Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2012; 8(2):258-264. doi:10.7150/ijbs.3278 This issue Cite
Mini-review
An Updated Nomenclature for Keratin-Associated Proteins (KAPs)
1. Gene-Marker Laboratory, Faculty of Agriculture and Life Sciences, PO Box 84, Lincoln University, Lincoln 7647, New Zealand
2. Ruakura Research Centre, AgResearch Limited, Private Bag 3115, Hamilton 3240, New Zealand
3. Lincoln Research Centre, AgResearch Limited, Private Bag 8742, Lincoln 8140, New Zealand
4. HUGO Gene Nomenclature Committee (HGNC), EMBL-EBI, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1SD, UK
5. National Bureau of Animal Genetic Resources, PO 129, Karnal 132001, Haryana, India
6. Molecular Biology Laboratory, SARDI Livestock and Farming Systems, Roseworthy Campus, Roseworthy, SA 5371, Australia
7. College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China
8. College of Animal Science, Inner Mongolia Agricultural University, Hohhot 010018, China
Received 2011-7-18; Accepted 2011-12-21; Published 2012-1-12
Abstract
Most protein in hair and wool is of two broad types: keratin intermediate filament-forming proteins (commonly known as keratins) and keratin-associated proteins (KAPs). Keratin nomenclature was reviewed in 2006, but the KAP nomenclature has not been revised since 1993. Recently there has been an increase in the number of KAP genes (KRTAPs) identified in humans and other species, and increasingly reports of variation in these genes. We therefore propose that an updated naming system is needed to accommodate the complexity of the KAPs. It is proposed that the system is founded in the previous nomenclature, but with the abbreviation sp-KAPm-nL*x for KAP proteins and sp-KRTAPm-n(p/L)*x for KAP genes. In this system “sp” is a unique letter-based code for different species as described by the protein knowledge-based UniProt. “m” is a number identifying the gene or protein family, “n” is a constituent member of that family, “p” signifies a pseudogene if present, “L” if present signifies “like” and refers to a temporary “place-holder” until the family is confirmed and “x” signifies a genetic variant or allele. We support the use of non-italicised text for the proteins and italicised text for the genes.
This nomenclature is not that different to the existing system, but it includes species information and also describes genetic variation if identified, and hence is more informative. For example, GenBank sequence JN091630 would historically have been named KRTAP7-1 for the gene and KAP7-1 for the protein, but with the proposed nomenclature would be SHEEP-KRTAP7-1*A and SHEEP-KAP7-1*A for the gene and protein respectively. This nomenclature will facilitate more efficient storage and retrieval of data and define a common language for the KAP proteins and genes from all mammalian species.
Keywords: Keratin-associated protein (KAP) gene (KRTAP), nomenclature, genetic variation, diversity, species.
Introduction
Hair and wool fibres typically consist of three major structural components: the cuticle, the cortex and the central medulla. Approximately 90% of the cortical cells contain longitudinally arrayed keratin intermediate filaments (IFs), consisting of keratin (K) proteins. These filaments have a matrix surrounding them, that contains the keratin-associated proteins (KAPs), which cross-link with the IFs through extensive disulfide bonding [1]. The role of the KAPs in IF assembly into arrays is considered to be crucial and therefore they most likely affect wool and hair attributes such as strength, inertness and rigidity [2].
The KAPs were originally best understood in sheep, reflecting the then economic importance of wool and the preponderance of wool protein biochemistry undertaken from the mid-twentieth century. Recently our understanding of the KAPs has advanced significantly, with the advent of the large-scale whole-genome sequencing of human KAP genes. Moreover, extensive genetic variation is now being described in the KAP genes (KRTAPs) from sheep [3-6] and humans [7,8]. However sequence homology comparison between the KRTAPs from different species adds a new complexity to KAP naming, as obvious homologues are at times difficult to find. Whilst there may be genetic identity, there can also be differences in the nucleotide sequence and the chromosomal arrangement of the genes.
