Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2012; 8(4):459-469. doi:10.7150/ijbs.3821 This issue Cite
Research Paper
OLFML3 Expression is Decreased during Prenatal Muscle Development and Regulated by MicroRNA-155 in Pigs
1. State Key Laboratory for Animal Nutrition, Beijing 100193, P. R. China;
2. The Key Laboratory for Domestic Animal Genetic Resources and Breeding of Ministry of Agriculture of China, Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing 100193, P. R. China;
3. College of Animal Science and Technology, Northwest A & F University, No. 22 Xinong Road, 712100 Yangling, Shaanxi, P. R. China;
4. College of Animal Science and Technology, Northeast Agricultural University, Haerbin, 150030, P.R.China.
Received 2011-11-17; Accepted 2011-12-30; Published 2012-3-3
Abstract
The Olfactomedin-like 3 (OLFML3) gene has matrix-related function involved in embryonic development. MicroRNA-155 (miR-155), 21- to 23-nucleotides (nt) noncoding RNA, regulated myogenesis by target mRNA. Our LongSAGE analysis suggested that OLFML3 gene was differently expressed during muscle development in pig. In this study, we cloned the porcine OLFML3 gene and detected its tissues distribution in adult Tongcheng pigs and dynamical expression in developmental skeletal muscle (12 prenatal and 10 postnatal stages) from Landrace (lean-type) and Tongcheng (obese-type) pigs. Subsequently, we analyzed the interaction between OLFML3 and miR-155. The OLFML3 was abundantly expressed in liver and pancreas, moderately in lung, small intestine and placenta, and weakly in other tissues and postnatal muscle. There were different dynamical expression patterns between Landrace and Tongcheng pigs during prenatal skeletal muscle development. The OLFML3 was down-regulated (33-50 days post coitus, dpc), subsequently up-regulated (50-70 dpc), and then down-regulated (70-100 dpc) in Landrace pigs, while in Tongcheng pigs, it was down-regulated (33-50 dpc), subsequently up-regulated (50-55 dpc) and then down-regulated (55-100 dpc). There was higher expression in Tongcheng than Landrace in prenatal muscle from 33 to 60 dpc, and opposite situation from 65 to 100 dpc. Dual luciferase assay and real time PCR documented that OLFML3 expression was regulated by miR-155 at mRNA level. Our research indicated that OLFML3 gene may affect prenatal skeletal muscle development and was regulated by miR-155. These finding will help understanding biological function and expression regulation of OLFML3 gene in mammal animals.
Keywords: pig, OLFML3, microRNA-155, skeletal muscle, development.
Introduction
Olfactomedin-like 3 (OLFML3) is a member of the olfactomedin-like gene family, containing an olfactomedin(OLF) domain in the C-terminal region and a less well conserved coiled-coil domain in the N-terminal region [1]. Since olfactomedin was first cloned from frog olfactory tissue [2], more than 100 known OLF members have been discovered in various species ranging from Caenorhabditis elegans to Homo sapiens [3]. Many OLF members have been demonstrated to play important roles in various physiological processes. Myocilin, a member of OLF family, is similar to OLFML3 in structure, and the mutations of its protein are closely associated with primary open angle glaucoma and the identification will aid in early diagnosis [4-8]. Noelin plays an important role in neural development [9, 10]. The OLFML3 gene, also known as HNOEL-iso and hOLF44 in humans [11], ONT3 in mice [12] and ONT1 in chicken and Xenopus [13, 14], was dropped into subfamily VII on the basis of phylogenetic analysis [3]. Furthermore, the OLFML3 gene may have important roles in embryonic development [3], however, to our knowledge, the functional properties and regulation mechanism of OLFML3 gene remained unclear.
The gene expressions are mediated by many factors, such as transcription factors, microRNA and methylation. MicroRNAs (miRNAs) are evolutionary conserved small noncoding RNAs of 21- to 23- nucleotides that regulate genes expression by inhibiting translation of target messenger RNA (mRNA) transcripts binding to the 3′-untranslated regions (3′UTRs), leading to mRNA cleavage, translational repression, or both [15]. In recent years, miRNAs have emerged as critical regulators of gene expression. They regulate 30% of all animal genes in a variety of biological processes, including development, immunity, cell differentiation, apoptosis, metabolism and signal transduction [16-21]. Many studies documented that miRNAs play important roles in mammalian skeletal muscle development [22-26]. Bioinformatics prediction showed that OLFML3 was regulated by miR-155. MiR-155, a typical multifunctional miRNA, has been proved to be associated with cell cycles [27] and may accelerate cell proliferation [28] and myogenic differentiation [29].
