Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2012; 8(2):159-170. doi:10.7150/ijbs.8.159 This issue Cite
Research Paper
The Olfactory Co-receptor Orco from the Migratory Locust (Locusta migratoria) and the Desert Locust (Schistocerca gregaria): Identification and Expression pattern
1. China Agricultural University, Key Lab for Biological Control of the Ministry of Agriculture, Beijing 100193, China.
2. University of Hohenheim, Institute of Physiology, 70593 Stuttgart, Germany.
Received 2011-10-11; Accepted 2011-11-17; Published 2011-12-13
Abstract
In locusts, olfaction plays a crucial role for initiating and controlling behaviours, including food seeking and aggregation with conspecifics, which underlie the agricultural pest capacity of the animals. In this context, the molecular basis of olfaction in these insects is of particular interest. Here, we have identified genes of two orthopteran species, Locusta migratoria and Schistocera gregaria, which encode the olfactory receptor co-receptor (Orco). It was found that the sequences of LmigOrco and SgreOrco share a high degree of identity to each other and also to Orco proteins from different insect orders. The Orco-expressing cells in the antenna of S. gregaria and L. migratoria were visualized by in situ hybridization. Orco expression could be assigned to clusters of cells in sensilla basiconica and few cells in sensilla trichodea, most likely representing olfactory sensory neurons. No Orco-positive cells were detected in sensilla coeloconica and sensilla chaetica. Orco expression was found already in all nymphal stages and was verified in some other tissues which are equipped with chemosensory hairs (mouthparts, tarsi, wings). Together, the results support the notion for a decisive role of Orco in locust olfaction.
Keywords: locust, olfaction, Orco, gene expression, in situ hybridization.
Introduction
The polyphagous grasshoppers Locusta migratoria and Schistocerca gregaria are serious pests in agriculture in wide parts of Africa, the Middle East and Asia. The pest status of the insects is based on the aggregation to destructive swarms and the continuous tapping for new food resources [1]. In locust, behaviours underlying aggregation and food seeking heavily depend on volatile compounds emitted from conspecifics or plants, respectively [2]. These chemical signals are mainly detected by olfactory sensory neurons (OSNs) on the antennae [3, 4], which project their chemoreceptive dendrites into morphologically different types of cuticular hair structures (olfactory sensilla). Based on their morphology and cell numbers different types of antennal olfactory sensilla are discriminated: sensilla basiconica housing 20-50 OSNs and sensilla trichodea containing 1-3 OSNs [5, 6]. In addition, sensory cells in sensilla coeloconica have been found to respond to odorants [3].
Research over the past 25 years has led to the identification of proteins involved in odorant recognition and olfactory signal transduction in insects. Members of large families representing diverse odorant-binding proteins (OBPs) and distinct chemosensory proteins (CSPs) are considered to mediate the transfer of odorants across the aqueous sensillum lymph towards the OSNs [7-10]. Each of the OSNs express one particular odorant receptor (OR) gene selected from a large OR gene repertoire. This OR is inserted into the dendritic membrane of OSNs [11, 12], where it can interact with volatile compounds [13-15]. In addition to a distinct OR-subtype each OSN also expresses the common olfactory receptor co-receptor (Orco) [16], formerly named OR83b in Drosophila and OR2 in moth and other insects [17]. Orco is supposed to form heteromers with the particular OR [18, 19] and functions as a cation channel, which is opened upon OR activation [18, 20].
Until now, orthologues of the Orco gene have been identified only in insect species belonging to two groups within the class Insecta: in the Endopterygota (ants, bees, beetles, moths, flies), also referred to as holometabolous insects [21, 22] and in the Hemipteroid Assemblage (aphids, bugs, lice), which comprises hemimetabolous species [23, 24]. These two sister groups are viewed to form a single monophyletic division within the Neoptera group of winged insects (Pterygota) and to share a common ancestor distinct from other Neoptera divisions, which all are hemimetabolous [25]. Among these the orthoptera (grasshoppers, crickets) are phylogenetically clearly separated from the Endopterygota and the Hemipteroid Assemblage [26]. This distant phylogenetic relationship could imply, that olfactory genes of orthopteran insects, like the locust L. migratoria and S. gregaria, may be only very distantly related to those of insects from the Endopterygota and the Hemipteroid Assemblage. Consequently, homology based cloning strategies to identify olfactory genes of locust based on sequence information from insects of the Endopterygota and the Hemipteroid Assemblage may be difficult to apply. In support of this notion, the first OBP of L. migratoria could only be identified upon isolation of the protein from antenna and N-terminal sequencing; indeed the three identified locust OBPs show only between 10 - 30% sequence identity to OBPs from insects of the Endopterygota and the Hemipteroid Assemblage [27, 28]. This phylogenetic distance could in fact be the reason why an Orco gene has not been identified from the hemimetabolic Orthoptera. To approach the question to what extend Orco genes may be conserved over long phylogentic periods the orthopteran species Locusta migratoria and Schistocerca gregaria were investigated. Information about Orco of these species may also have some immediate implications for novel strategies to control the locusts.
Materials and Methods
Insect rearing and tissue collection
Locusta migratoria and Schistocerca gregaria insects were obtained from local suppliers. Antennae of adult animals and different stages of L. migratoria were dissected, immediately frozen in liquid N2 and stored at -70°C until RNA isolation. In the same way mouth part, tarsus and wing tissues from adult L. migratoria were prepared.
