide with the single transmembrane region in BxPrx. (D) A survey of the available ATP-binding cassette transporter genes in animals including nematodes.
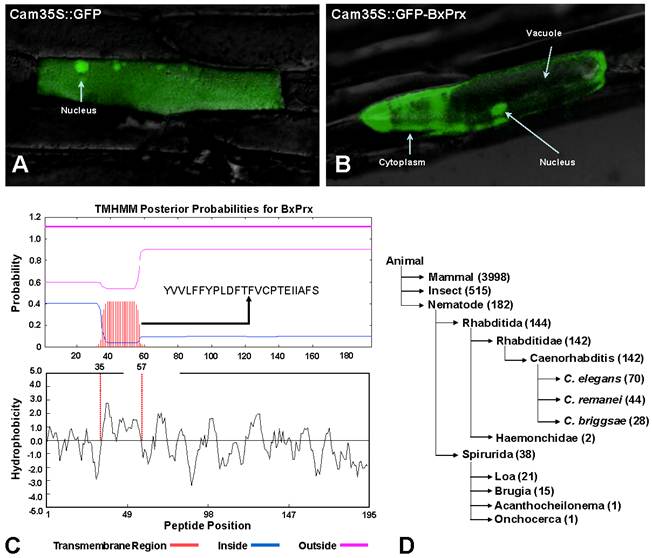
The in silico secretome study supports the cellular localization of BxPrx in the onion epidermal cells. The estimated NN-score of BxPrx as a secretome is 0.597 (above the default threshold of 0.5), which means that BxPrx is a non-classical secretable protein. These results indicate that BxPrx, a leader-less peptide, can be secreted and enters the cytoplasm without a leading signal peptide. A transmembrane helix composed of 23 hydrophobic amino acids provided a plausible transmembrane mechanism for the leader-less BxPrx (Fig. 3C). In addition, surveying GenBank shows that ATP-binding cassette transporters (ABC-transporter) are commonly found in animals including all nematodes for which sequence information is available (Fig. 3D). ABC transporters typically share a highly conserved ATPase domain and provide energy for transportation and other biological processes. Therefore, it is germane to speculate that transmembrane helix and ABC transporters could potentially facilitate the transmembrane activity of the leader-less BxPrx in B. xylophilus.
Recombinant BxPrx: optimization and validation
Three concentrations of IPTG (1, 2, and 5 mM) and two different incubation temperatures (20 and 37 °C) were examined to maximize the cytosolic induction of recombinant pET-28a-BxPrx (Fig. 4). The optimal induction conditions were determined to be 1 mM IPTG and 20 °C for 16 hrs because 1) the induction of recombinant BxPrx by IPTG showed no significant improvement beyond 1 mM concentration (Fig. 4A), and 2) the yield of soluble BxPrx recombinant protein was greatly reduced at 37 °C, although the overall production of BxPrx recombinant protein was significantly increased at a higher incubation temperature. Under optimal conditions, 100 ml of bacterial culture produced 0.949±0.025 g of precipitate. After a single step purification procedure (Ni-NTA agarose), it generated 1.233±0.067 mg of purified BxPrx protein (Fig. 4B).
Recombinant protein expression, purification and validation. (A) Optimal concentration of IPTG for the induction of recombinant BxPrx. 1, 2, 3, and 4 are the protein expressions with blank pET-28a plasmid induced by 5, 2, 1, and 0 mM of IPTG, separately; 5, 6, 7, and 8 are the protein expressions of recombinant BxPrx induced by 5, 2, 1, and 0 mM of IPTG, respectively. (B) Recombinant BxPrx expression after induced by 1mM IPTG at 20 oC for 16 hrs. (1) without IPTG, (2) crude, (3) supernatant after sonication, (4) resuspended pellet after sonication, (5) 1st flow-through after initial protein loading (6) flow-through after washing solution, and (7-9) the 2nd, 3rd, 10th fraction of the eluted BxPrx. (C) Mass spectrometry test of the purified BxPrx. 5 peaks (highlighted with arrows) in the mass spectrum matched with 5 peptide segments (Inset, highlighted in red) of the BxPrx protein in B. xylophilus. (D) Specific activity assay of recombinant BxPrx. Control, without BxPrx; Two different substrates, hydrogen peroxide (H2O2) and cumene hydroperoxide (C9H12O2), respectively, were used for the activity assay of recombinant BxPrx.