The nomenclature system for KAPs has not been reviewed since the proposition and adoption of the system of Rogers and Powell. This was first proposed in 1993 [9] and explained in detail in 1997 [10]. While this system has served us well until now; the differences now being realised between species, the extensive genetic variation now being documented in some KRTAPs and some misuse or misinterpretation of the nomenclature, suggests it is time for a revision.
Past and current nomenclatures for keratin-associated proteins (KAPs)
The earliest attempts to classify keratins had their origins in the methods used to separate wool proteins. In 1934 these proteins were divided into two extractable classes: those with a lower sulphur content than whole wool and those with a higher sulphur content, otherwise known as SCMK-A and SCMK-B respectively [11]. This division was based on the fractional “salting-out” of s-carboxymethylated proteins. Subsequently the former group became the intermediate filament proteins, while the latter group became the KAPs. The advent of amino acid analysis enabled a further sub-division of this high sulphur class (SCMK-B) into high (HS) and ultra-high sulphur (UHS) proteins, and this based on whether their cysteine content was above or below 30 mol% [12,13]. Amino acid analysis also had further impact on our knowledge of the KAPs with the discovery of a third class of proteins in wool, one that proved to be rich in glycine and tyrosine, so-called high glycine-tyrosine proteins (HGT) [14,15].
Subsequent attempts to fractionate the HS group of proteins and identify sub-components led to further improvement in our understanding of this class of proteins and also a proliferation of new protein names. The use of fractional precipitation with ammonium sulphate solutions resulted in the definition of two fractions SCMK-B1 and SCMK-B2 [16], with subsequent sub-fractionation of the B2 group into a further three components (their names now shortened to B2A, B2B and B2C) by chromatography [17]. In parallel with these studies, column electrophoresis was used to fraction the HS components, one component, SCMK-BIII, being split by gel filtration into two new HS protein families: BIIIA and BIIIB [18,19].
The HGT were sub-divided into the Type I and II sub-classes by ion-exchange chromatography [20], the former being of moderate percentage of glycine and tyrosine, and comprising two components C2 and F [21]. In contrast, the Type II family proteins, which contain a higher percentage of these two amino acids, were thought to contain up to 10 individual components [10,22], although only one has been fully sequenced to date [23]. Finally, although their existence had been known about for some time, members of proteins from the UHS group were identified, the first cuticle UHS proteins being sequenced in 1990 and 1994 [24,25] and cortical UHSs in 1994 and 1995 [26,27].
The increasing diversity of the KAPs, coupled with their non-uniform naming, led Rogers and Powell [10] to suggest a nomenclature for the KAP proteins and genes using the abbreviation KAPm.nxpL for the protein and KRTAPm.nxpL for the gene. In subsequent iterations of this system the gene name became italicised, although use of this convention is only sporadic in the literature. In the Rogers and Powell [10] system, “m” denotes a family or unique protein, “n” denotes a component number, “x” denotes a variant, “p” denotes a pseudogene and “L” stands for “like”. This nomenclature divides the KAPs of all species into families and further into family members based on similarities in their amino acid sequences. Historically then, what was originally called SCMK-B became SCMK-B2, then HS-B2A and then KAP1.1 for the protein and KRTAP1.1 for the gene. Somewhat strangely through this time, the HS, UHS and HGT classification system persisted, perhaps reflecting that the abbreviations gave some indication of the type of protein being described, although in the last few years the discovery of KAPs that contain moderate amounts of cysteine and glycine has made this older classification system even more inadequate.
Problems encountered with the current nomenclature
A variety of things have arisen since 1997, which leads us to propose that the KAP/KRTAP nomenclature is revised and adjusted. Firstly the 2006 release of a consensus nomenclature for the mammalian keratins [28] identified the need to adhere to guidelines proposed by the Human Genome Nomenclature Committee (HGNC), whose prominence in the area of nomenclature grew following the sequencing of the human genome. Consistency with the recommendations of this organisation seems sensible, especially in the context of what is known about KAPs/KRTAPs. While the Rogers and Powell [10] system was useful, the protein nomenclature included a term “p” for a pseudogene, a term that could only really describe a non-expressed or faulty form of a gene and not a protein. The gene nomenclature was also somewhat confusing as “p” and “L” probably should not be present together. There is also some confusion over the use of punctuation in gene names, with both full stops and hypens being used between the m (family) and n (constituent) in the nomenclature and with seemingly little consistency or pattern.
