10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2011; 7(6):902-911. doi:10.7150/ijbs.7.902 This issue Cite
Research Paper
Autosomal Interactions and Mechanisms of Pyrethroid Resistance in House Flies, Musca domestica
1. Department of Entomology and Plant Pathology, Auburn University, Auburn AL 36849, USA
2. Genomics Laboratory, Auburn University, Auburn, AL 36849, USA
3. Current address: College of Forestry, Northeast Forestry University, Harbin, China
4. Current address: College of Plant Protection, Southwest University, Chongqing, China
5. Current address: Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
6. Current address: Department of Life-Science, Northwest A&F University, Shaanxi, China
Received 2011-6-24; Accepted 2011-7-22; Published 2011-7-27
Abstract
Five BC1 lines and 16 house fly mass-cross homozygous lines were generated from crosses of the pyrethroid resistant ALHF (wild-type) and susceptible aabys (bearing recessive morphological markers on each of five autosomes) strains. Each of the resulting homozygous lines had different combinations of autosomes from the resistant ALHF strain. Levels of resistance to permethrin were measured for each line to determine the autosomal linkage, interaction and, possibly, regulation in pyrethroid resistance of house flies. Results indicated that factors on autosome 4 are not involved in the development of resistance in house flies, while factors on autosomes 1, 2, 3 and 5 play important roles in pyrethroid resistance. The sodium channel gene has been mapped on autosome 3 and multiple cytochrome P450 genes overexpressed in resistant ALHF house flies have been genetically mapped on autosome 5, suggesting that P450 mediated detoxification and sodium channel-mediated target site insensitivity located on autosomes 3 and 5, respectively, are major factors related to resistance development in house flies. However, neither the factors on autosome 3 or 5 alone, nor the factors from both autosomes 3 and 5 combined could confer high levels of resistance to pyrethroid. In addition, strong synergistic effects on resistance was obtained when autosomes 1 and 2 interact with autosome 3 and/or 5, suggesting that the trans factors on autosomes 1 and 2 may interact with factors on autosomes 3 and 5, therefore, playing regulatory roles in the development of sodium channel insensitivity- and P450 detoxification-mediated resistance.
Keywords: House fly lines, pyrethroid resistance, autosomes, Genetic crossing, Cytochrome P450s, Sodium Channels
Introduction
THE HOUSE FLY, Musca domestica, is one of the most serious pests affecting livestock and poultry facilities in the United States and worldwide. House flies are the vectors of more than 100 human and animal intestinal diseases, including bacterial, protozoan, helminthic, and viral and rickettsial infections [1]. Furthermore, house flies have shown remarkable abilities to develop resistance to the insecticides used against them in that many strains are now cross-resistant to unrelated classes of insecticides [2-6].
Increased metabolic detoxification [5, 7, 8] and decreased target site sensitivity of the insect nervous system [9-11] are two of the major mechanisms involved in the development of insecticide resistance in house flies. The products of three gene families, cytochrome P450 monooxygenases (P450s), hydrolases, and glutathione S-transferases (GSTs), have been implicated in the metabolic detoxification of insecticides. Transcriptional up-regulation and/or increased amplification of these detoxification genes are known to decrease the dose available at the target site, resulting in the loss of insecticide's effectiveness and the development of insecticide resistance in insects [12-24]. Point mutations of target sites such as sodium channels, GABA receptors, and acetylcholinesterases (AChEs) also reduce the nervous system's response to insecticides (i.e., target-site insensitivity), rendering the dose ineffective at the target proteins and, in turn, resulting in insecticide resistance [25-30]. Decreased target site sensitivity to pyrethroids, known as knockdown resistance (kdr), in house flies has been linked to insensitivity of the para-type sodium channel gene (Vssc1) resulting from point mutations in the sodium channel of house flies [11, 29, 31, 32]. Among those mutations, the mutation of leucine to phenylalanine (L to F) in the sodium-channel gene has been extensively studied in pyrethroid and DDT resistant house flies [9-11]. The kdr phenotype has been genetically linked to autosome 3 of house flies [2, 33, 34]. Although an individual mechanism or gene could by itself confer resistance, there is a great deal of evidence indicating that the interactions among multiple resistance mechanisms or genes are responsible for high levels of insecticide resistance [2, 5, 35-43].
The house fly stain ALHF exhibits a high level of resistance to pyrethroids [5]. The major mechanisms of pyrethroid resistance in this strain include increased detoxification mediated by P450 monoxygenases and decreased sensitivity of voltage-gated sodium channels (kdr) [22-24, 41, 44]. Previous genetic studies of ALHF have linked pyrethroid resistance, mainly on autosomes 2, 3 and 5, with a minor role of factor(s) on autosome 1 [39]. Furthermore, multiple P450 genes, CYP6A5, CYP6A5v2, CYP6A36, CYP6A37, CYP4D4v2 and CYP6A38, that are known to be overexpressed or induced by permethrin in ALHF have been located on autosome 5 and the regulation of these P450 genes have been linked to autosomes 1 and 2 [22-24]. However, the precise nature of the interaction between the factors on these autosomes and the development of pyrethroid resistance in ALHF is unclear. The current study characterizes these autosomal interactions and their contributions, both individually and in combination, in the pyrethroid resistance of house flies. The possible involvement of autosomal interactions in the regulation of P450 mediated detoxification and target site insensitivity in insecticide resistance is also discussed.