RNA extraction and cDNA synthesis
Total RNA was isolated from frozen tissues using Trizol reagent (Invitrogen, Germany) following the manufacturers protocols. Poly (A)+ RNA was isolated from 100 μg of total RNA with oligo (dT)25 magnetic dynabeads (Dynal, Germany) according to the suppliers specifications, with final elution in 30 μL of water. Poly (A)+ RNAs were transcribed into cDNAs in a reaction (total volume 20 μL) containing 250 ng mRNA solution from the dynabeads preparation, 4 μL first strand buffer (250 mM Tris pH 8.3, 375 mM KCl, 15 mM MgCl2), 1 μL 10 mM dNTP mix, 1 μl RNaseout, 2 μl DTT (0.1M),1 μL oligo-dT18 primer (500 ng μl-1) and 1 μL Superscript II reverse transcriptase (Invitrogen). The cDNA synthesis was performed for 50 min at 50 °C followed by 15 min at 70 °C.
Cloning of Orco sequences from L. migratoria and S. gregaria
Based on two conserved amino acid stretches (AIKYWV and VCQQCQK) found in all identified Orco primary structures, a pair of degenerated primers (Orco-deg sense: 5' GCNATHAARTAYTGGGT 3'; Orco-deg antisense: 5'TTYTGRCAYTGYTGRCAYAC 3') was designed and used to amplify homologues nucleic acid sequences from antennal cDNA of L. migratoria and S. gregaria, respectively. For identifying L. migratoria Orco sequences, 1 μL from a cDNA synthesis reactions, primed by using the Orco-deg antisense primer, were employed in 50 μl standard PCRs with Titanium Taq polymerase (Clontech, USA) and 100 pmol of each degenerated sense and antisense primer. For amplification of S. gregaria Orco sequences an oligo-dT primed cDNA was used as template.
PCR conditions used were: 1 min 40 s at 95°C, then 19 cycles with 95°C for 30 s, 55°C for 40 s and 68°C for 1 min, with a decrease of the annealing temperature by 0.5°C per cycle. Subsequently, 19 further cycles at the condition of the last cycling step (45°C annealing temperature) were performed followed by incubation for 7 min at 68°C. PCR products were run on 1.2% agarose gels and visualized by ethidium bromide staining. DNA bands of the expected length range were gel purified by Gene clean (MP Biomedicals, Heidelberg, Germany), and subcloned into the pGem-T vector (pGem-T vector system; Promega, Wis., USA). PCR products were analysed on an ABI310 sequencing system using vector and cDNA derived primers and the BIG dye cycle sequencing kit (v3.1; Applied Biosystems, Foster City, Ca, USA).
Rapid amplification of 5'cDNA ends (5'RACE)
To get the 5´end, S. gregaria Orco sequence RACE-PCR was performed using the GeneRacer Kit (Invitrogen) according to the manufacturer's manual. Based on the partial S. gregaria Orco sequence obtained by using degenerate primers, specific primers for 5'RACE-PCR were designed:
Sgre5'race: 5' CTGGCACACGATCTGCACGAAAGT 3';
Sgre5'race nested: 5' GCCCACGAACCTGACGACGTGCTTGT 3'.
Touchdown PCRs were performed as follows: 2 min at 95°C, then 20 cycles with 95°C for 30 s, 65°C for 1 min and 68°C for 1 min, with a decrease of the annealing temperature by 0.5°C per cycle. Subsequently, 19 further cycles at the condition of the last cycling step (55°C) were performed, followed by incubation for 10 min at 68°C. For nested PCR, the initial annealing temperature was changed to 60°C, the other conditions were the same as above. Primer concentrations in all reactions were 2 pmol μl-1. The finally obtained RACE-PCR products were cloned and sequenced as described above. Based on the 5' end of the SgreOrco sequence primers were designed to amplify an N-terminal LmigOrco sequence. LmigOrco-NT sense: 5'GCTGATCCGCATGGTGCAGTA 3', LmigOrco-NT antisense: 5' TCACGGTGGTGGTGAGCAT 3'. PCR condition were used the same as in the initial PCR. After obtaining a further upstream part of the LmigOrco sequence, 5' Race PCR was performed in the same way as with the SgreOrco sequence-specific primers as follows, LmigOrco5'race: 5' GCCTTGGAGCGCACCGCGAAGTAG 3', LmigOrco5'race nested: 5' GCCGCCGCTGTACTCGAGCATCC 3'.
Expression of LmigOrco in different tissues and developmental stages
Template cDNAs were transcribed from total RNA isolated from different tissues and antennae of different nymphal stages (instars 1-5) and adults of L. migratoria using oligo-dT18 primer (500 ng μl-1) as described above. RT-PCR experiments were performed employing gene-specific primers. To amplify L. migratoria Orco sequences the primer pair LmigOrco, antisense: 5' TTGGCACTGCTGACATACGAT 3' and sense: 5'GATCAAATACTGGGTCGAGCG 3' was used. For testing the integrity of the cDNA preparation primers used for the L. migratoria actin gene (accession number AY344445) were applied. LmigAct antisense: 5'CTGTTTGCCTTTGGGTTC3'; LmigAct sense: 5'GACAATGGCTCTGGTATGTG 3'. PCR reactions were performed under the following thermal program: 94°C for 1 min 30 s; 30 cycles of 94°C for 30 s, 50°C for 40 s, 72°C for 1 min 30 s; followed by one cycle at 72°C for 7 min. PCR products were run on 1.2% agarose gels and visualized by ethidium bromide staining.