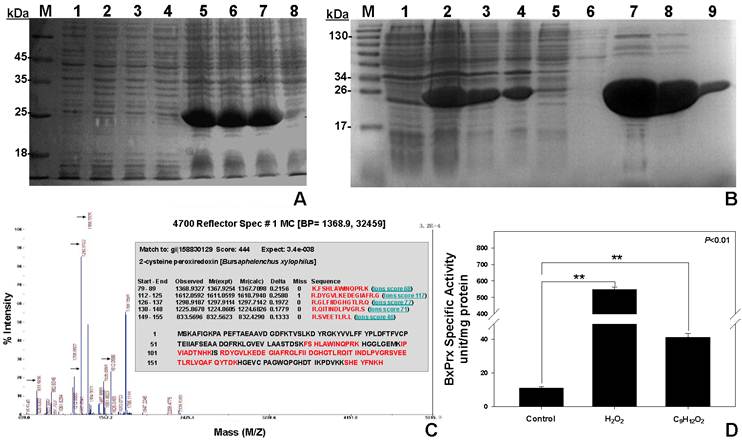
The identity of heterogeneously expressed protein was validated by the mass spectrometry analysis (Fig. 4C). The purified recombinant protein exhibited high sequence homology with 5 peptide fragments from BxPrx. The specific activity of BxPrx was measured at 547.2±15.2 units/mg protein and 41.1±2.3 units/mg protein to reduce hydrogen peroxide (H2O2) and cumene hydroperoxide (C9H12O2), respectively, with a negative control of 11.1±0.8 units/mg protein (Fig. 4D). Recombinant BxPrx showed highly specific activity toward hydrogen peroxide.
Immunoblot analysis and immunohistolocalization study
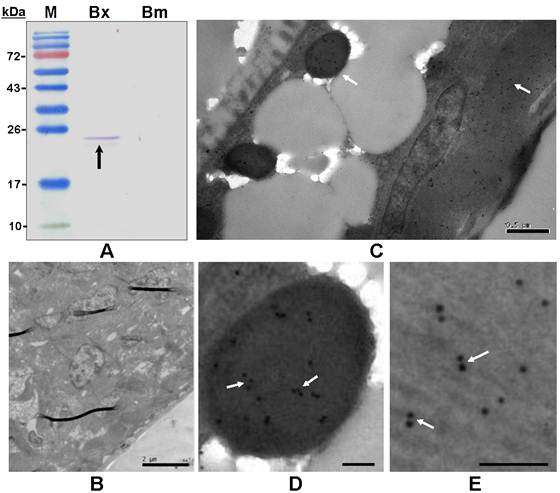
The polyclonal antiserum raised against the recombinant BxPrx recognized the target BxPrx as a single band, but only detected the protein extracted from B. xylophilus not B. mucronatus (Fig. 5A). This result suggested that the Prx2 in B. mucronatus either has a low homology with BxRrx or is expressed at significantly lower concentrations. Bursaphelenchus. mucronatus is closely related to B. xylophilus, but only the latter one causes pine wilt disease. The differences in Prx2 structure and expression profile may lead to the distinctive differences in pathogenicity between the two species. Immunohistochemistry was carried out to determine the distribution of BxPrx in tissues (Fig. 5B-E). The results showed that BxPrx was abundantly expressed in B. xylophilus and the protein was predominantly localized under the cuticle surface (Fig. 5C), i.e., in the particles inside the body (Fig. 5D) and the muscle cells under the body surface (Fig. 5E). The negative control with preimmune serum showed no detectable binding in B. xylophilus samples (Fig. 5B).