Materials and Methods
House fly strains
Two house fly strains, ALHF and aabys, were used as parental strains in this study. ALHF was originally collected in 1998 from a poultry farm in Alabama. This strain exhibited a high level of resistance after subsequent selection with permethrin for six generations and has been annually selected with permethrin to maintain its resistance level [5]. aabys is an insecticide-susceptible strain with recessive morphological markers ali-curve (ac), aristapedia (ar), brown body (bwb), yellow eyes (ye), and snipped wings (sw) on autosomes 1, 2, 3, 4, and 5, respectively. The aabys strain was originally obtained from Dr. J. G. Scott (Cornell University).
Genetic crosses
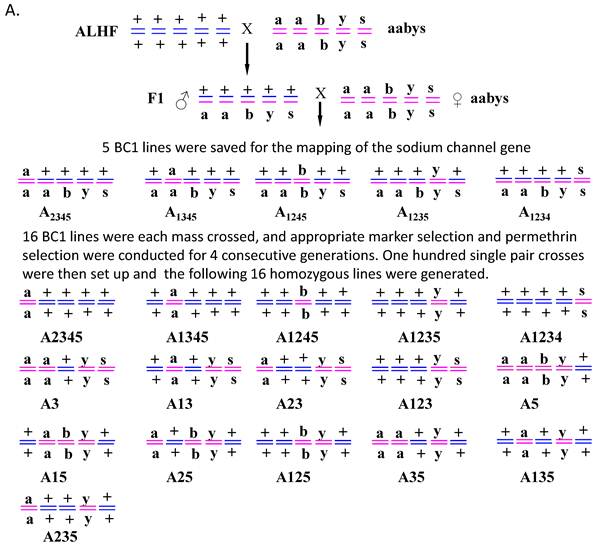
Five backcross (BC1) lines were genetic isolated via genetic crosses of ALHF and aabys house flies as described by Liu and Yue [39]. Briefly, virgin females were collected within 8 h of emergence. Reciprocal crosses of ALHF and aabys were performed and F1 males were back-crossed to aabys females that were isolated every 8 h. Five phenotypes were saved from the back-cross generation (BC1) with the genotypes of: ac/ac, +/ar, +/bwb, +/ye, +/sw (A2345); +/ac, ar/ar, +/bwb, +/ye, +/sw (A1345); +/ac, +/ar, bwb/bwb, +/ye, +/sw (A1245); +/ac, +/ar, +/bwb, ye/ye, +/sw (A1235); and +/ac, +/ar, +/bwb, +/ye, sw/sw (A1234) (Fig. 1). Since crossing over does not or very rarely occurs in male flies [43], the presence of a mutant phenotype indicated that the respective autosome with a mutant-type marker was derived from aabys. The genotype of BC1 for each line was homozygous for the recessive mutant allele from aabys and heterozygous for the dominant wild-type alleles from ALHF.
Sixteen homozygous house fly lines of ac/ac, ar/ar, +/+, ye/ye, sw/sw (A3); ac/ac, ar/ar, bwb/bwb, ye/ye, +/+ (A5); +/+, ar/ar, +/+, ye/ye, sw/sw (A13); ac/ac, +/+, +/+, ye/ye, sw/sw (A23); +/+, ar ar, bwb/bwb, ye/ye, +/+ (A15); ac/ac, +/+, bwb /bwb, ye/ye, +/+ (A25); ac/ac, ar/ar, +/+, ye/ye, +/+ (A35); +/+, +/+, +/+, ye/ ye, sw/ sw (A123); +/+, +/+, bwb /bwb, ye/ye, +/+ (A125); +/+, ar/ar, +/+, ye/ye, +/+ (A135); ac/ac, +/+, +/+, ye/ye, +/+ (A235); +/+, +/+, bwb/bwb, +/+, +/+ (A1245); +/+, + /+, +/+, ye/ye, +/+ (A1235); +/+, +/+, +/+, +/+, sw/sw (A1234); +/+, ar /ar, +/+, +/+,+/+ (A1345); ac/ac, +/+, +/+, +/+, +/+ (A2345) were derived from mass crosses of BC1 lines for four generations. Briefly, for each generation, flies were sorted for appropriate phenotypes and selected with permethrin at a dose that killed approximately 70% of the treated flies. One hundred single-pair crosses were then set up for each line as described by Liu [44]. Families of each single pair cross that showed only individuals of the desired phenotype for three consecutive generations were combined and then selected with permethrin for three generations. The name of each line was assigned according to the autosomes bearing wild-type markers from ALHF. For instance, the A2345 strain had wild-type markers on autosomes 2, 3, 4, 5 from ALHF and the mutant marker on autosome 1 from aabys.