HGNC suggests punctuation should be avoided, the exception being its use in defining groups of related genes and in this respect, in our proposed nomenclature we attempt to define a system that should lead to greater consistency in naming.
Two other and more substantive issues have also emerged, the increasing diversity of the genes from different species, and the genetic variation there-in. We feel that these matters need to be better accommodated in the nomenclature.
To date, more than 100 KAP genes have been isolated from a range of mammalian species including sheep, humans, mice and rabbits. These genes have been placed in 27 families, each comprising 1-12 members [29-32]. In the human genome, the 89 functional KRTAPs identified have been placed into 25 families (Table 1), although Wu et al [33] suggest up to 122 functional (n=101) and pseudo (n=21) genes based on analysis of sequences lodged in databases. These genes are clustered into five domains on three different human chromosomes (Table 2). In an analysis of eight species, Wu et al [33] have suggested that humans have a similar number of genes to other primates, but that rodents have an expanded repertoire.
While there appears to be conservation at the sequence level across species [33], it is still not clear as to which of these genes are expressed and where and when this expression occurs. For example, except for families 16, 22, 25 and 27, all of the KRTAPs are expressed in the human hair follicle [29, 30].
In sheep, only 13 functional KRTAPs from 7 families have been reported to date and homologues for the other human genes have not been identified yet (Table 1). This is probably, in part, a result of the limited amount of research undertaken on sheep KRTAPs in the last decade.
KAP genes identified to date in human and sheep
| Family | Category1 | Known family members2,3,4 | |
|---|---|---|---|
| Human | Sheep | ||
| KAP1 | HS | 4 (2)a,b | 3 (3)i-l,5 |
| KAP2 | HS | 5+1a | 06 |
| KAP3 | HS | 3+1a | 2+1 (1)m,n |
| KAP4 | UHS | 11+1 (10)a,c | 2o |
| KAP5 | UHS | 12+2d | 3+1 (1)p,q |
| KAP6 | HGT | 3e | 1 (1)r,s |
| KAP7 | HGT | 1e | 1 (1)n,t,u |
| KAP8 | HGT | 1+2e | 1 (1)t,u,v |
| KAP9 | UHS | 7+1a | |
| KAP10 | HS | 12+1f,g | |
| KAP11 | HS | 1f | |
| KAP12 | HS | 4+1f,g | |
| KAP13 | HS | 4+2f | |
| KAP15 | HS | 1f | |
| KAP16 | HS | 1a | |
| KAP17 | UHS | 1a | |
| KAP19 | HGT | 7+4f | |
| KAP20 | HGT | 2f | |
| KAP21 | HGT | 2+1f | |
| KAP22 | HGT | 1f | |
| KAP23 | HS | 1h | |
| KAP24 | HS | 1h | |
| KAP25 | HS | 1h | |
| KAP26 | HS | 1h | |
| KAP27 | HS | 1h | |
| Total | 88+17 (12) | 13+2 (9) | |
1 HS: ≤30 mol % cysteine; UHS: >30 mol % cysteine; HGT: 35-60 mol % of glycine and tyrosine.
2 Membership is based on the identification of the gene, and not the protein.
3 Numbers after “+” represent pseudogenes, and numbers of family members having genetic variation reported are shown in the brackets.
4 Superscript letters indicate the references from which the genetic information was derived: a[35], b[7], c[29], d[40], e[41], f[42], g[43], h[30], i[44], j[3], k[36], l[4], m[45], n[37], o[26], p[25], q[5], r[23], s[6], t[46], u[38] and v[39].
5 While there are currently only three genes reported in the literature for KAP1, there are four known protein sequences.