Immunoblot analysis and immunohistolocalization of BxPrx in B. xylophilus. (A) Immunoblot analysis of a polyclonal antiserum raised against recombinant BxPrx to total proteins extracted from B. xylophilus and B. mucronatus, respectively. M, Molecular marker; Bx, B. xylophilus; Bm, B. mucronatus. A single band was detected in B. xylophilus, but not in B. mucronatus. (B-E) Immunohistolocalization of BxPrx in B. xylophilus using preimmune serum from a healthy rabbit as a negative control (B). BxPrx antiserum is found to bind to particles (D) and muscle fiber cells (E) under the cuticle of B. xylophilus (C). D and E are the close-up imagines of C. The scale bars of B-E are 2 µm, 0.5µm, 0.1µm and 0.1µm respectively.
Discussion
BxPrx gene discovery
Prx2 is a common theme in parasitic and free-living nematodes, and its primary structure is highly conserved. The alignment of BxPrx with Prx2 from other species demonstrates that BxPrx has two highly conserved cysteine functional sites for antioxidant activity. The N-terminal cysteine is referred to as peroxidatic cysteine (Cp), and the resolving cysteine (Cr) is located at the C-terminus. Cp is the primary site of enzyme oxidation which can attack peroxides that is subsequently oxidized to a cysteine sulfenic acid (Cp-SOH) [18]. Then, the reduction of the enzyme to its original active reduced state involves thioredoxin and Cr [31, 32]. The antioxidant catalytic reaction of BxPrx is assisted by the catalytic triad composed of threonine, cysteine, and arginine. In the BxPrx primary structure, arginine is about 80 amino acids away from threonine and cysteine, but they are in close proximity in the tertiary structure to initiate the catalytic reaction. In the following catalytic reaction, deprotonation of the catalytic cysteine (49 Cys) thiol group is facilitated by H- bonding with the threonine hydroxyl group, and the positive charge of the arginine side chain. Therefore, the conserved catalytic triad is critical to the antioxidant activity of BxPrx; the destruction of this tertiary structure could lead to the loss of antioxidative activity.
BxPrx also has two conserved motifs of Gly-Gly-Leu-Gly (GGLG) near the N-terminus and Tyr-Phe (YF) in the C-terminal arm. GGLG and YF are common features of Prx2s, and they can detect the fluctuation of hydrogen peroxide levels in the environment [33, 34]. The GGLG motif is a part of the ATP binding site in the complex structure of peroxiredoxin [33] and is located in close proximity to Cp in the BxPrx tertiary structure, which could provide a convenient energy supply for the catalytic reaction. The catalytic redox reaction of peroxiredoxin is an ATP-dependent reaction. As a H2O2 sensitive motif, GGLG can react quickly and supply energy for the initiation of an antioxidant reaction for Prx2 when H2O2 levels increase. Therefore, GGLG potentially plays a critical role in the antioxidative mechanism of BxPrx. On the other hand, a YF-containing C-terminal helix is observed to be located above the active site in the tertiary structure of BxPrx to prevent unfolding, which will cause a longer reaction of the active site Cr. Consequently, Prx is prone to inactivation when challenged with high levels of H2O2, i.e., Prx is sensitive to increasing H2O2 [34]. Therefore, by having two sensitive motifs for H2O2, BxPrx should also play a role in the signal transmission of B. xylophilus, in terms of sensing the ROS defense of the host plant and detoxifying ROS as an antioxidant enzyme.