Amplification of the sodium channel gene fragment in the house fly
Males and females of each house fly strains and lines had their genomic RNA extracted for each experiment using the acidic guanidine thiocyanate-phenol-chloroform method [17]. Three replications were performed, each on a different day, for a total of 60 individual house flies from each strain or line. The first strand complementary DNA (cDNA) was synthesized with SuperScript II reverse transcriptase (Invitrogen) and an antisense 5'-anchored oligo(dT) primer (5'-TAATACGACTCACTATAGGGAGATTTTTTTTTTTTTTTT-3' [45]. The sodium channel cDNA fragments where the L-to-F mutation resides were amplified by the primer pair Kdr (HF) F3 (5'-TCATCTTTGCCGTGATGGGAATGC-3') and Kdr (HF) R3 (5'-AATGATGATGTCACCATCCAGCTG-3') as described by Xu [42]. The amplicons generated using this primer pair spanned the intron/exon boundaries in order to avoid amplification of the genomic DNA, thus preventing genomic DNA contamination. The PCR solution with JumpStartTM Taq DNA Polymerase (Sigma), cDNA template, and the primer pair was heated to 95°C for 2min, followed by 40 cycles of PCR reaction (94°C 30s, 60°C 30s and 72°C for 2min) and a final extension of 72°C for 10 min. The PCR products were subjected to single nucleotide polymorphism (SNP) determination and sequencing.
Genetic isolation of house fly lines. (A). Diagrammatic representation of the genetic isolation of 16 house fly lines generated by crossing resistant ALHF and susceptible aabys strains. House fly lines were named according to the autosomes bearing the wild-type marker derived from ALHF. (B). The morphological phenotype of each of house fly strains or lines.

SNP determination for kdr allelic expression variation in house flies, Musca domestica
The frequency and heterozygosity of the kdr allele at the L-to-F site in each of the house fly strains and lines were investigated by SNP determination using an ABI Prism SNaPshot Multiplex Kit and analyzed on the ABI Prism® 3100 Genetic Analyzer using Genemapper software according to the manufacturer's instructions (Applied Biosystems). A sodium channel cDNA fragment from each of the individuals was used as template. The primer for the SNP determination was designed according to the sequence immediately upstream of the kdr allele. Three replications of the SNP determination were carried out using the same cDNA for each individual, with different preparations of the PCR templates. To confirm that our PCR products were in fact sodium channel gene fragments, the PCR products from each sample were sequenced.
Insecticide bioassays
Topical applications were performed on each of the strains and lines by dropping 0.5-µl of insecticide (dissolved in acetone) onto the thoracic notum of 2-3-day old female flies [5]. Each bioassay consisted of 20 flies per dose and four or five doses that resulted in >0 and <100% mortality. Control groups received acetone alone. Treated flies were placed in 6 oz. Sweetheart ice cream cups (Sweetheart Cup Co., Owings Mills, MD) along with a piece of dental wick saturated in 15% sugar water. Mortality was assessed after 24 h. Flies that did not move were scored as dead. All tests were performed at 25±2°C and replicated at least three times on different days for each of two consecutive generations. Bioassay data were pooled and probit analysis was conducted using POLO probit analysis software. Significant differences in the resistance levels of the house fly lines were based on nonoverlapping 95% confidence intervals [44].
Results
Genetic linkage of the sodium channel gene in house flies
To investigate the autosomal interaction with target site insensitivity in the insecticide resistance of ALHF house flies, we conducted a linkage study of the sodium channel gene in house flies by characterizing the allelic expression variation at the L-to-F locus in the sodium channel gene in the parental strains of ALHF and aabys, the F1 generation, and five BC1 lines, A2345, A1345, A1245, A1235, and A1234, each of which was homozygous for one autosome bearing a recessive morphological marker from the aabys strain and heterozygous for the other four autosomes (i.e., containing autosomes from both ALHF and aabys strains). Our results indicated that all the resistant ALHF individuals expressed the T allele (Table 1), as expected based on our previous study [42]; all individuals of the susceptible aabys strain expressed the C allele; and all individuals of F1 progeny expressed both C and T alleles, corresponding to their heterozygous nature, at the RNA level (Table 1). The house fly BC1 line of A1245 with the genotype +/ac, +/ar, bwb/bwb, +/ye, +/sw, bearing a recessive morphological marker from autosome 3 of aabys, which was homozygous for autosome 3 from aabys and heterozygous for the other 4 autosomes (from both aabys and ALHF), expressed only the C allele in all individuals tested. House fly BC1 lines A2345, A1345, A1235 or A1234, which had recessive morphological markers on autosomes 1, 2, 4, or 5 from aabys and was thus homozygous for autosomes 1, 2, 4, or 5, respectively, from aabys, expressed both alleles (C/T). These findings strongly indicate that the sodium channel gene is located on autosome 3. This result is consistent with previous reports of the genetic linkage of knockdown resistance in house flies [2, 33, 34].