6 No ovine genes have been identified, despite three protein sequences having been reported [10,47].
Chromosome locations of KAP gene families in humans and sheep
| Species | Chromosome location | Gene families | References | ||
|---|---|---|---|---|---|
| Human | 11p15.5 | KAP5 | [40] | ||
| 11q13.4 | KAP5 | [40] | |||
| 17q21.2 | KAP1, 2, 3, 4, 9, 16, 17 | [35] | |||
| 21q22.1 | KAP6, 7, 8, 13, 15, 19, 20, 21, 22, 24, 25, 26, 27 | [30, 41] | |||
| 21q22.3 | KAP10, 12 | [42] | |||
| Sheep | 1 | KAP6, 7, 8 | Web reference1 | ||
| 11 | KAP1, 3 | Web reference1 | |||
| 21 | KAP5 | Web reference1 | |||
The known sheep genes are clustered into three domains on three chromosomes (Table 2). Given the similarities in the chemical make-up and structure of wool and human hair, and the similarity of the individual genes or clustered families (Table 2), it is expected that more individual KRTAPs and families will be found in the sheep genome. Equally it would not be unreasonable to expect that the overall number of putative KRTAPs might be higher than previously thought. While the discovery of more and more KAP genes is not in itself any reason to change the nomenclature system, it does highlight the need to revise its suitability, especially in light of the issues described below.
Impact of species of origin on KAP/KRTAP classification
We believe that the species of origin of any KAP protein or gene sequence needs to be stated in any classification system. Homology comparisons between species reveals that KAP proteins of the same name (and therefore one would assume family), may actually have low inter-species homology, but never-the-less tend to cluster with other family members from the same species. For example, sheep KAP1-3 does not share a high homology with KAP1-3 from either humans or mice, but is more similar to other sheep KAP1 family members (Fig 1). All the sheep KAP1 family members tend to cluster, and all the human KAP1 family members tend to cluster, on sequence-based comparison (Fig. 1), and a similar phenomenon can also be seen for the KAP3, KAP4 and KAP5 families (data not shown). Consequently it can be difficult to assign a new sequence to a family, or constituent group within that family, especially if that sequence is one of the first obtained from a species.
Phylogenetic relationship of KAP1 family members from different species. The phylogenetic tree is constructed using DNAMAN (version 5.2.10, Lynnon BioSoft, Vaudreuil, Canada) and is based on the predicted amino acid sequences of the following known KAP1-n genes (with the GenBank accession numbers in bracket): HUMAN-KRTAP1-1 ((NM_030967), HUMAN-KRTAP1-3 (NM_030966), HUMAN-KRTAP1-4 (AC007455), HUMAN-KRTAP1-5 (NM_031957), SHEEP-KRTAP1-1 (X01610), SHEEP-KRTAP1-3 (X02925), SHEEP-KRTAP1-4 (X01610), MOUSE-KRTAP1-3 (NM_001085526), MOUSE-KRTAP1-4 (BC150831), MOUSE-KRTAP1-5 (BC147095) and RAT-KRTAP1-6 (NM_001025135). The tree was constructed using 1000 bootstrap duplications. The numbers at the forks indicate the bootstrap confidence values and only those equal to or higher than 50% are shown. Branch lengths are proportional to genetic distance.
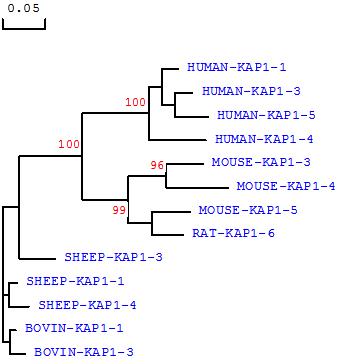
The assignment of KAP/KRTAP family membership is already in some instances not supported by homology (Fig. 1), and some assignments seem strange. For example, Liu et al [34] recently reported the presence of a Capra hircus KAP16-6 gene, although it has very low sequence homology with any human KAP genes, including the published human KAP16-1 gene sequence (GenBank accession number AC003958; [35]), and in the absence of evidence of there being a KAP16-2, KAP16-3, KAP16-4 or KAP16-5 gene in humans. Hence we believe that at the very least, the species of origin of any given sequence needs to be clearly identified, although the need to identify species might be less important in literature if a publication is focussed on a single species and no conclusions that may have implications across species are drawn.