The phylogenetic analysis showed that Prx2 in mammals, arthropods and nematodes originated from the same root, which suggests that Prx2 could play a similar function in all these organisms. Among them, BxPrx had the shortest genetic distance to a Prx2 in Ascaris suum in the constructed NJ tree with bootstrap value of 58 on the root branch of the two Prxs; in MP tree, the bootstrap value on the root branch of the two Prxs is 46, so it is cutoff when values over 50 are shown. Ascaris suum (a large roundworm) is a parasitic nematode that causes ascariasis in pigs. Although the hosts of the two parasites belong to different taxonomic groups, the two parasites have similar transmission pathways and have similar life histories in their respective hosts. Bursaphelenchus xylophilus is transmitted by insect vectors such as the Japanese pine sawyer beetle, and A. suum also utilizes a dung beetle as an intermediary for distribution. They are both migratory parasites and they can move within the host to feed. Similar transmission pathways and living conditions result in similar parasite-host relationships, which suggests a similar function for their antioxidant Prx.
Transmembrane mechanism of the leader-less BxPrx
Based on an array of web-based bioinformatics analyses, BxPrx, a leader-less peptide, is predicted to have transmembrane capabilities. The outcome of this in silico prediction was validated by both nuclear- and immunohistolocalization studies. Unlike classical secretion via the endoplasmic reticulum (ER)/Golgi Body pathway for proteins with signal peptides, non-classical secretion of leaderless proteins requires a structural accommodation (e.g., transmembrane regions within the leader-less peptides) or a carrier (transmembrane proteins such as the ABC transporter). In this study, a stretch of 23 consecutive hydrophobic amino acids assembles the only transmembrane helix on the OFR of BxPrx, which potentially can assist BxPrx transmission through the hydrophobic lipid bilayer of the cell membrane. A domain swap or mutation at the transmembrane helix would provide a definitive answer for the involvement of this transmembrane mechanism in B. xylophilus.
ABC transporters are members of a protein super-family that is one of the largest and most ancient families found in all extant phyla from prokaryotes to humans [35]. As transmembrane proteins, ABC transporters can carry a wide variety of substrates across extra- and intra-cellular membranes using the energy from ATP hydrolysis [36]. Moreover, ABC transporters, which are prevalent among all nematodes, were considered as a possible transmembrane mechanism for the ER/Golgi-independent leader-less secretion pathway in a parasitic flatworm, liver fluke Fasciola hepatica, especially for the secretion of the leader-less F. hepatica antioxidants such as Prx2 [37]. Therefore, we propose that leader-less BxPrx is secreted either by its transmembrane region or by a specific ABC transporter or by both mechanisms working together in B. xylophilus.
Characterization of recombinant BxPrx
Higher induction temperatures lead to a greater production of recombinant protein within a shorter time which typically results in the abnormal folding of a protein and the formation of an inclusion body. In this study a large quantity of active recombinant BxPrx was obtained by induction at optimal conditions (20 oC overnight with 1 mM IPTG). Zipfel et al [38] recombinantly expressed a Prx2 from the human parasitic nematode, Onchocerca volvulus. The detailed characterization of this recombinant Prx2 indicated that O. volvulus Prx2 predominantly expressed at the host-parasite interface, which was important for the infection by this parasitic nematode.
The different reactions on the immunoblots between B. xylophilus and B. mucronatus suggest differences at either the Prx2 genes or Prx2 expression profiles. Bursaphelenchus mucronatus, an indigenous species in China, is very similar to the morphology and biology of B. xylophilus [39]. However, B. mucronatus and B. xylophilus have distinctively different pathogenicities. Bursaphelenchus mucronatus has weak pathogenicity [40] and the disease prevalence of B. mucronatus is far less than that of B. xylophilus [3]. It was reported that the propagation of B. xylophilus was higher than B. mucronatus, and even higher under competitive conditions in laboratory cultures [39]. After more than two-decades of invasion, B. xylophilus has significantly increased its colonization capability and has replaced B. mucronatus in natural forest ecosystems in China [3]. This phenomenon might be the result of a differentially expressed gene associated with the pathogenicity and invasive capability of the two parasites. Most recently, a microRNA survey provided novel information on the regulation of growth, development, behavior, and stress response in B. xylophilus at the epigenetic level [25]. Combining genomic and epigenomic resources in B. xylophilus will shed light on the molecular underpinnings that govern the infection and distribution of B. xylophilus.