Linkage analysis and autosomal interaction in pyrethroid resistance of house flies
Linkage analysis of autosomal involvement in insecticide resistance was conducted by comparing the levels of insecticide resistance to permethrin in the house fly strains and 16 homozygous house fly lines described above. The LD50 values of permethrin for the parental resistant and susceptible strains of ALHF and aabys were 2,100 and 2 ng/fly, respectively (Table 2), revealing a 1100-fold resistance to permethrin in the ALHF strain. The resistance level of permethrin in the A1235 strain (flies with a recessive mutant marker on autosome 4 from aabys) was 950-fold (Table 2), which was similar to that in the parental ALHF strain (1100-fold), and there were no significant differences in resistance levels between A1235 and ALHF based on the overlapping 95% confidence intervals for the two strains [44]. These results indicate that factors on autosome 4 are not involved in the development of pyrethroid resistance in ALHF, which agrees with the findings of our previous study [39]. House fly lines with recessive morphological markers on autosomes 1, 2, 3 or 5 from aabys (A2345, A1345, A1245, or A1234, respectively) had significantly decreased levels of permethrin resistance compared with the resistant parental strain ALHF, exhibiting resistant ratios of 390, 210, 95, or 140, respectively, compared with the susceptible aabys strain (Table 2), implying that factors on autosomes1, 2, 3 and 5 play very important roles in the development of pyrethroid resistance in ALHF. These data suggest that pyrethroid resistance in ALHF is likely conferred by factors on autosomes 1, 2, 3, and 5 and indicate the presence of interactions among resistance factors on different autosomes in the ALHF strain, which has also been identified in the pyrethroid resistant LPR house fly strain [44, 46].
Allelic expression of L-to-F kdr locus in ALHF, aabys, F1 progeny and five BC1 (backcross) house fly lines of M. domestica.
| House Fly Strains/Lines | na | Phenotype | Nucleotide Changesb | ||
|---|---|---|---|---|---|
| CTT | C/TTT | TTT | |||
| aabys | 30 | mutant type | 30 | 0 | 0 |
| ALHF | 30 | Wild | 0 | 0 | 30 |
| F1 | 30 | N/A | 0 | 30 | 0 |
| A2345 | 30 | ali-curve | 0 | 30 | 0 |
| A1345 | 30 | aristapedia | 0 | 30 | 0 |
| A1245 | 30 | brown body | 30 | 0 | 0 |
| A1235 | 30 | yellow eyes | 0 | 30 | 0 |
| A1234 | 30 | snipped wings | 0 | 30 | 0 |
a The total number of tested flies.
b The nucleotides changed in the L-to-F kdr locus are underlined.
Toxicity of permethrin to ALHF, aabys, and 16 homozygous house fly lines of M. domestica.
| strain | df | na | χ2a | LD50 (95% CI)b | Slope (SE) | RR* |
|---|---|---|---|---|---|---|
| aabys | 3 | 280 | 2.1 | 2.0 (1.0-2.0) | 3.0 (0.3) | - |
| ALHF | 3 | 284 | 0.7 | 2100 (1600-2600) | 2.5 (0.3) | 1100 |
| A2345 | 3 | 315 | 3.0 | 780 (680-900) | 3.0 (0.3) | 390 |
| A1345 | 3 | 300 | 1.4 | 410 (300-570) | 3.5 (0.3) | 210 |
| A1245 | 3 | 300 | 6.7 | 190 (150-250) | 4.0 (0.4) | 95 |
| A1235 | 5 | 407 | 1.2 | 1900 (1400-2300) | 1.6 (0.2) | 950 |
| A1234 | 3 | 240 | 3.6 | 270 (180-500) | 2.7 (0.3) | 140 |
| A3 | 5 | 315 | 2.7 | 74 (62-89) | 2.1 (0.2) | 37 |
| A13 | 5 | 752 | 5.5 | 92 (80-110 ) | 2.7 (0.2) | 46 |
| A23 | 3 | 225 | 1.9 | 110 (84-150) | 1.9 (0.2) | 55 |
| A123 | 3 | 240 | 3.9 | 240 (160-460) | 2.8 (0.3) | 120 |
| A5 | 3 | 300 | 6.5 | 24 (17-37) | 2.7 (0.3) | 12 |
| A15 | 3 | 300 | 3.4 | 29 (23-37) | 2.9 (0.3) | 15 |
| A25 | 3 | 300 | 6.9 | 30 (21-44) | 2.9 (0.3) | 15 |
| A125 | 3 | 300 | 2.1 | 150 (110-220) | 3.7 (0.3) | 75 |
| A35 | 3 | 300 | 7.2 | 310 (230-430) | 3.8 (0.4) | 160 |
| A135 | 3 | 300 | 7.5 | 400 (300-560) | 4.1 (0.4) | 200 |
| A235 | 3 | 300 | 1.4 | 650 (540-820) | 2.2 (0.3) | 330 |
na : Number of house flies tested.
LD50 (95% CI)b: LD50 values in nanograms per house fly.
*RRs were calculated by dividing LD50 of house fly strains and lines by LD50 of aabys.
We compared the levels of resistance in house fly lines with different autosomal combinations with autosome 3 from ALHF, i.e., the lines A3, A13, A23, and A123. The toxicity of permethrin to these house lines followed a descending order of A3 > A13 > A23 > A123. The flies with all three autosomes 1, 2 and 3 of ALHF (A123) had the highest levels of resistance among these four lines and the flies with only autosome 3 from ALHF (A3) had the lowest level of resistance (Table 2). These results suggest that factors on autosomes 1 and 2 interact with autosome 3, thus playing a regulatory role in sodium channel insensitivity-mediated insecticide resistance. The fact that line A23 had a higher level of resistance than line A13 suggests that factor(s) on autosome 2 may have a greater effect on resistance than those on autosome 1.