Researchers finding new KAP or KRTAP sequences probably also need to be more cautious in assigning the sequence to a family. Accordingly we recommend that the “L” term for “like”, is used if any doubt exists at all as to the origin of the sequence, and that this term, which is also used in the Rogers and Powell [10] system, continues in the nomenclature recommended below.
Genetic variation in the KRTAPs
Studies of variation in KRTAPs are limited in most species, and knowledge of the various types of genetic variation is therefore also limited. Currently humans and sheep are the two best studied species and probably because of the recent emphasis on human genome discovery and the historic importance of wool in making textiles. It is however expected that as more genomes are sequenced and in multiple individuals, the need for a comprehensive nomenclature that accommodates genetic variation more effectively, will increase.
In humans, studies of KRTAP variation have only been carried out in Caucasian and Japanese individuals and have been restricted to the KAP1 [7] and KAP4 [8] families. Four previously identified and apparently different KAP1-n genes (KRTAP1-1A, KRTAP1-1B, KRTAP1-6 and KRTAP1-7) have been shown to be allelic variants of a single gene [7], while four other KAP1-n genes (KRTAP1-8A, KRTAP1-8B, KRTAP1-3 and KRTAP1-9) were revealed to be allelic variants of another KAP1-n gene [7]. Two or three allelic variants have also been reported for 10 of the 11 human KAP4-n genes [8].
In sheep, genetic variation has been reported for the KRTAP1 [3,4,36], KRTAP3 ([37], KRTAP5 [5,37], KRTAP6 [6,37], KRTAP7 [37,38] and KRTAP8 [38,39] families. Up to nine alleles have been reported for KRTAP1-3 and KRTAP1-4. It should be noted that the apparently higher degree of variation found in sheep, compared to humans, is possibly due to a greater number of genomes being screened.
The variation detected in the KAP genes includes single nucleotide substitutions and length variation. Variation in length is noted in both sheep and human KAP1-n genes [3,7], and human KAP4-n genes [8]. It appears to be the result of having a variable number of cysteine-rich repeated coding sequences and these have probably arisen by intragenic deletion and/or duplication of the repeated segments of the genes during evolution [8].
There is little understanding of how variation in human KAP genes affects hair structure or other keratinagous tissue. However genetic variation in each individual KAP family could be much higher than previously thought based on recent research in sheep [4-6,36,38], and this may underpin some of the variation in hair and wool characteristics. The ability of the nomenclature system to accommodate extensive genetic variation, or what more correctly might be called polymorphism, given the over-use of this word in describing less variable gene systems; is one key driver in our revised system. What is more, while our revised nomenclature is not dissimilar to that of Powell and Rogers [10] in accommodating genetic variation, we feel that the term in the name that denotes genetic or allelic variation should be at the end of the gene name, especially as we believe an increased emphasis on how this variation affects wool and hair traits will emerge with time.
The proposed revised nomenclature for mammalian KAPs/KRTAPs
Given the numerous KAP genes identified to date and high levels of diversity among the KAPs from different species of more distant phylogenetic relationship, a revised nomenclature needs to be both flexible to accommodate variation and informative. We propose that the current KAP nomenclature [10] can be easily modified to accommodate the HGNC guidelines to become a consensus system for all mammalian species, as follows:
sp-KAPm-nL*x = protein
sp-KRTAPm-n(p/L)*x = gene
In this nomenclature, “sp” is a unique letter-based code for different species described by the protein knowledge-based UniProt (www.uniprot.org/docs/speclist), for example, “HUMAN” for Homo sapiens, “SHEEP ” for Ovis aries, “BOVIN” for Bos taurus and “MOUSE” for mus musculus. This would typically only be used in publications and when necessary; “m” is a number identifying the family; “n” is a constituent of that family; “p” signifies a pseudogene if there is an obvious fault in the gene (e.g. presence of an unexpected stop codon), while “L” if present, signifies “like” and refers to a temporary “place-holder” until the family (or constituency within that family) is confirmed; “x” is an alphabetical letter signifying the variant or allele, but preferably at the level of an extended haplotype encompassing the promoter, 5' UTR, exons and introns and 3'UTR. “p” and “L” should probably not be used together as a pseudogene will not produce a protein by definition, and hence genetic homology with any other sequence is of limited value.