Tissue localization of BxPrx
The immunohistolocalization study with a polyclonal antiserum against recombinant BxPrx suggest that: 1) BxPrx is fairly abundant, 2) this antioxidant can be found in muscle cells and particles, and 3) it is widely distributed and concentrated underneath the cuticle of the nematode. This localization profile coincides with Prxs in other parasitic nematodes. A 2-Cys Prx from a plant parasitic nematode, the yellow potato cyst nematode Globodera rostochiensis, was found on the cuticle surface and in the stylet secretions of infective and post-infective juveniles [41, 42]. The other 2-Cys Prx was localized in the hypodermis and cuticle in the infective larvae of Onchocerca volvulus, the nematode pathogen causing onchocerciasis or 'river blindness' disease in Africa [38]. Based on the abundance, distribution, and localization profile of BxPrx, it is germane to speculate that the secretion of this antioxidant in the cuticle region could be an adaptive response to protect this plant parasitic nematode against the host immune responses.
Summary
Prx has been found in a wide range of organisms, including bacteria, plants, invertebrates and mammals [18], and has been shown to play important roles in parasite-host interactions. In this study, a 2-cysteine peroxiredoxin gene in B. xylophilus (BxPrx) was successfully cloned and its primary structure was thoroughly analyzed. BxPrx, a conserved Prx2 protein, has two conserved cysteine active sites, a catalytic triad, and two H2O2 sensitive motifs. The spatial configuration and the putative functions of these conserved domains were analyzed using a tertiary structure simulated by the SWISS-MODEL. Two plausible transmembrane mechanisms are discussed in light of the presence of this leader-less BxPrx in the cytoplasm and under the B. xylophilus cuticle. The induction of recombinant BxPrx is optimized, and the purified recombinant protein exhibited specific Prx activity neutralizing H2O2. The combined results from the gene discovery, protein localization, and recombinant protein characterization suggest that BxPrx potentially plays a key role in combating the oxidative burst engineered by the ROS defense system in host plants during the infection process. In short, BxPrx is a putative genetic factor facilitating B. xylophilus infestation of pine tree hosts.
Acknowledgements
We are grateful to Weiquan Liu (China Agricultural University) and Hongjing Hao (China Institute of Atomic Energy) for their technical assistance. We also thank Bingyan Xie (Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences) for the B. xylophilus isolates. Special thanks go to John J. Obrycki, Kenneth F. Haynes and Barbara J. Duncan (University of Kentucky) for their comments on an earlier draft. This research was funded by a National “973” project (Award #: 2009CB119204).
Author Contribution
ZL, YC, and YW conceived the experiments; ZL, XL, QZ, and XZ designed the study; ZL, QZ, and XZ analyzed the data; and ZL, QZ, and XZ wrote the manuscript. All authors read and approved the final version of manuscript.