We next compared the levels of resistance in house fly lines A5, A15, A25, and A125 with different autosomal combinations with autosome 5 from ALHF, in which multiple cytochrome P450s were located. The toxicity of permethrin to these house lines followed a descending order of A5 > A15 > A25 > A125 (Table 2). Flies with factors on autosomes 1, 2 and 5 of ALHF (A125) had higher levels of resistance than flies with factors on only autosome 5 of ALHF (A5), suggesting that factors on autosomes 1 and 2 interact with factors on autosome 5 and may play a regulatory function P450 detoxification-mediated insecticide resistance. Comparing the individual interactions of autosome 1 or 2 with autosome 5, once again factors on autosome 2 had a greater effect on autosome 5 in terms of insecticide resistance than those on autosome 1 (Table 2).
We further examined the synergism effect of autosome 1, or autosome 2, or a combination of autosomes 1 and 2 on the combined autosomes 3 and 5 from ALHF. We compared the levels of resistance in house fly lines with different autosomal combinations A35, A135, A235, and A1235, all of which obtained at least two autosomes (3 and 5) from the ALHF resistant strain, thus containing both sodium channel gene(s) and multiple cytochrome P450 genes from ALHF. A descending order of permethrin toxicity was observed in these house fly lines, with A35 > A135 > A235 > A1235 (Table 2). Flies with factors on all four autosomes 1, 2, 3 and 5 of ALHF (A1235) had higher levels of resistance compared with the flies with only factors on both autosomes 3 and 5 from ALHF (A35), suggesting that factors on autosomes 1 and 2 interact with factors on autosome 3 and 5 and may play a role in the regulation of both the P450 genes on autosome 5 and the sodium channel gene on autosome 3.
Contribution of autosomes and autosomal interactions in sodium channel gene expression
An increasing body of evidence from both molecular and pharmacological studies has pointed to the involvement of point mutations in voltage-gated sodium channels in kdr and kdr-like resistance in many medically or agriculturally important pest species, including house flies. Among these kdr mutations, the substitution of leucine to phenylalanine [L to F], histidine [L to H], or serine [L to S] in the 6th segment of domain II (IIS6) results from a single nucleotide polymorphism, which, in the case of house flies, is resulting from the change of codon CTT to TTT, has been clearly associated with increased resistance to pyrethroids and DDT [3-9]. In the current study, we mapped the sodium channel gene on autosome 3 and found that autosomes 1 and 2 interact with autosome 3 to enhance levels of pyrethroid resistance. To test whether the factors on autosomes 1 and 2 are involved in the sodium channel insensitivity, we investigated the expression of the L-to-F kdr mutation in different house fly strains and lines by assessing the RNA expression variation of the alleles at the L-to-F site of the sodium channel. In the susceptible aabys strain, 100% (30/30) of the tested individuals expressed the C allele (susceptible). In the resistant strain, ALHF, 100% (30/30) of the tested individuals expressed the T allele (Tables 1 and 3). In the house fly lines with autosome 3 from ALHF, i.e., A2345, A1345, A1235, A1234, A3, A23, A13, A123, A35, A135, and A235, 100% (30/30) tested individuals of each expressed the resistance T allele, whereas in the house fly lines with autosome 3 derived from aabys, i.e., A5, A15, A25, A125 and A1245, 100% (30/30) expressed the C allele (Table 3). This result indicates that those individuals whose chromosome 3 was derived from the resistant ALHF strain expressed the T kdr allele at the L-to-F locus of the sodium channel, as in their parent resistant ALHF strain, regardless of variations in the factors on the other chromosomes. In contrast, in flies with a recessive morphological marker (brown body) on chromosome 3 derived from aabys, all the tested individuals expressed the susceptible C allele, just as in the aabys strain. This result indicates that there may be no trans-regulation factors on autosomes 1, 2, 4 or 5 that influence the L-to-F kdr allelic expression at the L-to-F locus of the sodium channels on autosome 3.
Allelic expression of L-to-F kdr locus in ALHF, aabys, and 16 homozygous house fly lines of M. domestica.
