Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2012; 8(3):328-343. doi:10.7150/ijbs.3517 This issue Cite
Research Paper
Gene Expression Profiling of the Cephalothorax and Eyestalk in Penaeus Monodon during Ovarian Maturation
1. The Queensland Department of Primary Industries and Fisheries, Animal Science, Bribie Island, Queensland 4507, Australia.
2. School of Land, Crop and Food Sciences, The University of Queensland, St Lucia, Queensland 4072, Australia.
3. Faculty of Science, Health and Education, University of the Sunshine Coast, Maroochydore, Queensland 4558, Australia.
Received 2011-9-14; Accepted 2012-1-30; Published 2012-2-9
Abstract
In crustaceans, a range of physiological processes involved in ovarian maturation occurs in organs of the cephalothorax including the hepatopancrease, mandibular and Y-organ. Additionally, reproduction is regulated by neuropeptide hormones and other proteins released from secretory sites within the eyestalk. Reproductive dysfunction in captive-reared prawns, Penaeus monodon, is believed to be due to deficiencies in these factors. In this study, we investigated the expression of gene transcripts in the cephalothorax and eyestalk from wild-caught and captive-reared animals throughout ovarian maturation using custom oligonucleotide microarray screening. We have isolated numerous transcripts that appear to be differentially expressed throughout ovarian maturation and between wild-caught and captive-reared animals. In the cephalothorax, differentially expressed genes included the 1,3-β-D-glucan-binding high-density lipoprotein, 2/3-oxoacyl-CoA thiolase and vitellogenin. In the eyestalk, these include gene transcripts that encode a protein that modulates G-protein coupled receptor activity and another that encodes an architectural transcription factor. Each may regulate the expression of reproductive neuropeptides, such as the crustacean hyperglycaemic hormone and molt-inhibiting hormone. We could not identify differentially expressed transcripts encoding known reproductive neuropeptides in the eyestalk of either wild-caught or captive-reared prawns at any ovarian maturation stage, however, this result may be attributed to low relative expression levels of these transcripts. In summary, this study provides a foundation for the study of target genes involved in regulating penaeid reproduction.
Keywords: Penaeus monodon, prawn, cephalothorax, eyestalk, ovarian maturation, gene expression, microarray.
Introduction
A major constraint to sustainable aquaculture of penaeid prawns is the current industry reliance on wild-caught broodstock. Dependence on wild-caught broodstock stems, for the most part, from reproductive dysfunction in captive-reared females. The precise physiological factors inhibiting maturation and spawning in captive-reared females remains unknown, although the inability of these animals to undergo final ovarian maturation has been well documented. Currently, eyestalk ablation, which involves the surgical removal of one or both eyestalks, remains the only method widely utilised by industry to induce ovarian maturation in both wild-caught and captive-reared females. A better understanding of the hormonal regulation of prawn reproduction would enable the development of methods for manipulation of reproduction to the exclusion of eyestalk ablation. Such methods may also help eliminate reliance on wild-caught broodstock.
Accordingly, endocrine control of prawn reproduction is an area that has received considerable attention, with emphasis placed on the roles of cephalothorax and eyestalk neuropeptides and hormones, the sesquiterpene methyl farnesoate, and to a lesser extent, the biogenic amines and vertebrate-type steroid hormones. The neurosecretory system of the eyestalk is known to consist of a cluster of peptidergic neurons located in the medulla terminalis X-organ (MTXO) and their globular axonic terminals that constitute the sinus gland (SG), a neurohemal organ that releases peptide hormones into the hemolymph [1]. The effect of eyestalk ablation has been attributed to a reduction in the level of the gonad-inhibiting hormone (GIH) normally present in the neurosecretory cells of the eyestalk. However, the procedure presumably also produces animals with other hormonal imbalances and consequential detrimental effects including high mortality. The cephalothorax contains target organs involved in reproduction including the hepatopancreas, the mandibular organ and the Y-organ.
One critical process in ovarian maturation that requires further study is the MTXO-SG regulation of vitellogenesis. Vitellogenesis involves the production of vitellogenin - the female-specific high density lipoprotein and precursor to the lipo-glycolcarotenopoprotein vitellin (the major egg yolk protein). Vitellogenesis also requires the accumulation of vitellogenin/vitellin and other proteins, carbohydrates, lipids, vitamins and minerals by the oocytes [2]. In crustaceans, vitellogenesis is generally understood to be negatively regulated by the GIH, which is produced and released from neurosecretory sites within the eyestalk. The MTXO-SG also secretes a variety of neuropeptides additional to GIH including crustacean hyperglycaemic hormone (CHH), moult-inhibiting hormone (MIH), mandibular organ-inhibiting hormone, and the chromatophorotropins: pigment dispersing hormone and red pigment concentrating hormone. The vast majority of neuropeptides signal through cell-surface guanosine-protein coupled receptors (GPCRs). These neuropeptide-receptor systems control a variety of physiological functions, for instance the numerous involuntary movements of internal ducts that are precisely timed for transporting reproductive materials [3]. Meanwhile, arrestins are important components for desensitization of GPCR cascades that mediate neurotransmission [3].
It is unclear whether neuropeptide hormone levels in the eyestalk are regulated primarily at the transcriptional or translational level, or via levels of secretion of stored products to the hemolymph, or whether their involvement in these processes is modulated chiefly via various mechanisms at the target tissue. Several studies on a variety of crustaceans have examined transcript and neuropeptide levels during reproduction and moulting cycles. In the crayfish Homarus americanus, GIH mRNA in the XO is at low levels at immature stage compared with pre-vitellogenic and vitellogenic stages [4]. CHH-A levels are highest at pre-vitellogenic stage, while CHH-B levels are highest at pre-vitellogenic stage and mature stage. Meanwhile, neuropeptide levels in the SG are not different in total GIH levels, and CHH levels are highest at the pre-vitellogenic stage. Hemolymph GIH levels are at their lowest during the vitellogenic stage compared with other stages, while hemolymph CHH levels are highest at mature stage. Together, these results provide strong evidence to suggest that CHH-A and -B may initiate vitellogenesis, while GIH prevents onset of vitellogenesis. Further investigation into the biological activities of these neuropeptides at the target tissue (Y-organ) via bioassays at the tissue, receptor and cellular levels, determined that intracellular signalling mechanisms may provide the most important level of moult control. In support of this, the level of an MIH mRNA appears not to change significantly in the eyestalk of Marsupenaeus japonicus during the moult-cycle [5].
Oligonucleotide microarrays have proven to be an excellent tool for studying gene expression in a holistic manner. In this study, ovarian maturation stage-specific differential gene expression was performed on the P. monodon cephalothorax and eyestalk (ES) by utilising custom oligonucleotide microarrays in both wild-caught and captive-reared animals. Here we report the identification of genes that are differentially expressed during ovarian maturation and between wild-caught and captive-reared animals. This will prove an important step towards explaining physiological differences observed during reproduction.