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Aikawa T, Kikuchi T, Kosaka H. Population structure of Bursaphelenchus xylophilus within single Pinus thunbergii trees inoculated with two nematode isolates. Forest Pathol. 2006;36:1-13
2. Kanzaki N, Futai K. A PCR primer set for determination of phylogenetic relationships of Bursaphelenchus species within the xylophilus group. Nematology. 2002;4:35-41
3. Cheng X, Cheng F, Xu R. et al. Genetic variation in the invasive process of Bursaphelenchus xylophilus (Aphelenchida: Aphelenchoididae) and its possible spread routes in China. Heredity. 2008;100:356-365
4. Zhao L, Wei W, Kang L. et al. Chemotaxis of the pinewood nematode, Bursaphelenchus xylophilus, to volatiles associated with host pine, Pinus massoniana, and its vector Monochamus alternatus. J Chem Ecol. 2007;33:1207-1216
5. Dwinell LD. The pinewood nematode: regulation and mitigation. Annu Rev Phytopathol. 1997;35:153-166
6. Shi J, Luo Y, Wu H. et al. Impact of the invasion by Bursaphelenchus xylophilus on forest growth and related growth models of Pinus massoniana population. Acta Ecol Sin. 2008;28:3193-3204
7. Futai K. Ecological studies on the infection sources of pine wilt (I). Population dynamics of pine wood nematodes in the withered stem of Japanese red pine. Bull Kyoto Univ Forests. 1986;57:1-3
8. Suzuki K, Kiyohara T. Influence of water stress on development of pine wilting disease caused by Bursaphelenchus lignicolus. Eur J Forest Pathol. 1978;8:97-107
9. Mori Y, Miyahara F, Tsutsumi Y. et al. Relationship between resistance to pine wilt disease and the migration or proliferation of pine wood nematodes. Eur J Plant Pathol. 2008;122:529-538
10. Hanawa F, Yamada T, Nakashima T. Phytoalexins from Pinus strobus bark infected with pinewood nematode, Bursaphelenchus xylophilus. Phytochemistry. 2001;57:223-228
11. Baker CJ, Orlandi EW. Active oxygen in plant pathogenesis. Annu Rev Phytopathol. 1995;33:299-321
12. Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251-275
13. LoVerde PT. Do antioxidants play a role in schistosome host-parasite interactions? Parasitol Today. 1998;14:284-289
14. Sies H. Strategies of antioxidant defense. Eur J Biochem. 1993;215:213-219
15. Chandrashekar R, Tsuji N, Morales TH. et al. Removal of hydrogen peroxide by a 1-cysteine peroxiredoxin enzyme of the filarial parasite Dirofilaria immitis. Parasitol Res. 2000;86:200-206
16. Chae HZ, Chung SJ, Rhee SG. Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem. 1994;269:27670-27678
17. McGonigle S, Curley GP, Dalton JP. Peroxiredoxins: a new antioxidant family. Parasitol Today. 1998;14:139-145
18. Netto LES, Chae HZ, Kang SW. et al. Removal of hydrogen peroxide by thiol-specific antioxidant enzyme (TSA) is involved with its antioxidant properties. J Biol Chem. 1996;271:15315-15321
19. Bruchhaus I, Richter S, Tannich E. Removal of hydrogen peroxide by the 29 kDa protein of Entamoeba histolytica. Biochem J. 1997;326:785-789
20. Manevich Y, Sweitzer T, Pak JH. et al. 1-Cys peroxiredoxin overexpression protects cells against phospholipid peroxidation-mediated membrane damage. Proc Natl Acad Sci USA. 2002;99:11599-11604
21. Bryk R, Griffin P, Nathan C. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature. 2000;407:211-215
22. Kim HS, Pak JH, Gonzales LW. et al. Regulation of 1-cys peroxiredoxin expression in lung epithelial cells. Am J Resp Cell Mol Biol. 2002;27:227-233
23. Fujii J, Ikeda Y. Advances in our understanding of peroxiredoxin, a multifunctional, mammalian redox protein. Redox Rep. 2002;7:123-130
24. Kikuchi T, Aikawa T, Kosaka H. et al. Expressed sequence tag (EST) analysis of the pine wood nematode Bursaphelenchus xylophilus and B. mucronatus. Mol Biochem Parasitol. 2007;155:9-17
25. Huang Q, Cheng X, Mao Z. et al. MicroRNA discovery and analysis of pinewood nematode Bursaphelenchus xylophilus by deep sequencing. PLoS ONE. 2010;5:e13271