In the present study, we first cloned and analyzed porcine OLFML3 gene and investigated its spatial distribution in adult Tongcheng pigs and dynamical expression patterns during skeletal muscle development (22 stages) from Landrace (lean-type) and Tongcheng (obese-type) pigs by real-time polymerase chain reaction (PCR). Subsequently, dual luciferase reporter assay and real-time PCR were used to explore the interaction between OLFML3 and miR-155 in PK15 and PIEC cells. The facts suggested that OLFML3 gene may affect prenatal skeletal muscle development and was regulated by miR-155 at mRNA level in pig.
Materials and methods
Animal samples and data collection
All animals were treated according to the animal procedures defined by national and local animal welfare bodies, and all animal work was approved by the Hubei Province, P. R. China for Biological Studies Animal Care and Use Committee.
Twelve tissues, including heart, liver, spleen, lung, kidney, stomach, fat, longissimus dorsi, small intestines, pancreas, placenta and uterus were collected from three adult Tongcheng pigs for spatial expression analysis. After copulation with the boar of each corresponding breed, the pigs were then sacrificed at a commercial slaughterhouse at the selected stages. Skeletal muscle at twelve prenatal developmental stages ( 33, 40, 45, 50, 55, 60, 65, 70, 75, 80, 90 and 100 days post coitus (dpc)) and ten postnatal stages ( 0, 9, 20, 30, 40, 60, 80, 120, 160 day and adult) from Landrace pigs and Chinese indigenous Tongcheng pigs were collected for dynamical expression analysis. All the tissue samples were harvested, immediately frozen in liquid nitrogen, and then stored at -80℃ until used.
Identification and isolation of porcine OLFML3 gene
From our LongSAGE library of swine prenatal skeletal muscle [30, 31], a differently expressed tag (GGACTCCAGATCTTGAG) that potentially represented the OLFML3 gene was selected for further analysis. We converted this tag sequence into its corresponding 3′cDNA by using the GLGI (generation of longer cDNA fragments from SAGE tags for gene identification) method [31]. Primers (GLGI-F, GLGI-R, Table 1) were used to amplify the sequences, which were then subjected to basic local alignment search tool (BLAST) searches for gene identification. To validate the GLGI result and obtain the 3′UTR, SMART RACE technology (Clontech, Palo Alto, CA, USA) was used to clone porcine OLFML3 transcripts containing the 3′-end. For first-strand cDNA synthesis and the porcine OLFML3 gene sequence obtained earlier, a gene-specific primer was designed for the 3′-RACE PCR (3′-GSP, Table 1). Reverse transcriptase and PCR were carried out according to the instruction manual of the SMART RACE cDNA amplification kit. The 3′-RACE products were gel-purified prior to sequencing.
Porcine expression sequence tags (ESTs) were obtained by using the basic local alignment search tool (BLAST) algorithm (http://blast.ncbi.nlm.nih.gov/) and the human mRNA sequence of OLFML3 (olfactomedin-like 3, GenBank accession no. NM_020190.2). The porcine ESTs that shared more than 80% sequence identity with human mRNA were selected and assembled using the Seqman program (DNAStar, Inc., Madison, WI, USA). Primer pairs (CDS-F, CDS-R, Table 1) were designed from the assembled EST contigs by using Primer 5.0 software to amplify the porcine OLFML3 cDNA sequence from longissimus dorsi of Tongcheng pigs. PCR was performed in a volume of 10 µL containing l µL 10 × PCR buffer (Mg2+ plus), 0.8 µL 2.5 mM dNTP mix, 0.3 µL of each PCR primer (100 ng/µL), 0.1 µL 5 U r-Taq DNA polymerase (Fermentas, Vilnius, Lithuania), 1 µL cDNA and 6.5 µL sterile water. The PCR reaction conditions were 5 min at 95 °C, followed by 26 cycles at 94℃ for 30 s, 60℃ for 30 s, 72℃ for 1 min, and followed by a final extension of 72℃ for 5 min. The PCR products were purified and subsequently cloned into the pMD18-T vector (Trans, Beijing, China) and seeded on Ampicillin plates. Minipreps were obtained from colonies grown in 1 ml LB-Amp broth for sequencing.