Materials and Methods
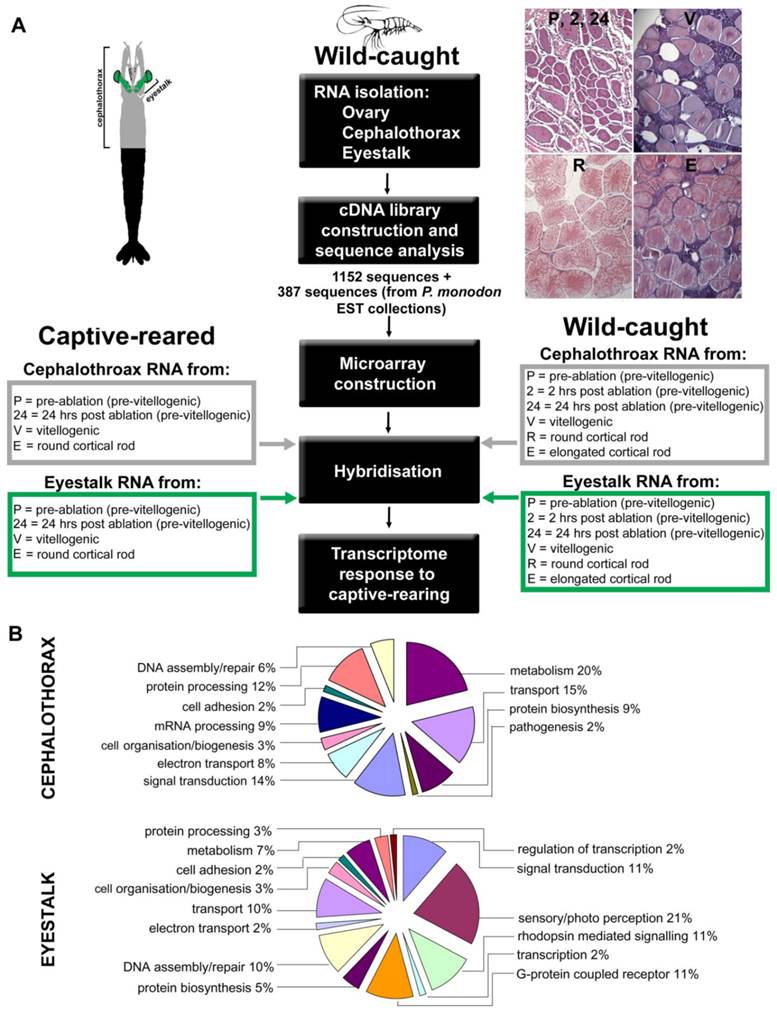
Sample collection. Wild-caught adult female P. monodon (116.75 ± 17.99 g) were obtained from commercial hatchery suppliers in Innisfail, Queensland and air freighted to the Queensland Department of Primary Industries and Fisheries (QDPIF), Bribie Island Aquaculture Research Centre (BIARC), Woorim, Queensland. Captive-reared adult female P. monodon (114.5 ± 19.6 g) were obtained from the Australian Institute of Marine Science (AIMS), Townsville, Queensland, following grow-out (pond) culture at BIARC. Animals were stocked in 5 tonne tanks with flow-through seawater heated to 26oC and acclimated for 7 days whilst fed fresh diet (squid and mussels) twice daily. All animals were moult staged according to extent of epidermal retraction. For wild-caught animals, tissue samples were collected at various ovarian maturation stages based on in vivo observation of maturing ovaries, as described by Duronslet et al., (1975) [6], for subsequent classification by histological analysis of developing oocytes as follows: whole ovaries, cephalothorax and ESs, (containing the MTXO-SG complex) were collected from un-ablated inter-moult females (immature ovaries) euthanized in saline ice slurry, snap frozen in liquid nitrogen and stored at -80°C until processing. Additionally, seventy inter-moult females were unilaterally eyestalk ablated to induce ovarian maturation, eye- and carapace-tagged and maintained for a further 7 days as above. Captive-reared animals were sampled across several moult cycles after ablation. In vivo ovarian maturation stage, moulting, mortality and other behaviour were recorded daily. Animals were sampled at 2 h and 24 h post ablation (immature ovaries) and further sampled during this period with tissues of interest collected as outlined above (including remaining eyestalk) from random animals representing each of 4 in vivo ovarian maturation stages: immature, early maturing, late maturing and mature. Gonadosomatic index (GSI) was also calculated (ovarian weight expressed as a percentage of total body weight) for all samples. All wild-caught animals were sampled during the first moult cycle after ablation.
Histological characterisation of ovarian samples. Small pieces (100 mg) of the middle ovarian lobes from specimens at selected ovarian maturation stages were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) solution (pH 7.2) overnight, washed with PBS at room temperature, dehydrated in ethanol series, embedded in paraffin, sectioned (6µm) and stained with either haematoxylin and eosin for detection of acidophilic and basophilic substances, Periodic Acid Schiff (PAS) for detection of carbohydrates (glycoproteins) or Luxol Blue for detection of phospholipids, or alternatively frozen and cut with cryostat and stained with Oil Red O for detection of simple lipids, as described by Bell & Lightner (1988) [7]. Ovarian stages used in the present study were characterized largely as determined by Tan Fermin & Pudadera (1989) [8] with some modifications as follows: previtellogenic (P) stage - the ovary contains only oogonia and basophilic previtellogenic oocytes at chromatin nucleolus and perinucleolus stage (GSI 1.7-2.9); vitellogenic (V) stage - in addition to the presence of oogonia and basophilic previtellogenic oocytes, the ovary contains yolk accumulating oocytes, the ooplasm of which is full of eosinophilic (acidophilic) yolk substances and also stains positive to PAS and Luxol blue indicating presence of glycoproteins and phospholipids respectively. Large globules in the ooplasm are also notable which stain positive with Oil Red O indicating simple lipids (GSI 3.5-6.5); round cortical rod (R) stage - staining affinities of oocytes are similar to those described for V stage ovaries with the addition of the appearance of round cortical rods (CRs) developing radially at the peripheral cortex of those oocytes containing yolk substances (GSI 7.7-10.5); elongated cortical rod (E) stage - staining affinities of oocytes are similar to those described for R stage ovaries except CRs are elongated and extended towards the nucleus (GSI 6.0-14.0).
RNA isolation. Total RNA was isolated from small pieces (100mg) of the middle ovarian lobes, whole cephalothoraxes and whole eyestalks (initially ground under liquid nitrogen using mortar and pestle) from prawns of interest, using TRIZOL reagent as recommended by the manufacturer (Invitrogen Life Technologies, Carlsbad, CA, USA). The samples were used for synthesis of complementary DNA (cDNA), creation of cDNA libraries and for construction and screening of microarrays. Concentration and purity of the RNA were determined using a spectrophotometer (GeneQuant Pro, GE Healthcare UK Ltd., Buckinghamshire, England) with 260 and 280 nm readings. RNA quality was assessed for all samples by visualisation on denaturing formaldehyde RNA gels (protocol recommended by Qiagen, Valencia, CA, USA) using ethidium bromide staining.
cDNA library creation and sequence analysis. Three P. monodon cDNA libraries were created in our laboratory from total RNA isolated from ovary, eyestalk (containing X-organ/Sinus gland complex) and whole cephalothorax (containing target organs including hepatopancreas, Y-organ and mandibular organ) collected from wild-caught animals. Briefly, reverse transcription of total RNA (pooled in equal part from three animals from each ovarian maturation stage) was conducted using the SMART IV Polymerase Chain Reaction (PCR) cDNA synthesis kit (Clonetech Laboratories, Inc., Mountain View, CA, USA). First strand cDNA synthesis was conducted using Deoxyribonucleotide triphosphate (dNTP) mix, SMART IV oligonucleotide and CDS III/3' PCR primer with Powerscript reverse transcriptase. After determining optimal number of cycles in order to reduce the likelihood of producing redundant cDNA libraries, single stranded cDNA served as template for PCR based cDNA amplification using dNTP mix, 5' PCR primer, CDS III/3' PCR primer with Advantage II polymerase. Complementary DNA was then purified using the QIAquick™ PCR purification Kit (Qiagen, Valencia, CA, USA). To ensure efficient ligation into pGEM-T easy vector (Promega, Madison, WI, USA) a standard A-tailing procedure was conducted. Complementary DNA size fractionation was then conducted to separate larger cDNA species using CHROMA SPIN™ columns (Clontech Laboratories, Inc., MountainView, CA, U.S.A) according to the manufacturer's protocol. Ligations were then conducted and subsequent transformation into XL10 Gold ultracompetent cells (Stratagene, La Jolla, CA, USA) according to the manufacturer's protocol.
Clones were sequenced using the 5' SMART PCR primer as sequencing primer. Sequences were edited (removal of poor sequence, vector and poly-A sequence) and then assembled into contiguous over-lapping sequence alignments (contigs) using Sequencher (Gene Codes Corporation, Ann Arbor, MI, USA). The sequences were annotated with the name of the highest basic local alignment search tool (BLAST) [9] score from an analysis of GenBank entries by the BLASTx and BLASTn procedures. Further putative functional annotation was assigned to each of the sequences by conducting a BLASTx similarity search using the GOanna annotation tool (http://agbase.msstate.edu/GOAnna.html). Protein domains were identified for selected sequences from the Pfam database (http://pfam.sanger.ac.uk).