The use of KAP for the protein and KRTAP for the gene is consistent with the keratin nomenclature where K is used for a protein and KRT is used for the associated gene.
To allow adequate time for transition and capture historically useful information, we suggest that in future, historic names might be bracketed after the first mention of the gene or protein, such as KRTAP1-1(B2A), for the next few years (see old terminology in Powell and Rogers [10]).
Concluding remarks
This nomenclature preserves the widely used and broadly referred to system proposed by Rogers and Powell [10], but with the important addition of a species identifier to constitute an informative KAP naming system and a minor change in the order of terms in the nomenclature. Accordingly, the new nomenclature should have minimal impact on current publications and databases, but if used correctly should facilitate more informed discussion about the KAP genes and proteins. We strongly would urge authors to use the L-term until such time as the location of the gene can be confirmed on the chromosome and its similarity to a sequence of known classification confirmed, or if they harbour any concerns about the family of origin of the protein or gene they are describing. It may be appropriate for scientists working on KAP genes and proteins to communicate more freely with each other and also with organisations like HGNC to insure that the nomenclature is used appropriately, while also being regularly revised to accommodate any future findings about the KAP genes and proteins.
Acknowledgements
This work was financially supported by FRST (C10X0710: Keeping New Zealand Wool Products at the Cutting Edge through Enhanced Wool Quality) and the Lincoln University Gene-Marker Laboratory. The Wool Research Organisation of New Zealand Inc and New Zealand Wool Industry Charitable Trust Postgraduate Scholarship to HG is acknowledged.
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Marshall RC, Orwin DFG, Gillespie JM. Structure and biochemistry of mammalian hard keratin. Electron Microsc Rev. 1991;4:47-83
2. Parry DAD, Steinert PM. Intermediate filament structure. New York: Springer-Verlag. 1995
3. Rogers GR, Hickford JGH, Bickerstaffe R. Polymorphism in two genes for B2 high sulfur proteins of wool. Anim Genet. 1994;25:407-415
4. Gong H, Zhou H, Hickford JGH. Polymorphism of the ovine keratin-associated protein 1-4 (KRTAP1-4) gene. Mol Biol Rep. 2010;37:3377-3380
5. Gong H, Zhou H, Plowman J. et al. Analysis of variation in the ovine ultra-high sulphur keratin-associated protein KAP5-4 gene using PCR-SSCP technique. Electrophoresis. 2010;31:3545-3547
6. Gong H, Zhou H, Hickford JGH. Diversity of the glycine/tyrosine-rich keratin-associated protein 6 gene (KAP6) family in sheep. Mol Biol Rep. 2011;38:31-35
7. Shimomura Y, Aoki N, Schweizer J. et al. Polymorphisms in the human high sulfur hair keratin associated protein 1, KAP1, gene family. J Biol Chem. 2002;277:45493-45501
8. Kariya N, Shimomura Y, Ito M. Size polymorphisms in the human ultrahigh sulphur hair keratin-associated protein 4, KAP4, gene family. J Invest Dermatol. 2005;124:11111-11118
9. Rogers GE, Powell BC. Organization and expression of hair follicle genes. J Invest Dermatol. 1993;101:50S-55S
10. Powell BC, Rogers GE. The role of keratin proteins and their genes in the growth, structure and properties of hair. In: (ed.) Jollès P, Zahn H, Höcker H. Formation and Structure of Human Hair. Basel: Birkhäuser Verlag. 1997:59-148