Primers information in experiments.
| Primer symbol | Primer sequence(5′-3′) | Binding region | Size(bp) | PCR(TM)(℃) |
|---|---|---|---|---|
| 5′UTR-F | GCTTTGAACCCTTGATATTG | 5′UTR | 60.0 | |
| 5′UTR-R | CCGACCACGACAAAAGGACC | Exon 1 | ||
| CDS-F | GCTGGCCGCACGACTGCACG | Exon 1 | 1687 | 61.0 |
| CDS-R | GGGCACTGAAGGGAGAAAGG | 3′UTR | ||
| Intron1-F | GCTGGCCGCACGACTGCACG | Exon 1 | 946 | 60.0 |
| Intron1-R | AGCGGCAGCATCTTGTTCTT | Exon 2 | ||
| Intron2-F | CGGGAGGCGCTGAGAACTG | Exon 2 | 681 | 58.0 |
| Intron2-R | GGTGAAGTCACGCAGCCTT | Exon 3 | ||
| GLGI-F | CATGGGACTCCAGATCTTGAG | 60.9 | ||
| GLGI-R | ACTATCTAGAGCGGCCGCTT | |||
| 3′-GSP | GCGGGGGTGGTGACTTGGAGAACACT | Exon 3 | ||
| miR-155-F | TAAAAAGGACAGGAATACA | 653 | 60.0 | |
| miR-155-R | CAAGTCTTCAGATACAATG | |||
| EXP-F | TTCCCAAGGCTGCGTGAC | Exon 3 | 210 | 60.0 |
| EXP-R | CACCACTGTGCGGTTTGC | Exon 3 | ||
| GAPDH-F | GGGCATGAACCATGAGAAGT | 233 | 60.0 | |
| GAPDH-R | AAGCAGGGATGATGTTCTGG | |||
| ssc-miR-155-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTACCCCTA | |||
| ssc-miR-155-F | GGAGGTTAATGCTAATTGTGATAG | 65 | 56.0 | |
| ssc-miR-155-R | CTCAACTGGTGTCGTGGAGTC | |||
| U6-F | GCTTCGGCAGCACATATACTAAAAT | 89 | 56.0 | |
| U6-R | CGCTTCACGAATTTGCGTGTCAT | |||
| 3′-UTR-F | GGGAGCTGGTCTTTGTTTT | 3′UTR | 485 | 60.0 |
| 3′-UTR-R | TTTGCAGAGGATTTTCCTT | 3′UTR | ||
Bioinformatics analysis
The TargetScan program (Version 5.1) (http://www.targetscan.org/) and PicTar WEB INTERFACE (http://pictar.mdc-berlin.de/cgi-bin/PicTar_vertebrate.cgi?species=vertebrate&from_data=target predictions for all human microRNAs based on conservation in mammals (human, chimp, mouse, rat, dog)) were used to scan potential miRNA target sites for the OLFML3 gene with the default parameters [32]. The results showed that OLFML3 gene was targeted by only miR-155 and contains a match with nucleotides 2 to 7 of the miRNA with the target sequences (G: U base-pairing was tolerated) in both human and mouse. Furthermore, there is remarkable conservation of the OLFML3 3'UTR sequence from Xenopus tropicalis to human within and around the predicted miR-155 target site, and the seed sequences of miR-155 are also highly evolutionarily conserved across multiple species. Consequently, the OLFML3 gene containing the miR-155 binding site within its 3'UTRs in pig was further analyzed.
Plasmid construct
A fragment of 485 bp encompassing the entire OLFML3 3'UTR was cloned from Tongcheng pig genomic DNA using gene-specific primers (3′-UTR-F, 3′-UTR-R, Table 1) and inserted downstream of the Renilla Luciferase ORF in the psiCHECK-2 vector (Promega, Madison, WI, USA) using the NotI and XhoI restriction sites. For detecting the fold change of OLFML3 mRNA, the Renilla luciferase ORF in the psiCHECK-2 vector was replaced with the OLFML3 CDS with NheI and XhoI sites. The sequence of porcine miR-155 was obtained according to miRBase (http://www.mirbase.org/) (ssc-mir-155, MI0015907) [33]. The approximately 653bp sequence, which included the miR-155 hairpin precursor and its native flanking sequence, were amplified by PCR from Tongcheng pig genomic DNA (ssc-miR-155-F, ssc-miR-155-R, Table 1) and subcloned into the pcDNA3.1 (+) (Invitrogen, Carlsbad, CA, USA) expression vector. All sequences were confirmed by DNA sequencing.
Cell culture
PK15 (porcine kidney epithelial cell line) and PIEC (porcine iliac endothelial cell line, Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, P. R. China) were cultured in Dulbecco's modified Eagle's medium (DMEM) with high glucose (Gibco, Invitrogen, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (Gibco, Invitrogen, Carlsbad, CA, USA) and 1% glutamine, in a 37°C incubator with 5% CO2.
Luciferase assay
PK15 and PIEC cells were seeded at 8 × 104 cells per well in 24-well plates (Corning Incorporated, NY, USA), 24 h after the plating, cells were transfected by FuGENE® HD Transfection Reagent (Roche, Basel, Switzerland) according to the manufacturer's instructions. In each well, 100 ng psiCHECK-2-3'UTR or mutant and 400 ng pcDNA3.1 (+)-miR-155 or pcDNA3.1 (+) vector (control) were cotransfected. After transfection for 36 h, cells were harvested by the addition of 500 μL passive lysis buffer. Renilla and firefly luciferase activities in the cell lysate were measured with the Dual Luciferase assay system (Promega, Madison, WI, USA) in a TD-20/20 luminometer (Turner Biosystems, Sunnyvale, CA, USA) and the Renilla luciferase signal was normalized to the firefly luciferase signal. The normalized Renilla luciferase activity was compared with the control, miR-155 and mutant using the Student's t-test (p<0.05).