Microarray construction. A total of 601 sequences were selected from the three cDNA libraries created for this study together with an additional 387 sequences obtained from P. monodon Expressed Sequence Tag (EST) collections created from ovary, hepatopancrease and eyestalk sourced from publicly available databases (GenBank http://www.ncbi.nlm.nih.gov) for inclusion as probes on the microarrays. These included key transcripts encoding known eyestalk neuropeptides including several crustacean hyperglycemic hormone (CHH) family members from P. monodon where available and from other penaeids where not, as follows: P. monodon CHH-1 (AY346378.1) -2 (AF104931.1), Litopenaeus vannamei moult-inhibiting hormone (MIH) (S73824.1), P. monodon sinus gland peptide (PmSGP)-I (AF104386.1) -II (AF104387.1) -III (AF104388.1) -IV (AF104389.1) -V (AF104390.1), L. vannamei pigment-dispersing hormone (PDH)-1 (Y11723.1) -2 (Y11722.1) and Marsupenaeus japonicus PDH-3 (AB247562.1). Oligonucleotide microarrays were produced by CombiMatrix corporation (Mukilteo, USA) using the CustomArray™ 4 × 2K platform which contains four identical, independent 2240-feature microarrays on each slide. Between two and five oligonucleotide probes were designed and incorporated on each microarray for each of the selected transcripts.
Target preparation and microarray hybridisation. Ovarian RNA samples from nine wild-caught animals representing six ovarian maturation stages (P, 2, 24, V, R, E) were used in microarray hybridisations. Similarly, RNA samples from three captive-reared animals representing four maturation stages (P, 24, V, E) were used in microarray hybridisations (Fig. 1A). For wild-caught animals, samples from each ovarian maturation stage were pooled into groups of four and five, enabling two hybridisations. For captive-reared animals, samples from each ovarian maturation stage from all three animals were pooled enabling one hybridisation for each stage. Importantly, as the four stages for captive-reared animals were (1) pre-ablation pre-vitellogenic (P), (2) post-ablation pre-vitellogenic (2 for 2 h post ablation, 24 for 24 h post ablation), (3) post-ablation vitellogenic (V), (4) post-ablation vitellogenic with cortical rods (E), this arrangement allowed for 2 samples of captive-reared pre-vitellogenic and 2 samples of captive-reared vitellogenic, thereby enabling t-tests between samples, while also allowing analysis across the whole 4 stages via cluster analysis. All hybridisations were single channel hybridisations conducted using equal amounts of RNA pooled from each individual.
Reverse transcription of total RNA, RNA amplification by in vitro transcription and amplified antisense RNA (aRNA) labelling were conducted using the RNA ampULSe amplification and labelling kit (KREATECH Biotechnologies, Amsterdam, Netherlands) according to manufacturer's instructions. Briefly, reverse transcription reactions were performed using T7 Oligo(dT) primer with Arrayscript reverse transcriptase. Second strand synthesis was then performed and cDNA purified using cDNA filter cartridges and used as template for aRNA synthesis in in vitro transcription reactions. aRNA was then purified using aRNA filter cartridges and yield and quality of aRNA assessed by measuring the absorbance at 260 nm and 280 nm using a spectrophotometer (GeneQuant Pro, GE Healthcare UK Ltd., Buckinghamshire, England). 1 µg of non-modified aRNA was labelled non-enzymatically using the Universal Linkage System (ULS) technology coupled with Cy5 fluorochrome. Unbound ULS label was removed using included KREApure columns and aRNA fragmentation conducted using RNA fragmentation reagents (Applied Biosystems/Ambion, Austin, TX, USA) according to manufacturer's instructions. Labelled aRNA was then hybridised to the microarray slides according to manufacturer's instructions. Briefly, slides were assembled with supplied hybridisation caps in hybridisation clamps and re-hydrated prior to use by adding nuclease-free water and incubating at 65°C for 10 min. Pre-hybridisation steps (30 min) and hybridisations (16 h) were conducted in 30µl volumes using hybridisation solution containing 6× saline-sodium phosphate-EDTA (SSPE), 0.05% Tween-20, 20mM EDTA, 5×Denhardt's solution, 100ng/µl denatured salmon sperm DNA, 0.05% SDS, and 6×SSPE, 0.05% Tween-20, 20mM EDTA, 25% deionized formamide, 100 ng/µl denatured salmon sperm DNA, 0.04% SDS and 1 µg of target aRNA respectively, with microarrays loaded onto a rotisserie in a hybridisation oven and incubated at 45°C. Microarrays were then washed according to manufacturer's instructions and scanned wet using supplied imaging solution with LifterSlips™ (Thermo Fisher Scientific Inc., Waltham, MA, USA) applied. After imaging, microarrays were stripped for subsequent re-hybridisation up to three times using the CombiMatrix CustomArray stripping kit (CombiMatrix Corporation, Mukilteo, USA) according to manufacturer's instructions.
Microarray data analysis. Microarray slides were scanned using a fluorescent microarray scanner (GenePix 4000B, MDS Analytical Technologies, Toronto, Canada). Microarray Imager (CombiMatrix Corporation, Mukilteo, USA) was then used to create spot intensity reports, generate gene ID mapping files and assign gene identification. Final intensity reports were retrieved as raw spot intensities in tab-delimited files. The data set is deposited in the Gene Expression Omnibus database (accession nos. GSE31862) at the following site: http://www.ncbi.nlm.nih.gov/geo. Final intensity reports were then used for further data analysis using data mining software Acuity 4.0 (MDS Analytical Technologies, Toronto, Canada). Data were pre-processed and normalised by applying background and floor correction, combining replicates (means) and global means normalisation. Data was grouped and classified using hierarchical and non-hierarchical clustering analyses. Genes displaying significant differential expression were identified via Two-Sample Students t-Tests.
Results
Overview of P. monodon cephalothorax and eyestalk EST collection
A total of 1152 clones were sequenced from the three cDNA libraries constructed from cephalothorax, eyestalk and ovary (Fig. 1A). A description of the sequence content of the cephalothorax and eyestalk libraries when the sequences were edited and assembled is given in Table 1 (ovary is described elsewhere). Percentage representations from the hierarchical Gene Ontology (GO) mapping: Biological Process is displayed in Fig. 1B. Transcripts encoding proteins involved in metabolism (including lipid, carbohydrate, chitin and nucleic acid metabolism) featured highly in the cephalothorax library. In the cephalothorax library, further abundant transcripts included those involved in transport (such as oxygen, iron and lipid transport) as well as transcripts involved in signal transduction and protein processing. In eyestalk, transcripts known to be involved in sensory/photo perception and signalling were abundant (21%), as were transcripts encoding proteins involved in DNA assembly/repair and transport (both 10%). No transcripts were identified which displayed significant BLAST homology to known reproductive neuropeptides.
Summary of Black tiger prawn Penaeus monodon EST collection from eyestalk and cephalothorax.
| Tissue | Eyestalk | Cephalothorax | Total |
|---|---|---|---|
| Total ESTs sequenced | 384 | 384 | 768 |
| Singletonsα | 136 | 150 | 286 |
| Contigsα | 18 | 18 | 36 |
| ESTs with E = 10-4β | 27 | 37 | 64 |
| GO annotationβ | 49 | 78 | 127 |
| No similarityδ | 100 | 91 | 191 |
Notes: αBased on total number of ESTs after editing. βBased on scans of the SwissProt + SpTrEMBL combined databases using the BlastX algorithm with default parameters. δBased on scans of the GenBank EST database using the BlastN algorithm with default parameters.
Experimental design and gene ontology to determine prawn transcriptome response to captive rearing. (A) Diagram outlines the experimental setup for microarray analysis. For hybridisation, RNA was obtained from cephalothorax and eyestalk at various ovarian stages of wild-caught and captive-reared animals. Representative ovary sections at the reproductive stages are shown, stained with haematoxylin and eosin. (B) Abbreviated Gene Ontology annotations for cephalothorax and eyestalk from the Black tiger prawn Penaeus monodon cDNA libraries. Shown are percentage representations from the hierarchical GO mapping: Biological Process.