11. Goddard DR, Michaelis L. Derivatives of keratin. J Biol Chem. 1934;112:361-371
12. Gillespie JM, Reis PJ. The dietary-regulated biosynthesis of high-sulphur wool proteins. Biochem J. 1966;98:669-677
13. Gillespie JM, Broad A. Ultra-high sulphur proteins in the hairs of the Artiodactyla. Aust J Biol Sci. 1972;25:139-145
14. Stein WH, Moore S. Chromatography of amino acids on starch columns; separation of phenylalanine, leucine, isoleucine, methionine, tyrosine, and valine. J Biol Chem. 1948;176:337-365
15. Harrap BS. The conformation of a soluble wool keratin derivative. Aust J Biol Sci. 1963;16:231-240
16. Gillespie JM. The isolation and properties of some soluble proteins from wool. V. The isolation of the high-sulphur protein SCMKB2. Aust J Biol Sci. 1963;16:241-251
17. Lindley H, Elleman TC. The preparation and properties of a group of proteins from the high-sulphur fraction of wool. Biochem J. 1972;128:859-867
18. Swart LS, Haylett T, Joubert FJ. Studies on the high sulfur proteins of reduced Merino wool. II. The isolation of a homogeneous protein. Text Res J. 1969;39:912-917
19. Haylett T, Swart LS, Parris D. Studies on the high-sulphur proteins of reduced Merino wool. Amino acid sequence of protein SCMBK-IIIB3. Biochem J. 1971;123:191-200
20. Gillespie JM, Darksus RL. Relation between the tyrosine content of various wools and their content of a class of proteins rich in tyrosine and glycine. Aust J Biol Sci. 1971;24:1189-1197
21. Gillespie JM. The structural proteins of hair: isolation, characterisation and regulation of biosynthesis. In: (ed.) Goldsmith LA. Physiology, Biochemistry and Molecular Biology of the Skin, 2nd ed. Vol 1. New York: Oxford University Press. 1991:625-659
22. Powell BC, Rogers GE. Differentiation in hard keratin tissues: hair and related structures. In: (ed.) Leigh F, Watt RF, Lane B. Keratinocyte Handbook. Cambridge: Cambridge University Press. 1994:401-436
23. Fratini A, Powell BC, Rogers GE. Sequence, expression, and evolutionary conservation of a gene encoding a glycine/tyrosine-rich keratin-associated protein of hair. J Biol Chem. 1983;268:4511-4518
24. MacKinnon PJ, Powell BC, Rogers GE. Structure and expression of genes for a class of cysteine-rich proteins of the cuticle layers of differentiating wool and hair follicles. J Cell Biol. 1990;11:2587-2600
25. Jenkins BJ, Powell BC. Differential expression of genes encoding a cysteine-rich keratin family in the hair cuticle. J Invest Dermatol. 1994;103:310-317
26. Fratini A, Powell BC, Hynd PI. et al. Dietary cysteine regulates the levels of mRNAs encoding a family of cysteine-rich proteins of wool. J Invest Dermatol. 1994;102:178-185
27. Powell BC, Arthur J, Nesci A. Characterisation of a gene encoding a cysteine-rich keratin associated protein synthesised late in rabbit hair follicle differentiation. Differentiation. 1995;58:227-232
28. Schweizer J, Bowden PE, Coulombe PE. et al. A new consensus nomenclature for mammalian keratins. J Cell Biol. 2006;174:169-174
29. Rogers MA, Schweizer J. Human KAP genes, only half of it? Extensive size polymorphisms in hair keratin associated genes. J Invest Dermatol. 2005;124:vii-ix
30. Rogers MA, Winter H, Langbein L. et al. Characterization of human KAP24.1, a cuticular hair keratin-associated protein with unusual amino acid composition and repeat structure. J Invest Dermatol. 2007;127:1197-1204