In situ hybridization
Digoxigenin (DIG)-labeled antisense and sense probes of SgreOrco and LmigOrco were generated from linearized recombinant pGem-T plasmids using the T7/SP6 RNA transcription system following recommended protocols (Roche). Receptor RNA probes were subsequently fragmented to an average length of about 300 bp by incubation in carbonate buffer (80 mM NaHCO3 120 mM Na2CO3, pH 10.2) following the protocol of [29]. Antennae were dissected, embedded in freezing medium Tissue-Tek O.C.T. Compound (Sakura Finetek Europe, Zoeterwoude, The Netherlands) and rapidly frozen at -70°C. Sections (12 μm) were prepared at -24°C (Jung CM300 cryostat), thaw mounted on SuperFrost Plus slides (Menzel-Gläser, Braunschweig, Germany) and air dried for 15 min. After incubation at 4°C in 4% paraformaldehyde in 0.1 M NaCO3, pH 9.5 for 30 min, slides were transferred to 1×PBS (= 0.85% NaCl, 1.4 mM KH2PO4, 8 mM Na2HPO4, pH 7.1) for 1 min, 0.2 M HCl for 10 min and PBS with 1%Triton X-100 for 2 min. Finally, sections were washed two times for 30 s in 1×PBS, rinsed for 10 min in 50% formamide, 5×SSC (1×SSC= 150 mM NaCl, 15 mM Na-citrate, pH 7.0) and drained. For in situ hybridization, 100μl hybridization solution (50% formamide, 2×SSC, 10% dextran sulphate, 0.2 mg ml-1 yeast t-RNA, 0.2 mg ml-1 herring sperm DNA) containing DIG-labeled antisense or sense RNA was placed onto the tissue section. After adding a coverslip, slides were incubated in a humid box (50% formamide) at 55°C overnight. Posthybridization, slides were washed twice for 30 min in 0.1×SSC at 60°C, then treated with 1% blocking reagent (Roche) in TBS (100 mM TRIS, pH 7.5, 150 mM NaCl) with 0.03% Triton X-100 for 30 min at room temperature and incubated for 30 min with an anti-digoxigenin alkaline phosphatase conjugated antibody (Roche) diluted 1:500 in TBS, 0.03% Triton X-100, 1% blocking reagent. After washing three times for 5 min in TBS, 0.05% Tween, slides were rinsed in DAP-buffer (100 mM TRIS, pH 9.5, 100 mM NaCl, 50 mM MgCl2). Subsequently, hybridization signals were visualized using NBT (nitroblue tetrazolium) and BCIP (5-brom-4-chlor-3-indolyl phosphate).
Antennae were analyzed on a Zeiss Axioskope2 microscope (Zeiss, Oberkochen, Germany) equipped with Axiovision software. Pictures were arranged using appropriate graphic programs. Images were not altered except for adjusting the brightness or contrast for uniform tone within a single figure.
Sequence analysis
For transmembrane domain predictions the TMHMM program (Version2.0) was used [30]. The sequence alignment were obtained using ClustAl [31]. The unrooted neighbour joining tree of the amino acid sequences of Orcos from the various insect species were calculated with the MEGA-5 program [32].
Results
Identification of Orco sequences from locust
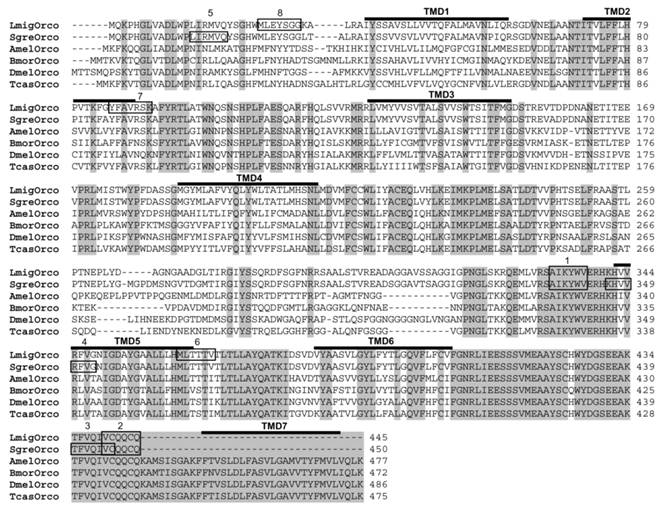
In this study, we set out to identify the olfactory co-receptor (Orco) of the two orthopteran L. migratoria and S. gregaria applying a RT-PCR-based strategy. Therefore, we designed degenerated primers based on sequences motifs, which are highly conserved among Orcos from moths, flies and bees representing the Lepidopteran, Dipteran and Hymenopteran orders, respectively. RT-PCR experiments with cDNA from the antennae of L. migratoria allowed an amplication of DNA fragments of the supposed size. Sequencing the PCR products and comparing the encoded amino acid sequence with Orco sequences from other insects revealed a relatively high sequence identity. Accordingly, the identified receptor sequence was named as L. migratoria Orco (LmigOrco) (Fig. 1).
Alignment of the amino acid sequences of LmigOrco and SgreOrco to selected Orco proteins of insects species from different insect orders. Amino acids identical in all sequences are marked with grey shading. Numbers to the right refer to the position of the last residue in a line in each Orco sequence. The abbreviations indicate (accession numbers in brackets): Amel = Apis melifera (XP_001121145); Dmel = Drosophila melanogaster (AAF52031); Bmor = Bombyx mori (AJ555487); Tcas = Tribolium castaneum (AM689911). Dashes indicate gaps introduced into the sequences for optimal alignment or missing amino acids at the C-terminus of the locust Orcos. The positions of putative transmembrane domains (TMD1 -TMD7), which were predicted based on DmelOrco, are indicated. The amino acid stretches corresponding to the cloning primers are boxed: 1 = Orco-deg sense, 2 = Orco-deg antisense, 3 = Sgre5'race, 4 = Sgre5'race nested, 5 = LmigOrco-NT sense, 6 = LmigOrco-NT antisense, 7 = LmigOrco5'race, 8 = LmigOrco5'race nested.