Microarray based differential gene expression
Cephalothorax. We examined ovarian maturation stage-specific differential gene expression in the cephalothorax of wild-caught animals. Gene expression profiles in the ovary among wild-caught and captive-reared animals throughout ovarian maturation are reported elsewhere.
Wild-caught cephalothorax - stage-specific profiles: Hierarchical cluster analysis was implemented. The resulting dendrogram, (Fig. 2), in accordance with Gap statistic analysis revealed two major clusters of genes with stage-specific expression in the wild-caught cephalothorax data set. Cluster 1 (Fig. 2A) contains 68 genes displaying elevated expression levels at early (pre-vitellogenic) stages. These include thrombospondin, shrimp ovarian peritrophin variants 1, 2 and 3, metallothionein, CP14 - a calcified cuticle protein, profilin, glycerol-3-phosphate transporter and 40 genes with no significant BLAST homology, therefore are presented as novel, some of which displaying very high expression levels. Cluster 2 contains 145 genes generally displaying elevated expression levels at later (vitellogenic) stages. To provide clear presentation of this large cluster, it is represented in figure 2, and described here, as its sub-clusters: 2a, 2b and 2c. Cluster 2a (Fig. 2B). This contains 87 genes displaying elevated expression levels at later (vitellogenic) stages with sub-clusters of genes (within cluster 2a) displaying either sustained elevated expression at later stages or displaying elevated expression at one or more of the later stages.
Dendrograms (hierarchical clustering) of microarray gene expression data in the cephalothorax of wild-caught Penaeus monodon during ovarian maturation: (A) cluster 1, (B) cluster 2a, (C) cluster 2b and (D) cluster 2c employing the Average Linkage method together with the Pearson Centered similarity metric. Gap statistic analysis, proposed by Tibshirani et al (2002) [37], using the output of the K-means (non-hierarchical) clustering algorithm was implemented to estimate the number of clusters in the data set. Columns represent six ovarian maturation stages: P = pre-ablation (pre-vitellogenic], 2 = 2 hrs post ablation (pre-vitellogenic), 24 = 24 hrs post ablation (pre-vitellogenic), V = vitellogenic, R = round cortical rod and E = elongated cortical rod). Rows represent gene expression for individual genes. Colours indicate gene expression level for individual genes (red = high expression, black = low expression).
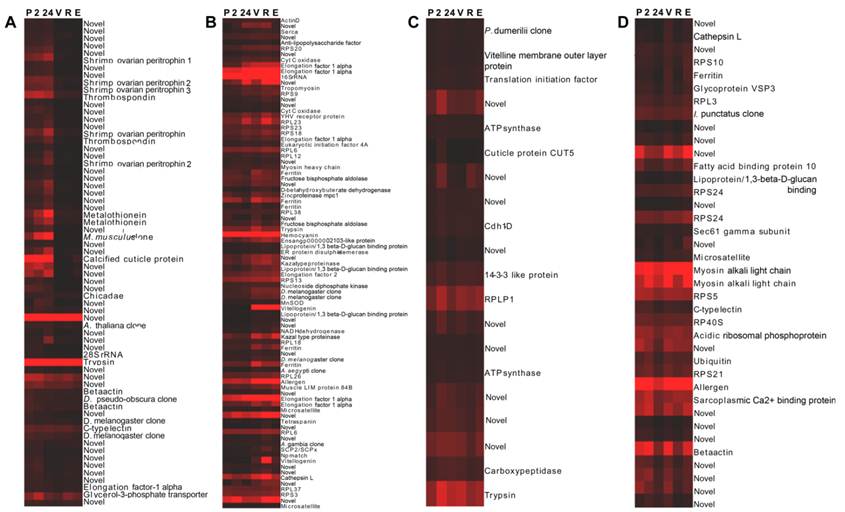
Genes include: (1) vitellogenin which displays sustained elevated expression at later stages, (2) SCPx/SCP2 - the sterol carrier protein-2/3-oxoacyl-CoA thiolase displaying elevated expression at later stages with a peak at R stage, (3) hemocyanin displaying comparatively high expression at all stages with a peak at V stage, (4) a chitinase (ensangp00000021035) displaying elevated expression at later stages with a peak at R stage, (5) the high density lipoprotein/1,3-beta-D-glucan-binding protein precursor up-regulated at later stages, (6) a kazal-type proteinase inhibitor up-regulated at later stages, (7) ferritin up-regulated at V stage, and (8) 25 novel genes, some displaying very high expression levels. Cluster 2b (Fig. 2C) contains 20 further genes generally displaying elevated expression at later stages with most genes, however, also displaying an expression peak at 24 h post-ablation. Genes include the vitelline membrane outer layer protein (VMO1). Cluster 2c (Fig. 2D) contains 38 further genes generally displaying elevated expression at later stages, however, also displaying reduced expression levels at 24 h post-ablation and again at R stage. Genes include a fatty acid binding protein and 15 novel genes, some displaying very high expression levels. Transcripts encoding the various ribosomal proteins are exclusive to cluster 2 (elevated expression at later stages). Cluster 2 also contains a greater number of transcripts encoding translational elongation factors compared with remaining clusters.
Wild-caught cephalothorax - differentially expressed genes: Genes displaying significant differential expression between collective early (pre-vitellogenic) stages and collective late (vitellogenic) stages in the wild-caught cephalothorax were identified via Two-Sample Students t-Tests (P<0.05). The results are listed in Table 2. Of the 56 transcripts identified as significantly differentially expressed, 24 transcripts displayed significant BLAST homology. An equal number of transcripts were identified as up-regulated at either pre-vitellogenic or vitellogenic stages.
Transcripts displaying statistically significant differential expression in Black tiger prawn Penaeus monodon microarray gene expression data in the cephalothorax of wild-caught animals during ovarian maturation.
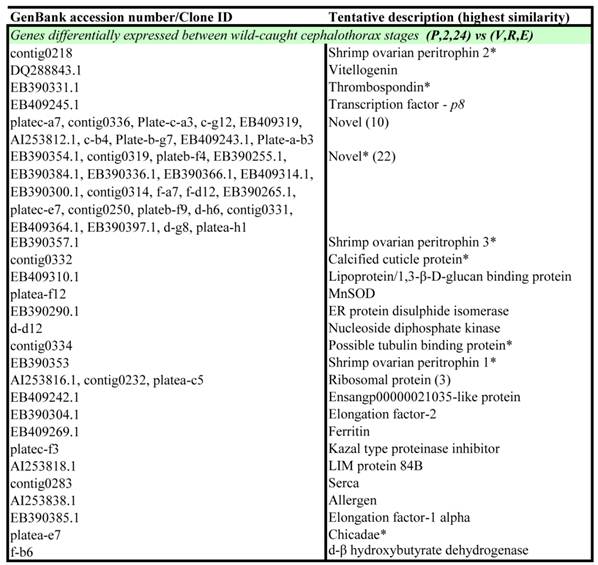
Notes: Transcripts were identified via Two-Sample Student's t-Tests with a p-value threshold of p = 0.05. P = pre-ablation (pre-vitellogenic), 2 = 2 hrs post ablation (pre-vitellogenic), 24 = 24 hrs post ablation (pre-vitellogenic), V = vitellogenic, R = round cortical rod and E = elongated cortical rod. All transcripts are up-regulated at the latter maturation stage unless indicated by an asterisk (*).
Eyestalk. To identify transcripts in the eyestalk with potential involvement in neuropeptide control of ovarian maturation, we examined ovarian maturation stage-specific differential gene expression in the eyestalks of both wild-caught and captive-reared animals. To identify possible physiological differences between wild-caught and captive-reared animals, we also examined differences in gene expression in the eyestalks between the two sources.