| House fly | na | Morphological Genotype | Nucleotide Changesb | ||
|---|---|---|---|---|---|
| CTT | C/TTT | TTT | |||
| aabys | 30 | ac/ac, ar/ar, bwb/bwb, ye/ye, sw/sw (mutant type) | 30 | 0 | 0 |
| ALHF | 30 | +/+, +/+, +/+, +/+, +/+ (wild type) | 0 | 0 | 30 |
| A2345 | 30 | ac/ac, +/+, +/+, +/+, +/+ | 0 | 0 | 30 |
| A1345 | 30 | +/+, ar /ar, +/+, +/+,+/+ | 0 | 0 | 30 |
| A1245 | 30 | +/+, +/+, bwb/bwb, +/+, +/+ | 30 | 0 | 0 |
| A1235 | 30 | +/+, + /+, +/+, ye/ye, +/+ | 0 | 0 | 30 |
| A1234 | 30 | +/+, +/+, +/+, +/+, sw/sw | 0 | 0 | 30 |
| A3 | 30 | ac/ac, ar/ar, +/+, ye/ye, sw/sw | 0 | 0 | 30 |
| A13 | 30 | +/+, ar/ar, +/+, ye/ye, sw/sw | 0 | 0 | 30 |
| A23 | 30 | ac/ac, +/+, +/+, ye/ye, sw/sw | 0 | 0 | 30 |
| A123 | 30 | +/+, +/+, +/+, ye/ ye, sw/ sw | 0 | 0 | 30 |
| A5 | 30 | ac/ac, ar/ar, bwb/bwb, ye/ye, +/+ | 30 | 0 | 0 |
| A15 | 30 | +/+, ar ar, bwb/bwb, ye/ye, +/+ | 30 | 0 | 0 |
| A25 | 30 | ac/ac, +/+, bwb /bwb, ye/ye, +/+ | 30 | 0 | 0 |
| A125 | 30 | +/+, +/+, bwb /bwb, ye/ye, +/+ | 30 | 0 | 0 |
| A35 | 30 | ac/ac, ar/ar, +/+, ye/ye, +/+ | 0 | 0 | 30 |
| A135 | 30 | +/+, ar/ar, +/+, ye/ye, +/+ | 0 | 0 | 30 |
| A235 | 30 | ac/ac, +/+, +/+, ye/ye, +/+ | 0 | 0 | 30 |
Discussion
The interactions of a number of resistance (R) genes, each of which is linked to different resistance mechanisms and confers only a small degree of resistance to a given compound in isolation, appeared to significantly increase the level of resistance to that compound [2, 37]. It has been suggested that insensitive target site genes would combine multiplicatively with a detoxification gene during the first stage of the detoxification process, and would then combine additively after saturation of the detoxification enzyme [37]. Comparison among the 16 homozygous house fly lines, each of which had one autosome from susceptible aabys and the remaining 4 from resistant ALHF parental strains, revealed that factors on autosomes 1, 2, 3, and 5 had an interactive effect on the enhanced levels of insecticide resistance. It is notable that strains A3 and A5, containing either autosome 3 or 5 derived from resistant ALHF flies, had only 37- and 12-fold resistance, respectively. Since multiple P450 genes have been mapped to autosome 5 of the house fly [22-24] and the sodium channel gene has been mapped on autosome 3, this finding indicates that the kdr factor (or other resistance factors) on autosome 3 and P450 factors on autosome 5 alone provide only a marginal level of resistance.
House fly lines containing autosomes 1 and 5, or 2 and 5 from ALHF, i.e. A15, and A25, enjoy increased resistance compared to house fly lines containing only autosome 5 from ALHF, suggesting that factors on autosome 1 and 2 play a role in the regulation of P450-mediated resistance in house flies. We also observed that house fly lines that included autosomes 1,2, and 5 from resistant ALHF flies (A125) had a synergistic effect on insecticide resistance compared with house fly lines with only autosomes 2 and 5 or 1 and 5 from ALHF, suggesting that the mechanism of P450 mediated detoxification alone could not confer the full level of resistance without the presence of other factors on both autosomes 1 and 2, and that the interaction of multiple factors and mechanisms is thus important for the development of high levels of resistance. The involvement of factors on autosomes 1 and 2 in insecticide resistance in the house fly has been reported in previous studies on resistance to insecticides [12, 38, 39]. In the pyrethroid resistant LPR strain, the overexpression of the P450 gene CYP6D1, which is responsible for the insect's high level of pyrethroid resistance, was mapped on autosome 1 and its overexpression found to be regulated by trans-regulatory factor (s) on autosome 2 [38]. In the organophosphate-resistant Rutgers house fly strain, the elevated level of the P450 CYP6A1 mapped on autosome 5 was also trans-regulated by factors on autosome 2 [12]. Furthermore, the factors controlling the overexpression of these P450 genes have been genetically linked to autosomes 1 and 2 [23, 24, 39]. The overexpression of CYP6A2 and CYP6A8 in Drosophila melanogaster resistant to DDT and malathion is regulated by a trans regulatory factor(s) on autosome 3 [47, 48]. The right arm of autosome 3 (3R) of Drosophila and autosome 2 of house flies are thought to be homologous. Plapp (1984) proposed that a “master gene” on autosome 2 in the house fly interacted with “minor” genes to regulate high levels of monooxygenase-mediated resistance to multiple types of insecticides. It is not clear whether these genes are regulated by the same trans regulatory factor(s) or whether this trans regulatory factor(s) on autosome 2 in the house fly is the “master gene” proposed more than twenty five years ago; this issue requires further investigation.
The findings reported here indicate that the factors on autosomes 1 and 2 are not involved in the L-to-F kdr allelic expression of the sodium channel in house flies. However, when house fly lines contained autosomes 1 and 3 or 2 and 3 from ALHF, the resistance in these lines increased, suggesting that factors on autosomes 1 and 2 play a role in the regulation of resistance conferred by the factors on autosome 3. Since only the L-to-F mutation in the sodium channel have been investigated in this study, we cannot exclude the possibility that the ALHF strain may also have kdr-related mutations on the sodium channel gene which, if present, might be regulated by factors on autosome 1 or 2. Nevertheless, a descending order of permethrin toxicity to house fly lines, such as A3>A13>A23>A123>>A1235, A5>A15>A25>A125>>A1235, or A35>A135>A235>A1235, may suggest another scenario, i.e., the trans-regulation factors on autosomes 1, 2 and, perhaps, 3 together play important roles on regulating P450 overexpression on autosome 5.