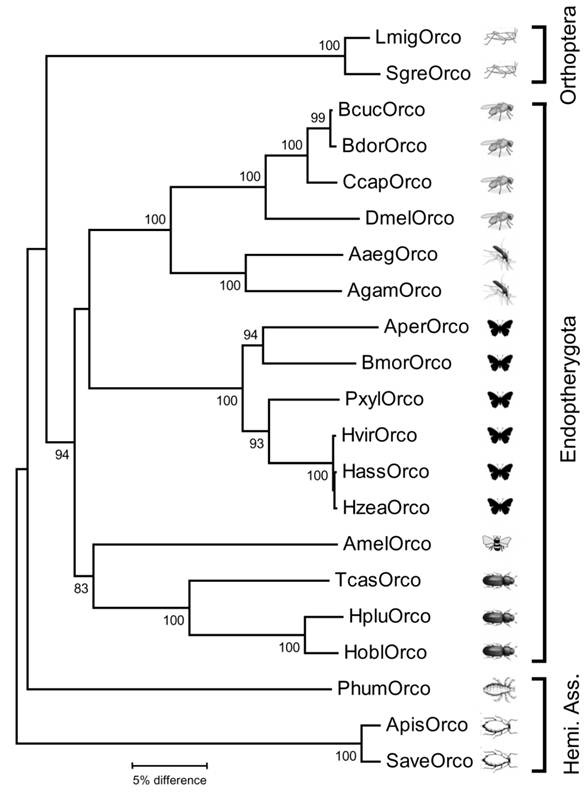
Neighbor joining tree of Orco sequences from insects belonging to different insect orders. The tree was constructed with MEGA5 based on a ClustAl alignment of the amino acid sequences shown in Fig. 1 and selected Orco sequences from other insects deposited in Genbank. Bootstrap support values are based on 1000 replicates; only support values above 80% are shown. Branch lengths are proportional. Abbreviations as in figure 1 and (accession numbers in brackets): Apis = Acyrthosiphon pisum (XP_001951646); Aaeg = Aedes aegypti (AAT01220); Agam = Anopheles gambiae (AY363725); Aper = Anthereae pernyi (AJ555486); Bcuc = Bactrocera curcurbitae (ADK97803); Bdor = Bactrocera dorsalis (ACC86853); Ccap = Ceratitis capitata (AAX14775); Hobl = Holotricha oblita (AEE69033); Hplu = Holotricha plumbea (ADM35103); Hvir = Heliothis virescens (AJ487477); Hzea = Helicoverpa zea (AAX14773); Hass = Helicoverpa assulta (ABU45983); Phum = Pediculus humanus (EEB12924); Pxyl = Plutella xylostella (BAG71421); Save = Sitobion avenae (ACT37280). Hemi. Ass. = Hemipteroid Assemplage.
Identification of antennal cells expressing LmigOrco and SgregOrco
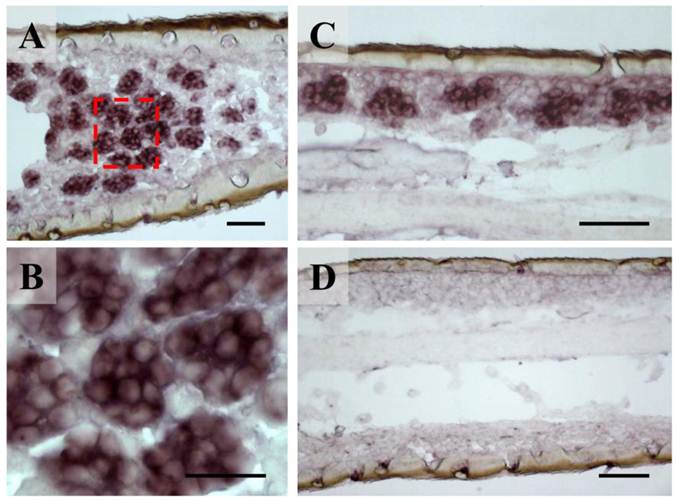
The unbranched but segmented locust antennae carry four morphologically distinguishable sensory sensilla types distributed over the complete flagellum in both sexes [5, 6]. The largest fractions are represented by the short olfactory sensilla basiconica, which house 20-50 OSNs and the sensilla coeloconica (pitted pegs) with 1-4 sensory cells, responding to chemo-, hygro- and thermostimulation [3, 5]. About five times lower in numbers are the long and slender olfactory sensilla trichodea with 1-3 OSNs and the sturdy sensilla chaetica usually housing up to 5 sensory neurons supposed to be involved in gustation or mechanosensation [5, 6]. To assess if the newly identified orthopteran Orco proteins are in fact expressed in antennal sensory cells, in situ hybridization experiments were performed. Horizontal sections through the antennae were incubated with DIG-labeled antisense RNA probes and labeled cells were subsequently visualized by employing color substrates. It was found that on sections from antennae of L. migratoria the Orco-specific riboprobe typically labeled cell clusters (Fig. 3A), which are arranged below the antennal surface (Fig. 3C). At higher magnification (Fig. 3B) the LmigOrco hybridization signals could be assigned to individual cells. In control experiments with a DIG-labeled Orco-specific sense RNA probe no labeling of cells was obtained (Fig. 3D).