Wild-caught eyestalk - stage-specific profiles: To identify groups of genes displaying stage-specific expression profiles in the eyestalk of wild-caught animals, hierarchical cluster analysis was implemented. The resulting dendrogram (Fig. 3) revealed six major clusters of genes with stage-specific expression in the wild-caught data set. Cluster I (Fig. 3A) contains 60 transcripts generally displaying elevated expression at pre-vitellogenic stages: pre-ES ablation (P), 2 hours post-ES ablation (2) and 24 hours post-ES ablation (24) compared with vitellogenic stages. However, some transcripts in cluster 1 displayed elevated expression at P and 2 stages and also at several vitellogenic stages. Transcripts include those encoding ferritin, myosin, tetraspanin, cyclophilin, rhodopsin, opsin, arrestin and several cuticle proteins. This cluster also contains 21 novel transcripts (no significant homology to previously characterised genes (BLASTx E-value >1 × 10-4). Cluster II (Fig. 3B) contains 13 transcripts displaying similar expression to those transcripts in cluster 1. Transcripts include further transcripts encoding rhodopsin and ferritin and several novel transcripts. Cluster III (Fig. 3C) contains 34 transcripts generally displaying elevated expression at 24 hours post-ES ablation stage. The majority of transcripts (28) in cluster III are novel. Cluster IV (Fig. 3D) contains 54 transcripts generally displaying reduced expression at 24 hours post-ES ablation stage, with some transcripts displaying elevated expression at vitellogenic stages. Transcripts include those encoding metallothionein, manganese superoxide dismutase, architectural transcription factor - p8 and several mitochondrial enzymes. Compared with other clusters, transcripts encoding ribosomal proteins and translation elongation factors are significantly more abundant in cluster IV. This cluster also contains 21 novel transcripts, some of which displayed very high expression levels. Cluster V (Fig. 3E) contains 22 transcripts generally displaying elevated expression at vitellogenic (V) stage and at various other vitellogenic stages, including 10 novel transcripts, some of which displayed very high expression levels. Cluster VI (Fig. 3F) contains 34 further transcripts generally displaying elevated expression at various vitellogenic stages with highest expression at round cortical rod (R) stage. Cluster 6 includes 21 novel transcripts.
Wild-caught eyestalk - differentially expressed genes: Genes displaying significant differential expression between ovarian maturation stages (in either wild-caught or captive-reared eyestalk samples) and between wild-caught and captive-reared eyestalk samples were identified via Two-Sample Students t-Tests (P<0.05). The results are listed in Table 3. Of the 55 transcripts identified as significantly differentially expressed during ovarian maturation in the eyestalk in either wild-caught or captive-reared animals, 24 transcripts displayed significant BLAST homology. Of the 24 transcripts identified as significantly differentially expressed in the eyestalk between wild-caught and captive-reared animals, 12 transcripts displayed significant BLAST homology. The majority of transcripts (21) identified as significantly differentially expressed between wild-caught and captive-reared animals displayed higher expression in captive-reared animals.
Discussion
We have utilised custom oligonucleotide microarrays to assess differential gene expression in specific stages of ovarian maturation, in the cephalothorax and eyestalk of wild-caught and captive-reared black tiger prawns P. monodon. In an effort to understand the cause of reproductive dysfunction in captive-reared animals, we have identified differences in gene expression between wild-caught and captive-reared animals. Here, we discuss those genes whose expression we found to be significantly altered between these two groups and are potentially relevant to oocyte maturation. Throughout this study, the term key transcripts is used to refer to transcripts which display significant homology (through BLAST) to genes previously identified as playing key roles in the process of ovarian maturation and other related metabolic processes.
We thought it appropriate to focus on transcripts that displayed significant BLAST homology to genes/proteins previously identified as involved in relevant biological processes. We further focussed on transcripts that displayed expression profiles of interest (ie. differences between wild-caught and captive-reared animals, and between animals with immature vs mature/maturing ovaries. For some transcripts, which displayed significant BLAST homology, we could identify no plausible link with the biology in question, and conversely, some transcripts that displayed interesting expression profiles were novel, as they displayed no BLAST homology and hence form the focus of further studies.
Transcripts displaying statistically significant differential expression in the eyestalk of wild-caught Black tiger prawns Penaeus monodon during ovarian maturation.
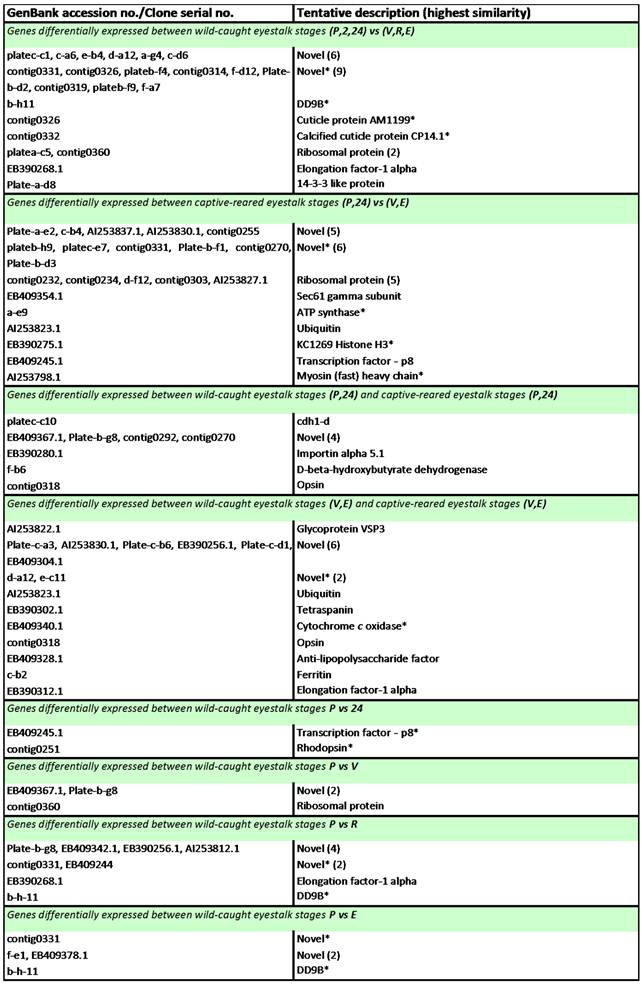
Notes: Transcripts were identified via Two-Sample Student's t-Tests with a p-value threshold of p ≤ 0.05. P = pre-ablation (pre-vitellogenic), 2 = 2 h post ablation (pre-vitellogenic), 24 = 24 h post ablation (pre-vitellogenic), V = vitellogenic, R = round cortical rod and E = elongated cortical rod. All transcripts are up-regulated at the latter stage unless indicated by an asterisk (*).
Dendrograms (hierarchical clustering) of microarray gene expression data in the eyestalk of wild-caught Black tiger prawns Penaeus monodon during ovarian maturation: Clusters I-VI (A-D) employing the Average Linkage method together with the Pearson Centered similarity metric. Gap statistic analysis, proposed by Tibshirani et al (2002) [37], using the output of the K-means (non-hierarchical) clustering algorithm was implemented to estimate the number of clusters in the data set. Columns represent six ovarian maturation stages (P = pre-ablation (pre-vitellogenic), 2 = 2 h post ablation (pre-vitellogenic), 24 = 24 h post ablation (pre-vitellogenic), V = vitellogenic, R = round cortical rod and E = elongated cortical rod). Rows represent gene expression for individual genes. Colours indicate gene expression level for individual genes (red = high expression, black = low expression).