31. Rogers MA, Langbein L, Praetzell-Wunder S. et al. Characterization and expression analysis of the hair keratin associated protein KAP26.1. Br J Dermatol. 2008;159:725-729
32. Rogers MA, Langbein L, Praetzel-Wunder S. et al. Human hair keratin-associated proteins (KAPs). Int Rev Cytol. 2006;251:209-263
33. Wu DD, Irwin DM, Zhang YP. Molecular evolution of the keratin associated protein gene family in mammals, role in the evolution of mammalian hair. BMC Evol Biol. 2008;8:241
34. Liu WJ, Fang Y, Fang GX. et al. Missense mutation of KAP16.6 gene on three goat breeds in China and their associations with Cashmere production traits. J Anim Vet Adv. 2009;8:2713-2718
35. Rogers MA, Langbein L, Winter H. et al. Characterization of a cluster of human high/ultrahigh sulfur keratin-associated protein genes embedded in the type I keratin gene domain on chromosome 17q12.21. J Biol Chem. 2001;276:19440-19451
36. Itenge-Mweza TO, Forrest RH, McKenzie GW. et al. Polymorphism of the KAP1.1, KAP1.3 and K33 genes in Merino sheep. Mol Cell Probes. 2007;21:338-342
37. McLaren RJ, Rogers GR, Davies KP. et al. Linkage mapping of wool keratin and keratin associated protein genes in sheep. Mamm Genome. 1997;8:938-940
38. Gong H, Zhou H, Plowman J. et al. Search for variation in the ovine KAP7-1 and KAP8-1 genes using PCR-SSCP screening. DNA Cell Biol. in press
39. Wood NJ, Phua SH, Crawford AM. A dinucleotide repeat polymorphism at the glycine- and tyrosine-rich keratin locus in sheep. Anim Genet. 1992;23:391
40. Yahagi S, Shibuya K, Obayashi I. et al. Identification of two novel clusters of ultrahigh-sulfur keratin-associated protein genes on human chromosome 11. Biochem Biophys Res Commun. 2004;318:655-664
41. Rogers MA, Langbein L, Winter H. et al. Characterization of a first domain of human high glycine-tyrosine and high sulfur keratin-associated protein (KAP) genes on chromosome 21q22.1. J Biol Chem. 2002;277:48993-49002
42. Rogers MA, Langbein L, Winter H. et al. Hair keratin associated proteins: characterization of a second high sulfur KAP gene domain on human chromosome 21. J Invest Dermatol. 2004;122:147-158
43. Shibuya K, Obayashi I, Asakawa S. et al. A cluster of 21 keratin-associated protein genes within introns of another gene on human chromosome 21q22.3. Genomics. 2004;83:679-693
44. Powell BC, Sleigh MJ, Ward KA. et al. Mammalian keratin gene families: organisation of genes coding for the B2 high-sulphur proteins of sheep wool. Nucleic Acids Res. 1983;11:5327-5346
45. Frenkel MJ, Powell BC, Ward KA. et al. The keratin BIIIB gene family: isolation of cDNA clones and structure of a gene and a related pseudogene. Genomics. 1989;4:182-191
46. Kuczek ES, Rogers GE. Sheep wool (glycine + tyrosine)-rich keratin genes. A family of low sequence homology. Eur J Biochem. 1987;166:79-85
47. Swart LS, Haylett T. Studies on the high-sulphur proteins of reduced merino wool. Amino acid sequence of protein SCMKB-3A3. Biochem J. 1973;133:641-654
Author contact
![]() Corresponding author: Jonathan G.H. Hickford, Gene-Marker Laboratory, Faculty of Agriculture and Life Sciences, PO Box 84, Lincoln University, Lincoln 7647, New Zealand. Email: jon.hickfordac.nz
Corresponding author: Jonathan G.H. Hickford, Gene-Marker Laboratory, Faculty of Agriculture and Life Sciences, PO Box 84, Lincoln University, Lincoln 7647, New Zealand. Email: jon.hickfordac.nz

 Global reach, higher impact
Global reach, higher impact