Expression of LmigOrco in the antenna of the migratory locust L. migratoria. In situ hybridizations were performed on horizontal tissue sections of male antennae with DIG-labeled antisense (A - C) or sense (D) RNA probes for LmigOrco. Signals were visualized using an anti-DIG antibody and color substrates. A, Hybridization signals in a segment of the filamentous locust antenna. B, Higher magnification of the area boxed in A showing a cluster of labeled cells. C, Hybridization signals can be assigned to clusters of cells bordering the antennal surface. D, No hybridization signals were obtained in ISH experiments with an Orco-specific sense RNA probe. Scale bars: 20 μm in A and B, 50 μm in C and D.
LmigOrco gene expression in cells of sensilla basiconica and sensilla trichodea. In situ hybridizations were performed using a DIG-labeled Orco-specific antisense RNA probes on sections of male antenna. A, shows a cluster of LmigOrco-positive cells under a short sensillum basiconicum. B, Three LmigOrco-expressing cells can be assigned to a longer sensillum trichodeum. C and D, No labeling of cells was detected under sensilla chaetica (C) or coeloconica (D). Scale bars: 10 μm.
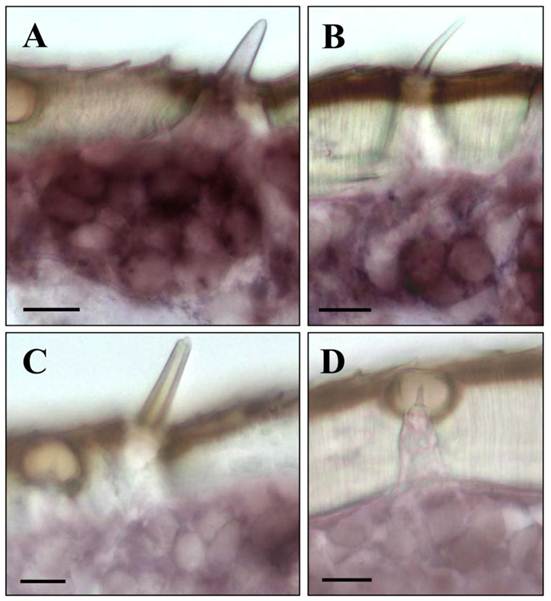
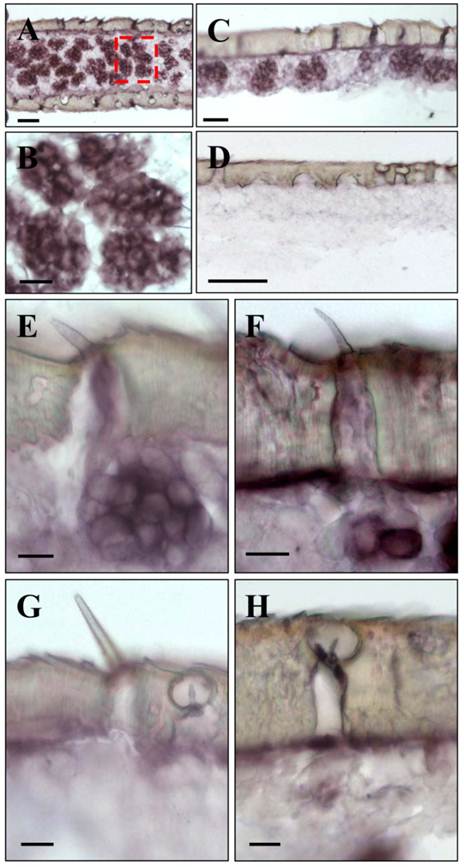
Upon a detailed inspection of sections at high magnification, the clusters of labeled cells could clearly be assigned to distinct sensory hairs which based on morphological criteria are identified as sensilla basiconica (Fig. 4A). In addition, a few positive cells were detected under sensory hairs identified as sensilla trichodea (Fig. 4B). In contrast, no specific labeling of cells was observed under sensilla chaetica (Fig. 4C) and sensilla coeloconica (Fig. 4D). The same results as depicted in Fig. 3 and Fig. 4 for male antenna were obtained in ISH-experiments with female antennal sections (not shown). This is in line with similar numbers and sensilla hair types on the male and female antenna [6, 35, 36]. Together these results indicate that LmigOrco is expressed in high numbers of cells located in basiconic sensilla and few cells housed in trichoid sensilla, but not in cells of sensilla coeloconica and sensilla chaetica. To compare the antennal topography of the Orco-expressing cells in L. migratoria and S. gregaria, in situ hybridisation experiments were performed with antennal sections of the desert locust Schistocerca using a DIG-labeled SgregOrco antisense probe. The results depicted in Fig. 5 indicate a labeling pattern quite similar to L. migratoria. On S. gregaria antennal sections clusters of labeled cells were visualized (Fig. 5A-C) and could be assigned to sensilla basiconica (Fig. 5E). In addition, some SgregOrco-positive cells were found in sensilla trichodea (Fig. 5F) whereas sensilla chaetica (Fig. 5G) sensilla coeloconica (Figs. 5G and 5H) and as well as sections probed with a DIG-labeled sense RNA probe of SgreOrco (Fig. 5D) were devoid of any labeled cell.