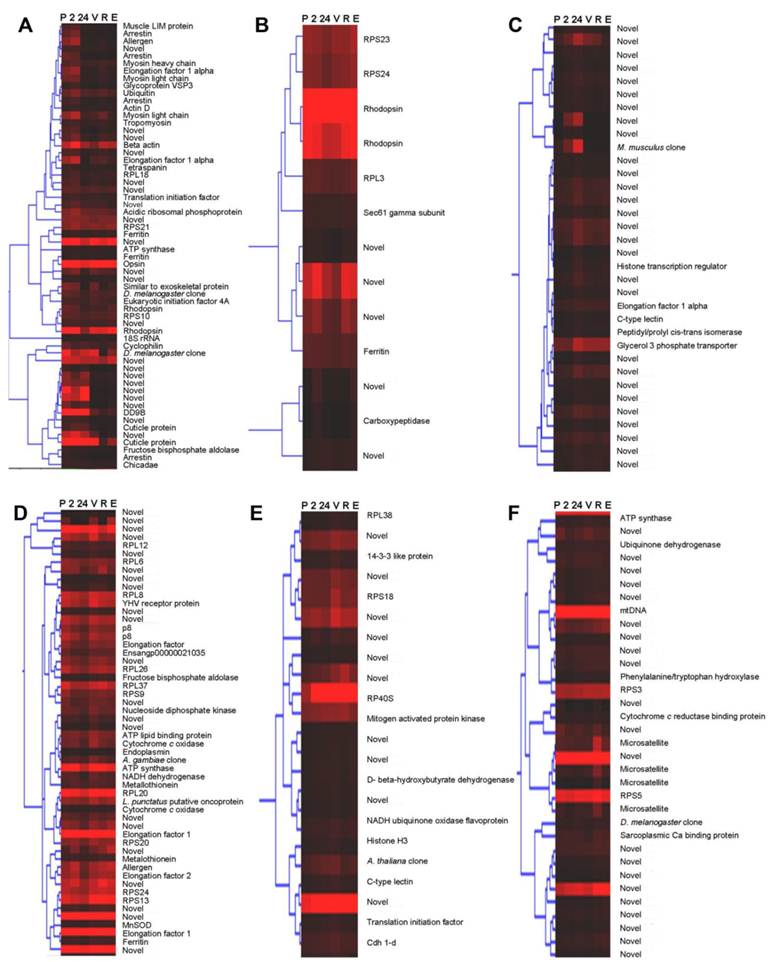
Clusters identified in the wild-caught cephalothorax data set contain a range of transcripts displaying elevated expression profiles at either collective pre-vitellogenic or vitellogenic stages. The identity of many of these transcripts reflects the dramatic shift in dynamics of energetic and metabolic processes towards mobilisation and biosynthesis of materials for ovarian maturation in tissues other than the ovary, including the hepatopancreas. Whereas, clusters identified in the wild-caught and captive-reared eyestalk data sets contain a range of transcripts identified as either expressed at similar levels at all maturation stages or displaying more transient expression profiles. Transcripts displaying BLAST homology to known genes/proteins while also displaying statistically significant differential expression (via Two-Sample Student's t tests (P<0.05)) between maturation stages, groups of stages, and animal source are discussed individually below.
We found that 1,3-β-D-glucan-binding high-density lipoprotein (pmβGBP-HDL) transcripts were up-regulated at vitellogenic compared with pre-vitellogenic stages in the wild-caught cephalothorax. βGBP-HDL has a bi-functional role in penaeids as a non-sex-specific HDL involved in lipid transport and as a pattern recognition protein (PRP) central to innate immunity [10]. The βGBP-HDL identified in Litopenaeus vannamei [11] is a lipoglycoprotein with ~50% lipid content, mainly phospholipids. βGBP-HDL transcripts have also been demonstrated to be predominantly expressed in the hepatopancreas by using RT-PCR [11]. Although most of the literature relevant to βGBP-HDL gene expression concerns its role as a PRP, βGBP-HDL gene expression has also been demonstrated to be influenced by starvation and lipid composition of diets in the hepatopancreas of juvenile L. vannamei [12]. Using density ultracentrifugation, it was demonstrated that the HDL fraction of ovarian homogenates in P. semisulcatus, comprising Vt and βGBP-HDL, is increased in mature compared with immature ovaries [13]. Up-regulation of βGBP-HDL gene expression in the cephalothorax at vitellogenic stages observed in this study provides further evidence of its involvement in delivery of lipid to developing oocytes.
A transcript with significant homology to the sterol carrier gene 2/3-oxoacyl-CoA thiolase (pmSCP2/SCPx) was isolated in the whole cephalothorax cDNA library. Whilst not identified as statistically significantly differentially expressed in t-Tests (P>0.05), the transcript displayed elevated expression levels at vitellogenic compared with pre-vitellogenic stages in the wild-caught cephalothorax. Conceptual protein domain analysis using the Pfam database confirmed that the incomplete P. monodon clone identified in this study contains the sterol-binding domain of SCP2. SCP2/SCPx is a fusion gene in many, but not all species, which encodes both the SCP2 and the SCPx with variations in their regulation occurring at the transcriptional and translational level [reviewed by [14]]. No complete nucleotide sequence exists yet for a crustacean and therefore the gene structure and mode of its regulation are currently unknown. In vertebrates, the SCP2 has many proposed roles in lipid metabolism and intracellular trafficking of cholesterol and other lipids, while the SCPx is involved in oxidation of branched-chain fatty acids FAs [14]. In vertebrates, the SCP2 has a range of ligand partners including Fatty Acids (Fas), fatty acyl CoAs, sterols and phospholipids [14], and therefore resolution of its full range of physiological functions is ongoing. SCP2 transcripts were detected in the ovary, gut, head and body in Aedes aegypti [15]. Based only on an EST identified in L. vannamei [16] the hepatopancreas remains so far the only site of SCP2 transcription identified in crustaceans. Up-regulation of SCP2/SCPx transcripts during vitellogenesis in this study is consistent with its possible involvement in processes associated with the mobilisation and metabolism of sterols towards both the accumulation of sterol in the ovaries and the production of reproductive steroid hormones. Given the magnitude of the accumulation of lipids in the ovary during vitellogenesis, and the multitude of ligand partners demonstrated for SCP2/SCPx, it is possible that SCP2/SCPx may also serve other key roles during ovarian maturation.
A transcript encoding a Kazal-type serine proteinase inhibitor (SPI) was identified as more highly expressed at vitellogenic compared with pre-vitellogenic stages in the wild-caught cephalothorax. SPIs are widely distributed in all multicellular organisms and play vital roles regulating many biological processes by limiting the level and extent of proteinase activity [17]. Kazal-type SPIs have been previously identified in the hepatopancreas and hemocytes [18] and of P. monodon. Recombinant SPI exhibited strong inhibitory activity against subtilisin in a tight-binding inhibition assay, suggesting a possible role as a defence component, and also against elastase, for which the inhibitory function is not known [18]. The elevated expression of kazal-type SPI transcripts at vitellogenic stages in the wild-caught cephalothorax in this study suggests a possible role for these proteins in regulating physiological processes associated with ovarian maturation. Also, a Manganese-superoxide dismutase (Mn-SOD) transcript encoding Mn-SOD was identified as more highly expressed at vitellogenic compared with pre-vitellogenic stages in the wild-caught cephalothorax. SODs are ubiquitous enzymes which provide the most important line of antioxidant defence systems against reactive oxygen species [19]. St. Clair et al., (1993) [20] also provide evidence of the importance of Mn-SODs ability to neutralise a cellular hyperoxidant state in processes of cellular differentiation. The elevated expression of transcripts at vitellogenic stages in the wild-caught cephalothorax in this study may be a reflection of the greater metabolic demand during ovarian maturation.
A total of 28 transcripts displayed higher expression levels at pre-vitellogenic stages compared with vitellogenic stages in the wild-caught cephalothorax. Of these transcripts, only six transcripts had been previously annotated, as identified through BLAST homology. These include several transcripts encoding TSP and SOP isoforms, a transcript encoding CP14 - a calcified cuticle protein and a transcript encoding profilin (chickadee). The roles of TSP and SOP outside the ovary in penaeids remain unclear. Expression of CP14 transcripts is likely moult cycle related. Kuballa et al., [21] have identified transcripts encoding a number of cuticular proteins displaying moult cycle stage-specific differential expression in the crab Portunis pelagicus. Profilins are ubiquitous proteins whose functions include sequestering actin monomers and inhibiting actin polymerization. They also have roles in cellular processes such as membrane trafficking, small-GTPase signalling and nuclear activities [22, 23]. The significance of the up-regulation of profilin transcripts at pre-vitellogenic stages in the wild-caught cephalothorax remains to be determined.