Acknowledgements
This project was supported by AAES Hatch/Multistate Grants ALA08-045 and ALA015-1-10026 to N.L. and Auburn University Biogrant 2008-Bio-Liu-03-08.
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Scott JG, Liu NN, Kristensen M. et al. A case for sequencing the genome of Musca domestica (Diptera: Muscidae). J Med Entomol. 2009;46:175-182
2. Scott JG, Georghiou GP. The biochemical genetics of permethrin resistance in the Learn-PyR strain of house fly. Biochem Genet. 1986;24:25-37
3. Pap L, Toth A. Development and Characteristics of resistance in the susceptible WHO/SRS house fly (Musca domestica) strain subjected to selection with beta-cypermethrin. Pestic Sci. 1995;45:335-349
4. Lee S, Zhang L, Shono T. Characteristics of the pyraclofos resistant strain of housefly. Appl Entomol Zool. 1996;31:119-125
5. Liu NN, Yue X. Insecticide resistance and cross-resistance in the house fly (Diptera: Muscidae). J Econ Entomol. 2000;93:1269-1275
6. Scott JG, Alefantis TG, Kaufman P. et al. Insecticide resistance in house flies from caged-layer poultry facilities. Pest Manag Sci. 2000;56:147-153
7. Taylor M, Feyereisen R. Molecular biology and evolution of resistance to toxicants. Mol Biol Evol. 1996;13:719-734
8. Scott JG. Cytochromes P450 and insecticide resistance. Insect Biochem Mol Biol. 1999;29:757-777
9. Ingles PJ, Adams PM, Knipple DC. et al. Characterization of voltage-sensitive sodium channel gene coding sequences from insecticide-susceptible and knockdown-resistant house fly strains. Insect Biochem Mol Biol. 1996;26:319-326
10. Williamson MS, Martinez TD, Hick CA. et al. Identification of mutations in the house fly para-type sodium channel gene associated with knockdown resistance (kdr) to pyrethroid insecticides. Mol Gen Genet. 1996;252:51-60
11. Soderlund DM. Sodium channels. In: (ed.) Gilbert LI, Iatrou K, Gill SS. Comprehensive Molecular Insect Science. Elsevier Pergamon. 2005:1-24
12. Carino FA, Koener JF, Plapp FW. et al. Constitutive overexpression of the cytochrome P450 gene CYP6A1 in a house fly strain with metabolic resistance to insecticides. Insect Biochem Mol Biol. 1994;24:411-8
13. Feyereisen R. Molecular biology of insecticide resistance. Toxicology Letters. 1995;82:83-90
14. Whyard S, Downe AE, Walker VK. Characterization of a novel esterase conferring insecticide resistance in the mosquito Culex tarsalis. Arch Insect Biochem Physiol. 1995;29:329-342
15. Liu NN, Scott JG. Phenobarbital induction of CYP6D1 is due to a trans acting factor on autosome 2 in house flies, Musca domestica. Insect Mol Biol. 1997;6:77-81
16. Liu NN, Scott JG. Increased transcription of CYP6D1 causes cytochrome P450-mediated insecticide resistance in house fly. Insect Biochem Mol Biol. 1998;28:531-5
17. Hemingway J, Karunaratne SH. Mosquito carboxylesterases: a review of the molecular biology and biochemistry of a major insecticide resistance mechanism. Med Vet Entomol. 1998;12:1-12
18. Small GJ, Hemingway J. Molecular characterization of the amplified carboxylesterase gene associated with organophosphorus insecticide resistance in the brown planthopper Nilaparvata lugens. Insect Mol Biol. 2000;9:647-653
19. Daborn PJ, Yen JL, Bogwitz MR. et al. A single P450 allele associated with insecticide resistance in global populations of Drosophila. Science. 2002;297:2253-6
20. Pridgeon JW, Liu NN. Overexpression of the cytochrome c oxidase subunit I gene associated with a pyrethroid resistant strain of German cockroaches, Blattella germanica. Insect Biochem Mol Biol. 2003;33:1043-8
21. Feyereisen R. Insect cytochrome P450. In: (ed.) Gilbert LI, Iatrou K, Gill S. Comprehensive Molecular Insect Science. Oxford. 2005:1-77
22. Zhu F, Liu NN. Differential expression of CYP6A5 and CYP6A5v2 in pyrethroid-resistant house flies, Musca domestica. Arch Insect Biochem Physiol. 2008;34:147-161
23. Zhu F, Feng J, Zhang L. et al. Characterization of two novel cytochrome P450 genes in insecticide resistant house flies. Insect Mol Biol. 2008;1:27-37
24. Zhu F, Li T, Zhang L. et al. Co-up-regulation of three P450 genes in response to permethrin exposure in permethrin resistant house flies, Musca domestica. BMC Physiology. 2008;8:1-13
25. Oppenoorth FJ. Biochemistry and genetics of insecticide resistance. In: (ed.) Kerkut GA, Gilbert LI. Comprehensive Insect Physiology, Biochemistry and Pharmacology. Pergamon Press, Oxford. 1985:731-773