SgreOrco gene expression in the antenna of the desert locust S. gregaria. In situ hybridization on sections of the antennae with DIG-labeled antisense (A - C and E - H) or sense (D) RNA probes for SgreOrco. A, Hybridization on a horizontal section of an antennal segment. B, Higher magnification of the area boxed in A showing several clusters of labeled cells. C, Clusters of labeled cells bordering the antennal surface. D, No hybridization signals were obtained with a sense RNA probe. E, LmigOrco-positive cells under a short sensillum basiconicum. F, Two SgreOrco-expressing cells can be assigned to a sensillum trichodeum. G and H, No labeling of cells was detected under a sensillum chaeticum (G) or sensilla coeloconica (H and G). A - E and G = female antenna; F and H, male antenna. Scale bars: 50 μm in A and D, 20 μm in B and C, 10 μm in E - H.
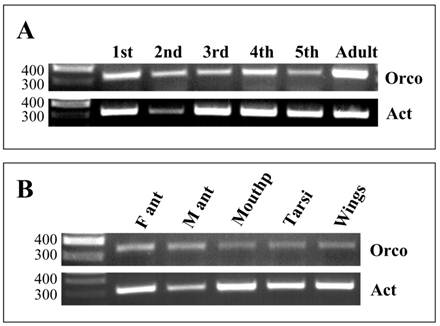
Expression of LmigOrco in different tissues and developmental stages of L. migartoria. RT-PCRs were performed using specific primer pairs and cDNAs prepared from the tissues indicated. A, RT-PCRs with cDNA prepared from the antenna of different nymphal stages (1st to 5th instar) and adults. B, Tissue specificity of LmigOrco expression. M ant, male antenna; F ant, female antenna; Mouthp, mouthparts. Amplification products were analysed on agarose gels and visualized by UV illumination after ethidium bromide staining. Based on the primer design the expected size of the PCR-products is 342 bp for LmigOrco and 314 bp for the actin control gene. The position of molecular weight markers (in bp) is indicated.
Expression of LmigOrco in developmental stages
To explore the onset and time course of Orco expression during development, different stages were analysed performing RT-PCR experiments with Orco-specific primers and cDNAs prepared from antennae of nymphal stages (1st to 5th instars) and adult animals (Fig. 6). Control PCR with primers for actin confirmed the integrity of the different cDNA preparations and for the 2nd instar template a band of slightly lower intensity. PCR experiments with Orco-specific primers provided bands of similar intensities with templates from all stages, from the 1st to 5th instar; however, the bands were considerably weaker than the band obtained with cDNA from antenna of adults (Fig. 6A).
Tissue distribution of Orco expression in locust
To compare the level of Orco expression in the two sexes and to assess if Orco may be expressed in other parts of the body, which have been shown to carry chemosensory sensilla [37-39], RT-PCR experiments were performed. cDNAs from male and female antennae as well as from mouthparts, tarsi and wings were analysed. The integrity of the cDNA templates was verified by primers for actin. Amplification products of the correct size and about the same band intensity were amplified in all cDNA preparations (Fig. 6B); the slightly weaker band for the male antennae possibly reflects a lower template concentration. With the primer pair specific for LmigOrco, PCR bands were obtained with antennal cDNA matching the size predicted from the cloned cDNA (Fig.6B). Comparing the intensity of the bands, no apparent differences in the band intensity was found between sexes. Also cDNA preparations from mouthparts, tarsi and wings led to PCR products of the correct size; these results indicate that transcripts of LmigOrco were also present in non-antennal tissues.
Discussion
In this study we identified genes encoding Orco proteins of the two locust species L. migratoria and S. gregaria. Hence, LmigOrco and SgreOrco are the first Orco genes identified for the insect order Orthoptera. Orthologs of the Orco gene have previously been reported in a variety of insect species representing different orders of the Endopterygota, including Diptera (flies and mosquitoes), Lepidoptera (moth), Hymenoptera (bees) and Coleoptera (beetles) [21, 22, 40], as well as in the Hemipteroid Assemblage, including Hemiptera (aphids) and Phthiraptera (lice) [23, 24], all of which are separated by millions of years on the evolutionary scale. Thus, it seems that the Orco gene is under a very high selective pressure to maintain the primary sequence of the encoded protein [22]. This view is supported by sequence alignments, which reveal a pronounced conservation of Orco; sequence identity is particularly apparent in the C-terminal end. In this context, it is interesting to note that some degree of sequence conservation is also apparent in the C-terminal end of the highly divergent sequences of insect odorant receptors [41-44]. This may indicate a crucial relevance of the C-terminal domain for both, the Orco and the OR proteins. The Orco protein is supposed to operate as a cation channel which is heteromerized with an OR protein [19, 33] and opens upon conformational changes of the odorant-activated OR [18]. In this functional interaction of the two proteins, the C-terminal region may play an important role.
The in situ hybridization experiments have shown that in both locust species, Orco is expressed in clusters of cells, which can be assigned to sensilla basiconica (Figs. 3 - 5). This type of chemosensory sensillum contains numerous neurons (20 - 50), however, it is presently unknown how many of these cells are olfactory sensory neurons. Since the olfactory co-receptor Orco is considered as a marker for olfactory cells, our finding indicated that many if not all of the sensory neurons found in sensilla basiconica are olfactory neurons. The finding that only a few hybridization signals can be allocated to sensilla trichodea (Fig. 5) is in line with the 1 - 3 sensory neurons which are housed in trichoid sensilla [3-5].