For this study, we chose to isolate RNA from whole intact eyestalks rather than dissect the MTXO-SG complex, given that its diffuse nature and proximity to retinal tissue increased the likelihood of irregular dissections. As such, transcripts detected in this study are likely to be expressed in other tissues besides the MTXO-SG including muscle, epidermis and tissues associated with sensory/photo perception. Key transcripts encoding known eyestalk neuropeptide hormones were not detected in wild-caught or captive-reared eyestalk samples at any ovarian maturation stage, suggesting low levels of transcript. However, several known and novel transcripts exhibited expression profiles suggestive of involvement in MTXO-SG function.
Transcripts encoding an arrestin family member were up-regulated at pre-vitellogenic stages [pre-ES ablation (P) stage and 2 hours post-ES ablation (2) stage] compared with later stages in the wild-caught ES. Although arrestins remain largely uncharacterised in crustaceans, it is well established that they form a family of proteins important for regulating the activity of GPCRs, with the different arrestins affecting the activity of their target GPCRs in different ways [24]. GPCRs form the largest family of cell-surface receptors, involved in numerous processes including hormonal system regulation, but also sensory/photo perception [25], and include receptors for biogenic amines. In particular, the biogenic amines dopamine and serotonin have been demonstrated to modulate release of various eyestalk neuropeptides. For example, dopamine has been shown to stimulate release of CHH from the eyestalk in the crab, Carcinus maenas [26]. Serotonin (5-hydroxytryptamine, 5-HT) injection has been demonstrated to induce ovarian maturation in penaeids [27, 28] and enhance release of CHH in the crayfish, Procambarus clarkia [29]. Ongvarrasopone et al., (2006) [30] have cloned a putative serotonin receptor from the ovary of P. monodon and further determined its expression in all tissues examined including the ES. It is possible that elevated arrestin levels at P and 2 stages in the wild-caught eyestalk indicate changes in GPCR related signalling activity at these stages, potentially associated with regulation of eyestalk neuropeptide release. These observations highlight the possibility that further functional characterisation of the various hormone receptors in the eyestalk, including their modes of regulation and probable coupling to excitation-release mechanisms in the neurosecretory cells of the eyestalk, may offer additional opportunities for manipulation of eyestalk neuropeptide levels. In addition, the results pertaining to arrestin and GPCR highlight further possibilities for manipulation of neuroendocrine functions.
A transcript encoding p8, an architectural transcription factor, was up-regulated at collective vitellogenic stages compared with collective pre-vitellogenic stages in the captive-reared but not the wild-caught ES, and also at pre-ES ablation (P) stage compared with 24 hours post-ES ablation (24) stage in the wild-caught ES. The p8 is a nuclear phosphoprotein related to the high mobility group class of proteins which function as architectural transcription factors, possessing considerable flexibility in regulating the expression of large numbers of genes [31]. Up-regulation at vitellogenic stages compared with pre-vitellogenic stages in the captive-reared eyestalk suggests p8s possible involvement in initiating expression of large numbers of genes in the eyestalk at vitellogenic stages, while also suggesting possible differences in transcriptional activity in the eyestalk between the two sources. Up-regulation in the wild-caught eyestalk at P stage compared with 24 stage also suggests possible reduced transcriptional activity in the eyestalk 24 hours post-ES ablation. Examination of the promoter regions of genes expressed in the eyestalk during ovarian maturation may further determine the extent of p8s involvement in these processes.
Up-regulation of ribosomal proteins and translation elongation factors at collective vitellogenic stages compared with collective pre-vitellogenic stages in both the wild-caught and the captive-reared eyestalk, and also at several vitellogenic stages compared with P stage in the wild-caught eyestalk, suggests increased protein biosynthesis in the eyestalk at vitellogenic stages. The significance of further transcripts displaying differential expression during ovarian maturation, which include transcripts encoding cell division, signalling and apoptosis proteins (14-3-3 like protein), ER protein translocation apparatus membrane proteins (sec 61 gamma subunit), actin-based motility proteins (myosin), as well as 26 novel transcripts, remains to be determined. This is also the case for further transcripts displaying differential expression between wild-caught and captive-reared eyestalk samples.
Incidental to this study, transcripts encoding opsin and rhodopsin, the GPCRs found in the photoreceptor cells of the retina [32] were up-regulated at both pre-vitellogenic and vitellogenic stages in the eyestalk of captive-reared compared with wild-caught animals and at pre-ES ablation stage compared with 24 h post-ES ablation stage in the eyestalk of wild-caught animals. Also, transcripts encoding two cuticle proteins were up-regulated at collective pre-vitellogenic stages compared with collective vitellogenic stages in the wild-caught ES. Commonality of these cuticular protein transcripts lies in their localization within the cuticle (established by Andersen, 1999 [33]), however, they display significant sequence variation and possess different cuticle protein domains. Differential expression of these cuticular transcripts, and indeed several other transcripts identified in this study, is likely modulated by moult-cycle stage rather than ovarian maturation stage. Kuballa et al., (2007) [21], demonstrated that transcripts encoding various cuticular proteins displayed moult-cycle stage specific differential expression in whole P. pelagicus, where differential expression occurred both between transcripts containing different domains and between transcripts containing the same domain. A transcript encoding DD9B - a further putative cuticular protein, identified as up-regulated in epithelial cells in post-moult Metapenaeus japonicus [34], was also up-regulated at collective pre-vitellogenic stages compared with collective vitellogenic stages in the eyestalk of wild-caught animals in this study.
Transcripts encoding known reproductive neuropeptide hormones were not detected in this study. This result could be attributed to low relative expression levels of these transcripts. In our laboratory, we have previously determined the inability to detect eyestalk neuropeptides when hybridising target tissue derived from whole animals to cDNA microarrays [21]. We had, however, anticipated that by targeting the eyestalk specifically, detectability of such rare transcripts would be improved. In this study we also utilised oligonucleotide arrays (synthesised 35-40 mers) rather than cDNA arrays. Several studies have demonstrated that the various oligonucleotide array platforms perform at least as well as cDNA arrays in detecting differential expression (eg., [35]). Moreover, Wang et al., (2003) [36] demonstrate that oligonucleotide probes and PCR amplicons perform equally well at detecting two-fold changes in rare transcript levels. Oligonucleotide arrays also have, in general, some advantages over cDNA arrays including higher uniformity of spots, higher specificity and more uniform melting temperatures. Amplification of signals on microarrays can be achieved using two basic methods: nucleic acid amplification prior to hybridisation and on-chip signal amplification post-hybridisation. Both methods potentially enable improved detection of rare transcripts. However, when using the former method, arrays are saturated at a lower target concentration as a result, while the latter method remains limited at the lower end of the dynamic range by the amount of signal detectable above background. A further explanation for the lack of neuropeptide transcripts identified in this study might be lack of adequate specificity/conservation of these sequences between P. monodon and the other species from which some heterologous sequences were obtained.
In summary, numerous transcripts displayed microarray based expression profiles of interest in the cephalothorax and eyestalk among wild-caught and captive-reared animals during ovarian maturation. In the cephalothorax, key transcripts include those encoding the 1,3-β-D-glucan-binding high-density lipoprotein and the sterol carrier gene 2/3-oxoacyl-CoA thiolase, both of which were up-regulated at vitellogenic stages indicative of their involvement in accumulation of lipids toward vitellogenesis. In the eyestalk, up-regulation of arrestin transcripts at pre-vitellogenic stages indicates probable modulation of GPCR activity at these stages, potentially associated with biogenic amine mediated release of reproductive neuropeptides. This observation highlights the prospects that improved understanding of hormone receptor regulation and function in the cephalothorax and eyestalk may, in time, provide opportunities for alternative methods of manipulation of neuropeptide levels. Additionally, transcripts encoding the architectural transcription factor p8 were up-regulated at vitellogenic stages, suggestive of its involvement in initiating expression of large numbers of genes at vitellogenic stages.