26. Narahashi T. Molecular and cellular approaches to neurotoxicology: past, present and future. In: (ed.) Lunt GG. Neurotox '88: Molecular Basis of Drug and Pesticide Action. New York. 1998:563-582
27. Bloomquist JR. Ion channels as targets for insecticides. Annu Rev Entomol. 1996;41:163-190
28. Zhu KY, Lee SH, Clark JM. A point mutation of acetylcholinesterase associated with azinphosmethyl resistance and reduced fitness in Colorado potato beetle. Pestic Biochem Physiol. 1996;55:100-108
29. Soderlund DM, Knipple DC. The molecular biology of knockdown resistance to pyrethroid insecticides. Insect Biochem Mol Biol. 2003;33:563-577
30. Buckingham SD, Sattelle DB. GABA receptors of insects. In: (ed.) Gilbert LI, Iatrou K, Gill S. Comprehensive Molecular Insect Science. Oxford, UK. 2005:107-142
31. Soderlund DM, Bloomquist JR. Molecular mechanisms of insecticide resistance. In: (ed.) Roush RT Tabashnik BE. Pesticide Resistance in Arthropods, Chapman and Hall. New York. 1990:58-96
32. Williamson MS, Denholm I, Bell CA. et al. Knockdown resistance (kdr) to DDT and pyrethroid insecticides maps to a sodium channel gene locus in the housefly (Musca domestica). Mol Gen Genet. 1993;240:17-22
33. Sawicki RM, Farnham AW. Genetic of resistance to insecticides of the SKA strain of Musca domestica, III location and isolation of the factors of resistance to dieldrin. Entomol Exp Appl. 1968;11:133-142
34. Farnham AW. Genetics of resistance of pyrethroid-selected houseflies, Musca domestica L. Pestic Sci. 1973;4:513-520
35. Georghiou GP. Isolation, characterization and re-synthesis of insecticide resistance factors in the housefly, Musca domestica. Proc 2nd Int Congr Pestic Chem. 1971;2:77
36. Frederick W, Plapp JR. The genetic basis of insecticide resistance in the house fly: evidence that a single locus plays a major role in metabolic resistance to insecticides. Pestic Biochem Physiol. 1984;22:194-201
37. Raymond M, Beyssat AV, Sivasubramanian N. et al. Amplification of various esterase B's responsible for organophosphate resistance in Culex mosquitoes. Biochem Genet. 1989;27:417-423
38. Liu NN, Scott JG. Genetic analysis of factors controlling elevated cytochrome P450, CYP6D1, cytochrome b5, P450 reductase and monooxygenase activities in LPR house flies, Musca domestica. Biochem Genet. 1996;34:133-148
39. Liu NN, Yue X. Genetics of pyrethroid resistance in the house fly (Diptera: Muscidae). Pestic Biochem Physiol. 2001;70:151-8
40. Vontas J, Blass C, Koutsos AC. et al. Gene expression in insecticide resistant and susceptible Anopheles gambiae strains constitutively or after insecticide exposure. Insect Mol Biol. 2006;14:509-521
41. Liu NN, Liu H, Zhu F. et al. Differential expression of genes in pyrethroid resistant and susceptible mosquitoes, Culex quinquefasciatus (S.). Gene. 2007;394:61-68
42. Xu Q, Liu H, Zhang L. et al. Resistance in the mosquito, Culex quinquefasciatus, and possible mechanisms for resistance. Pest Manag Sci. 2005;61:1096-1102
43. Gao J, Scott JG. Role of the transcriptional repressor mdGfi-1 in CYP6D1v1-mediated insecticide resistance in the house fly, Musca domestica. Insect Biochem Mol Biol. 2005;36:387-395
44. Liu NN, Scott JG. Genetics of resistance to pyrethroid insecticides in the house fly, Musca domestica. Pest Biochem Physiol. 1995;52:116-124
45. Tomita T, Scott JG. cDNA and deduced protein sequence of Cyp6D1: the putative gene for a cytochrome p450 responsible for pyrethroid resistance in house fly. Insect Biochem Mol Biol. 1995;25:275-283
46. Georghiou GP. Genetics of resistance to insecticides in houseflies and mosquitoes. Exp Parasitol. 1969;26:224
47. Dombrowski SM, Krishnan R, Witte MS. et al. Constitutive and barbital-induced expression of the Cyp6a2 allele of a high producer strain of CYP6A2 in the genetic background of a low producer strain. Gene. 1998;221:69-77
48. Maitra S, Dombrowski SM, Basu M. et al. Factors on the third chromosome affect the level of Cyp6a2 and Cyp6a8 expression in Drosophila melanogaster. Gene. 2000;248:147-156
Author contact
![]() Corresponding author: Dr. Nannan Liu, 301 Funchess Hall, Auburn University, Auburn, AL 36849-5413, USA. Phone: (334) 844-2661. Fax: (334) 844-5005. E-mail: liunannedu
Corresponding author: Dr. Nannan Liu, 301 Funchess Hall, Auburn University, Auburn, AL 36849-5413, USA. Phone: (334) 844-2661. Fax: (334) 844-5005. E-mail: liunannedu

 Global reach, higher impact
Global reach, higher impact