Orco-positive cells were not found in sensilla chaetica (Fig. 4). This result is in accordance with the view, that the sensory neurons of sensilla chaetica serve a mechanosensory or gustatory function [5]. Orco-positive cells were also lacking in coeloconic sensilla (Fig. 5) but interestingly this type of sensilla has been shown to respond to odorants [3]. Since Orco is only co-expressed with so-called classical ORs this apparent discrepancy could point to the possibility that a different type of odorant receptor, e.g. the so-called ionotropic receptors (IRs), is expressed in neurons of sensilla coeloconica. IR-receptors are structurally related to ionotropic glutamate receptors and several subtypes have been identified as candidate olfactory receptors in fruit flies and some other insects [45-47]. In fact, a recent study has provided evidence that in sensilla coeloconica of Drosophila sensory neurons do express IR-receptors [45].
Based on the semi-quantitative RT-PCR experiments similar levels of LmigOrco- transcripts were present in antennal cDNA preparations from male and female animals. This finding is in line with the fact, that there are no obvious differences in sensilla types and numbers between the two sexes [5, 36]. The evidence for Orco-transcripts also in cDNA preparations from mouthparts, tarsi and wings indicates that Orco is ectopically expressed in cells of non-antennal structures, which do comprise chemosensors. The physiological function of chemosensory sensilla on these body parts of L. migratoria is unknown. In fact, on the labial palps some sensilla basiconica have been found, which typically serve an olfactory function, but the majority of chemosensilla on this organ represent sensilla chaetica, which are supposed to have a gustatory function [37]. On the L. migratoria wings and tarsi only sensilla chaetica have been described, which are probably involved in contact chemoreception [38, 39]. Whether our findings may hint to the presence of some odor-responsive cells in these sensilla types or to a possible role of Orco in non-olfactory chemoreceptive cells is unclear, but is in line with recent reports which describe the presence of Orco-transcripts in the proboscis of the mosquito Anopheles gambiae and the moth Heliothis virescens [40, 41].
In a perspective view, due to the broad expression in the olfactory system of locusts, Orco may be considered as a potential target for a disruption of olfactory signaling. An efficient interference with the sense of smell would severely affect odorant- and pheromone-driven behaviors, which are critical for aggregation, feeding and reproduction of locust. In this regard, this study could be a starting point for research, which eventually may lead to novel olfactory-based approaches to control the agriculture pest capacity of locusts.
Data deposition
The Orco sequences reported in this paper have been deposited in GenBank under accession numbers: JN989549 (LmigOrco) and JN989550 (SgreOrco).
Acknowledgements
We are grateful to Nelli Dick for excellent technical assistance.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft and the NSFC (Natural Science Foundation of China) Project No. 30730012.
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Lomer CJ, Bateman RP, Johnson DL, Langewald J, Thomas M. Biological control of locusts and grasshoppers. Annu Rev Entomol. 2001;46:667-702
2. Hassanali A, Njagi PG, Bashir MO. Chemical ecology of locusts and related acridids. Annu Rev Entomol. 2005;50:223-45
3. Ochieng SA, Hansson BS. Responses of olfactory receptor neurones to behaviourally important odours in gregarious and solitarious desert locust, Schistocerca gregaria. Physiol Entomol. 1999;24:28-36
4. Cui X, Wu C, Zhang L. Electrophysiological response patterns of 16 olfactory neurons from the trichoid sensilla to odorant from fecal volatiles in the locust, locusta migratoria manilensis. Arch Insect Biochem Physiol. 2011;77:45-57
5. Ochieng SA, Hallberg E, Hansson BS. Fine structure and distribution of antennal sensilla of the desert locust, Schistocerca gregaria (Orthoptera: Acrididae). Cell Tissue Res. 1998;291:525-36
6. Jin X, Zhang SG, Zhang L. Ultrastructure of four types of antennal sensilla in Locusta migratoria manilensis (Insects, Orthoptera). J Agric Biotech. 2004;12:300-5
7. Pelosi P, Zhou JJ, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cell Mol Life Sci. 2006;63:1658-76
8. Leal WS. Proteins that make sense. In: (ed.) Blomquist G, Vogt R. Insect pheromone biochemistry and molecular biology; The biosynthesis and detection of pheromones and plant volatiles. London: Elsevier Academic Press. 2003 Ch15
9. Vogt RG. Biochemical diversity of odor detection: OBPs, ODEs and SNMPs. In: (ed.) Blomquist G, Vogt R. Insect pheromone biochemistry and molecular biology. The biosynthesis and detection of pheromones and plant volatiles. London: Elsevier Academic Press. 2003 Ch14
10. Zhou JJ. Odorant-binding proteins in insects. Vitam Horm. 2010;83:241-72
11. Gohl T, Krieger J. Immunolocalization of a candidate pheromone receptor in the antenna of the male moth, Heliothis virescens. Invert Neurosci. 2006;6:13-21
12. Dobritsa AA, van der Goes van Naters, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827-41
13. Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965-79
14. Wang G, Carey AF, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2010;107:4418-23
15. Carey AF, Wang G, Su CY, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464:66-71
16. Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703-14