Acknowledgements
We thank the Bribie Island Aquaculture Research Centre and the Australian Institute for Marine Science for providing the captive-reared animals, consumables and facilities used in this study and Mr Bill Izard for the supply of wild-caught prawn broodstock. We are grateful to Mrs Hazra Thaggard for her expert assistance and Dr Gay Marsden for her expert contribution towards the analysis of prawn reproductive stages. PB was a University of Queensland APA recipient and the PhD thesis is available at http://espace.library.uq.edu.au/view/UQ:231165. We gratefully acknowledge the financial support of The Fisheries Research and Development Corporation for grant 2005/205 to WRK and the University of the Sunshine Coast internal research grant URG08/03 to AE and WRK.
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Chang ES. Hormonal control of molting in decapod crustacea. Am Zool. 1985;25:179-85
2. Adiyodi RG, Subramonium T. Arthropoda-Crustacea. Reproductive Biology of Invertebrates. Adiyodi KG, Anilkumar G, eds. New York: John Wiley and Sons Ltd. 1983:443-95
3. DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. β-Arrestins and cell signaling. Annu Rev Physiol. 2007;69:483-510
4. De Kleijn DPV, Janssen KPC, Waddy SL. et al. Expression of the crustacean hyperglycaemic hormones and the gonad-inhibiting hormone during the reproductive cycle of the female American lobster Homarus americanus. J Endocrin. 1998;156:291-8
5. Ohira T, Watanabe T, Nagasawa H, Aida K. Molecular cloning of a molt-inhibiting hormone cDNA from the kuruma prawn Penaeus japonicus. Zoolog Sci. 1997:785-9
6. Duronslet MJ, Yudin IA, Wheeler RS, Clark WHJ. Light and fine structural studies of natural and artificially induced egg growth of penaeid shrimp. Proc Wld Maricult Soc. 1975;6:105-22
7. Bell TA, Lightner DV. A handbook of normal penaeid shrimp histology. Baton Rouge: World Aquaculture Society. 1988
8. Tan-Fermin JD, Pudadera RA. Ovarian maturation stages of the wild giant tiger prawn, Penaeus monodon Fabricius. Aquaculture. 1989;77:229-42
9. Altschul SF, Madden TL, Schaffer AA. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25:3389-402
10. Barracco MA, Duvic B, Soderhall K. The β-1,3-glucan-binding protein from the crayfish Pacifastacus leniusculus, when reacted with a b-1,3-glucan, induces spreading and degranulation of crayfish granular cells. Cell Tissue Res. 1991;266:491-7
11. Romo-Figueroa MG, Vargas-Requena C, Sotelo-Mundo RR. et al. Molecular cloning of a β-glucan pattern-recognition lipoprotein from the white shrimp Penaeus (Litopenaeus) vannamei: correlations between the deduced amino acid sequence and the native protein structure. Dev Comp Immunol. 2004;28:713-26
12. Muhlia-Almazán A, Sánchez-Paz A, García-Carreño F. et al. Starvation and diet composition affect mRNA levels of the high density-lipoprotein-β glucan binding protein in the shrimp Litopenaeus vannamei. Comp Biochem Physiol Part B: Biochem Mol Biol. 2005;142:209-16
13. Ravid T, Tietz A, Khayat M. et al. Lipid accumulation in the ovaries of a marine shrimp penaeus semisulcatus (de haan). J Exp Biol. 1999;202(Pt 13):1819-29
14. Gallegos AM, Atshaves BP, Storey SM. et al. Gene structure, intracellular localization, and functional roles of sterol carrier protein-2. Prog Lipid Res. 2001;40:498-563
15. Krebs KC, Lan Q. Isolation and expression of a sterol carrier protein-2 gene from the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2003;12:51-60
16. Zhao Z-Y, Yin Z-X, Weng S-P. et al. Profiling of differentially expressed genes in hepatopancreas of white spot syndrome virus-resistant shrimp (Litopenaeus vannamei) by suppression subtractive hybridisation. Fish Shellfish Immunol. 2007;22:520-34
17. Laskowski JM, Kato I. Protein inhibitors of proteinases. Ann Rev Biochem. 1980;49:593-626
18. Somprasong N, Rimphanitchayakit V, Tassanakajon A. A five-domain Kazal-type serine proteinase inhibitor from black tiger shrimp Penaeus monodon and its inhibitory activities. Dev Comp Immunol. 2006;30:998-1008
19. Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33:337-49
20. St. Clair DK, Oberley TD, Muse KE, St Clair WH. Expression of manganese superoxide dismutase promotes cellular differentiation. Free Radic Biol Med. 1994;16:275-82
21. Kuballa AV, Merritt DJ, Elizur A. Gene expression profiling of cuticular proteins across the moult cycle of the crab Portunus pelagicus. BMC Biol. 2007;10:45
22. Rawe VY, Payne C, Schatten G. Profilin and actin-related proteins regulate microfilament dynamics during early mammalian embryogenesis. Human reproduction (Oxford, England). 2006;21:1143-53
23. Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461-9
24. Ferguson SS. Evolving concepts in G protein-coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol Rev. 2001;53:1-24
25. Gurevich VV, Benovic JL. Visual arrestin interaction with rhodopsin. Sequential multisite binding ensures strict selectivity toward light-activated phosphorylated rhodopsin. J Biol Chem. 1993;268:11628-38
26. Luschen W, Willig A, Jaros PP. The role of biogenic amines in the control of blood glucose level in the decapod crustacean, Carcinus maenas L. Comp Biochem Physiol C Comp Pharm Toxic. 1993;105:291-6
27. Vaca AA, Alfaro J. Ovarian maturation and spawning in the white shrimp, Penaeus vannamei, by serotonin injection. Aquaculture. 2000;182:373-85
28. Wongprasert K, Asuvapongpatana S, Poltana P. et al. Serotonin stimulates ovarian maturation and spawning in the black tiger shrimp Penaeus monodon. Aquaculture. 2006;261:1447-54
29. Lee C-Y, Yang P-F, Zou H-S. Serotonergic regulation of crustacean hyperglycemic hormone secretion in the crayfish, Procambarus clarkia. Physiol Biochem Zool. 2001;74:376-82
30. Ongvarrasopone C, Roshorm Y, Somyong S. et al. Molecular cloning and functional expression of the Penaeus monodon 5-HT receptor. Biochim Biophys Acta. 2006;1759:328-39
31. Reeves R, Beckerbauer L. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim Biophys Acta. 2001;1519:13-29
32. Hargrave PA, McDowell JH. Rhodopsin and phototransduction: a model system for G protein-linked receptors. FASEB J. 1992:2323-31
33. Andersen SO. Exoskeletal proteins from the crab, Cancer pagurus. Comp Biochem Physiol A Mol Integr Physiol. 1999;123:203-11
34. Watanabe T, Persson P, Endo H, Kono M. Molecular analysis of two genes, DD9A and B, which are expressed during the postmolt stage in the decapod crustacean Penaeus japonicus. Comp Biochem Physiol B Biochem Mol Biol. 2000;125:127-36
35. Lee H-S, Wang J, Tian L. et al. Sensitivity of 70-mer oligonucleotides and cDNAs for microarray analysis of gene expression in Arabidopsis and its related species. Plant Biotechnol J. 2003;2:45-57
36. Wang H-Y, Malek R, Kwitek AE. et al. Assessing unmodified 70-mer oligonucleotide probe performance on glass-slide microarrays. Genome Biol. 2003;4:5
37. Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. Journal of the Royal Statistical Society: Series B (Statistical Methodology). 2002;63:411-23
Author contact
![]() Corresponding author: Prof. Abigail Elizur, Faculty of Science, Health and Education, University of the Sunshine Coast, Maroochydore, QLD 4558. Phone: +61 7 5459 4813. Fax: +61 7 5430 2889. Email: aelizuredu.au.
Corresponding author: Prof. Abigail Elizur, Faculty of Science, Health and Education, University of the Sunshine Coast, Maroochydore, QLD 4558. Phone: +61 7 5459 4813. Fax: +61 7 5430 2889. Email: aelizuredu.au.

 Global reach, higher impact
Global reach, higher impact