26. Kikuchi T, Jones JT, Aikawa T. et al. A family of glycosyl hydrolase family 45 cellulases from the pine wood nematode Bursaphelenchus xylophilus. FEBS Lett. 2004;572:201-205
27. Marchler-Bauer A, Lu S, Anderson JB. et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:225-229
28. Bendtsen JD, Nielsen H, von Heijne G. et al. Improved prediction of signal peptides: SignalP 3.0. Mol Biol. 2004;340:783-795
29. Haslekås C, Viken MK, Grini PE. et al. Seed 1-cystein Peroxiredoxin antioxidants are not involved in dormancy, but contribute to inhibition of germination during stress. Plant Physilol. 2003;133:1148-1157
30. Kwatia MA, Botkin DJ, Williams DL. Molecular and enzymatic characterization of Schistosoma mansoni thioredoxin peroxidase. J Parasitol. 2000;86:908-915
31. Knoops B, Loumaye E, Van Der Eecken V. Evolution of the peroxiredoxins. Flohé L, Harris J R. Peroxiredoxin Systems. New York: Springer. 2007:27-40
32. Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543-1552
33. Jönsson TJ, Johnson LC, Lowther WT. Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace. Nature. 2008;451:98-101
34. Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650-653
35. Jones PM, George AM. The ABC transporter structure and mechanism: perspectives on recent research. Cell Mol Life Sci. 2004;61:682-699
36. Davidson AL, Dassa E, Orelle C. et al. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317-364
37. Robinson MW, Menon R, Donnelly SM. et al. An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica. Mol Cell Proteomics. 2009;8:1891-1907
38. Zipfel PF, Schrum S, Bialonski A. et al. The peroxidoxin 2 protein of the human parasite Onchocerca volvulus: recombinant expression, immunolocalization, and demonstration of homologous molecules in other species. Parasitol Res. 1998;84:623-631
39. De Guiran G, Bruguier N. Hybridization and phylogeny of the pinewood nematode (Bursaphelenchus spp.). Nematologica. 1989;35:321-330
40. Mamiya Y, Enda N. Bursaphelenchus mucromus n. sp. (Nematoda: Aphelenchoididae) from pine wood and its biology and pathogenicity to pine trees. Nematologica. 1979;25:353-361
41. obertson L, Robertson WM, Sobczak M. et al. Cloning, expression and functional characterization of a peroxiredoxin from the potato cyst nematode Globodera rostochiensis. Mol Biochem Parasitol. 2000;111:41-49
42. Williamson VM, Gleason CA. Plant-nematode interactions. Curr Opin Plant Biol. 2003;6:327-333
Additional Tables and Figures
Primer sequences for the expression of recombinant BxPrx
| Primer ID | Primer Sequence (5′-3′) |
|---|---|
| BxPrx3S | TTYGTNTGYCCNACNGAR |
| BxPrx5R | NGCNGGRCANACYTCNCC |
| BxPrxSB | CGGGATCCATGTCCAAGGCTTTCATT* |
| BxPrxRS | ACGCGTCGACTTAATGTTTGTTGAAATA** |
“*”: BamH I recognition site is underlined . “**”: Sal I recognition site is underlined.
Construction of recombinant plasmid vectors. Recombinant plasmids of Cam35S-GPS-BxPrx (A) and pET-28a-BxPrx (B), respectively, were constructed by ligating the entire ORF of BxPrx into the Cam35S-GFP between restriction sites BamH I and Sal I.
Author contact
![]() Corresponding author: Dr. Qingwen Zhang, Department of Entomology, China Agricultural University, Yuanmingyuan West Road, Beijing 100193, China. Phone: 86-10-62733016 Fax: 86-10-62733016 Email: zhangqingwennet or Dr. Xuguo "Joe" Zhou, Department of Entomology, University of Kentucky, S-225 Agricultural Science Center North, Lexington, KY 40546-0091. Phone: 859-257-3125; Fax: 859-323-1120; Email: xuguozhouedu
Corresponding author: Dr. Qingwen Zhang, Department of Entomology, China Agricultural University, Yuanmingyuan West Road, Beijing 100193, China. Phone: 86-10-62733016 Fax: 86-10-62733016 Email: zhangqingwennet or Dr. Xuguo "Joe" Zhou, Department of Entomology, University of Kentucky, S-225 Agricultural Science Center North, Lexington, KY 40546-0091. Phone: 859-257-3125; Fax: 859-323-1120; Email: xuguozhouedu

 Global reach, higher impact
Global reach, higher impact