17. Vosshall LB, Hansson BS. A unified nomenclature system for the insect olfactory coreceptor. Chem Senses. 2011;36:497-8
18. Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002-6
19. Neuhaus EM, Gisselmann G, Zhang WY, Dooley R, Stortkuhl K, Hatt H. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nature Neurosci. 2005;8:15-7
20. Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007-11
21. Krieger J, Klink O, Mohl C, Raming K, Breer H. A candidate olfactory receptor subtype highly conserved across different insect orders. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2003;189:519-26
22. Jones WD, Nguyen TAT, Kloss B, Lee KJ, Vosshall LB. Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Current Biology. 2005;15:R119-R121
23. Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, Clark JM, Lee SH, Robertson HM, Kennedy RC, Elhaik E, Gerlach D, Kriventseva EV, Elsik CG, Graur D, Hill CA, Veenstra JA, Walenz B, Tubio JM, Ribeiro JM, Rozas J, Johnston JS, Reese JT, Popadic A, Tojo M, Raoult D, Reed DL, Tomoyasu Y, Kraus E, Mittapalli O, Margam VM, Li HM, Meyer JM, Johnson RM, Romero-Severson J, Vanzee JP, Alvarez-Ponce D, Vieira FG, Aguade M, Guirao-Rico S, Anzola JM, Yoon KS, Strycharz JP, Unger MF, Christley S, Lobo NF, Seufferheld MJ, Wang N, Dasch GA, Struchiner CJ, Madey G, Hannick LI, Bidwell S, Joardar V, Caler E, Shao R, Barker SC, Cameron S, Bruggner RV, Regier A, Johnson J, Viswanathan L, Utterback TR, Sutton GG, Lawson D, Waterhouse RM, Venter JC, Strausberg RL, Berenbaum MR, Collins FH, Zdobnov EM, Pittendrigh BR. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci U S A. 2010;107:12168-73
24. Smadja C, Shi P, Butlin RK, Robertson HM. Large gene family expansions and adaptive evolution for odorant and gustatory receptors in the pea aphid, Acyrthosiphon pisum. Mol Biol Evol. 2009;26:2073-86
25. Kristensen NP. Phylogeny of extant hexapods. In: CSRIO (ed.) The insects of Australia: A Textbook for Students and Research Workers. Melbourne: Melbourne University Press. 1991
26. The Tree of Life Web Project. Maddison DR, Schulz K-S. http://www.tolweb.org
27. Ban L, Scaloni A, D'Ambrosio C, Zhang L, Yahn Y, Pelosi P. Biochemical characterization and bacterial expression of an odorant-binding protein from Locusta migratoria. Cell Mol Life Sci. 2003;60:390-400
28. Yu F, Zhang S, Zhang L, Pelosi P. Intriguing similarities between two novel odorant-binding proteins of locusts. Biochem Biophys Res Commun. 2009;385:369-74
29. Angerer LM, Angerer RC. In situ hybridization to cellular RNA with radiolabelled RNA probes. In: (ed.) Wilkinson DG. In situ hybridization. Oxford: IRL Press. 1992 Ch2
30. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567-80
31. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673-80
32. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731-9
33. Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20
34. Lundin C, Kall L, Kreher SA, Kapp K, Sonnhammer EL, Carlson JR, Heijne G, Nilsson I. Membrane topology of the Drosophila OR83b odorant receptor. FEBS Lett. 2007;581:5601-4
35. Gao Q, Zhang S, Zhang L. Comparison of number and distribution of sensillum on different developmental antenna ol Locusta mogratoria manilensis. J Bejing University of Agriculture. 2007;22:29-33
36. Greenwood M, Chapman RF. Differences in numbers of sensilla on the antennae of solitarious abd gregarious Locusta migratoria L. (Orthoptera: Acrididae). Int J Morphol and Embryol. 1984;13:295-301
37. Jin X, Zhang SG, Zhang L. Expression of odorant-binding and chemosensory proteins and spatial map of chemosensilla on labial palps of Locusta migratoria (Orthoptera: Acrididae). Arthropod Struct Dev. 2006;35:47-56
38. Zhou SH, Zhang SG, Zhang L. The chemosensilla on tarsi of Locusta migratoria (Orthoptera: Acrididae): distribution, ultrastructure, expression of chemosensory proteins. J Morphol. 2009;270:1356-63
39. Zhou S-H, Zhang S-G, Zhang L. Expression of chemosensory proteins in hairs on wings of Locusta migratoria (Orthoptera: Acrididae). J Appl Entomol. 2008;132:439-50
40. Pitts RJ, Fox AN, Zwiebel LJ. A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector Anopheles gambiae. Proc Natl Acad Sci U S A. 2004;101:5058-63
41. Krieger J, Raming K, Dewer YME, Bette S, Conzelmann S, Breer H. A divergent gene family encoding candidate olfactory receptors of the moth Heliothis virescens. Eur J Neurosci. 2002;16:619-28
42. Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327-38
43. Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725-36
44. Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147-59
45. Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149-62
46. Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44-60
47. Olivier V, Monsempes C, Francois MC, Poivet E, Jacquin-Joly E. Candidate chemosensory ionotropic receptors in a Lepidoptera. Insect Mol Biol. 2011;20:189-99
Author contact
![]() Corresponding author: Prof. Dr. H. Breer, University of Hohenheim, Institute of Physiology, Garbenstrasse 30, 70599 Stuttgart, Germany. e-mail: breerde or Prof. Long Zhang, Ph. D., China Agricultural University, Key Lab for Biological Control, Ministry of Agriculture, Beijing 100193, China, e-mail: locustedu.cn.
Corresponding author: Prof. Dr. H. Breer, University of Hohenheim, Institute of Physiology, Garbenstrasse 30, 70599 Stuttgart, Germany. e-mail: breerde or Prof. Long Zhang, Ph. D., China Agricultural University, Key Lab for Biological Control, Ministry of Agriculture, Beijing 100193, China, e-mail: locustedu.cn.

 Global reach, higher impact
Global reach, higher impact