Real-time quantitative PCR
To detect whether the OLFML3 mRNA levels were affected by overexpression of miR-155, we seeded PK15 cells into 12-well plates; and after transfection for 48 h, total RNA was harvested and detected by real time PCR. The manipulation was performed in triplicate to confirm the result. The tissues distribution and dynamical expression of OLFML3 were also assayed by RT-PCR.
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA); 1ug total RNA was reverse transcribed in a final volume of 20 µl using the RevertAid First Strand cDNA Synthesis Kit (Fermentas, Vilnius, Lithuania). Gene-specific primers (EXP-F, EXP-R and GAPDH-F, GAPDH-R, Table 1) were designed for quantitative PCR. Each real-time PCR reaction was performed in a final volume of 20 µl reaction mixture containing SYBR Premix Ex Taq (2×), Rox Reference Dye Ⅱ (50×) (TaKaRa, Dalian, China), gene-specific primers, template cDNA and sterile water. The PCR amplification was performed on a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) under the following cycling conditions: 30 s at 95 °C, followed by 40 cycles at 95 °C for 5 s, 60 °C for 30 s and 72 °C for 34 s. RT-PCRs were performed in triplicate and porcine glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified as an internal control.
The overexpression levels of mature miR-155 were determined by stem-loop real-time PCR [34]. After the reverse transcription reaction, the level of miR-155 was detected using a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) under the following conditions: after an initial denaturation at 95 °C for 30 s, the amplification was performed by denaturation at 95 °C for 5 s, annealing and extension at 56 °C for 34 s for 40 cycles. The expression levels of U6 small nuclear RNA (U6 snRNA) were also determined by real-time PCR and were used to normalize the miR-155.
Gene expression levels were quantified relative to internal control expression using Gene Expression Macro software (Bio-Rad, Richmond, CA, USA), and the results were analyzed by the 2-△△Ct method [35].
Results
Sequence characterization of porcine OLFML3 gene
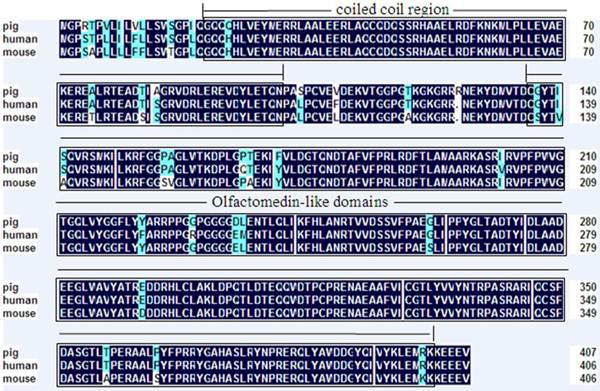
The sequence obtained from GLGI and 3′-RACE has 72% and 82% of identical to sequence from Homo sapiens OLFML3, respectively, suggested that the LongSAGE tag represented the swine OLFML3 gene. Compared with the available cDNA and DNA sequences of human and mouse OLFML3, a total of 2,872-bp genomic sequences, including three exons and two introns, were cloned in our experiment (GenBank accession no. JN699054). Sequence analysis revealed that porcine OLFML3 cDNA comprised a 1224-bp open reading frame (ORF) flanked by a 120-bp 5′UTR and 500-bp 3′UTR sequences, with a consensus ATTAAA polyadenylation signal 16 bp upstream of the poly (A) stretch. Porcine OLFML3 gene shows 86% and 87% identity with human (NM_020190.2) and mouse (NM_133859.2) counterparts, respectively. Further comparison revealed that each of the 5′-donor and 3′-acceptor splice sites conformed to the GT-AG rule (Table 2). Porcine OLFML3 was predicted to encode a polypeptide of 407 amino acid residues with a calculated molecular mass of 45.9 kD and an isolectric point of 6.6765. The predicted porcine OLFML3 polypeptide exhibits 94%, 94%, 92%, 94% and 65% similarity with human (NP_064575.1), cattle (NP_001068665.1), mouse (NP_598620.2), rat (NP_001101178) and gallus (NP_001035107.1) homologues, respectively. Amino acid sequence analysis also showed that the precursor protein of OLFML3 contains coiled coil region and Olfactomedin-like domain, similar to the domains of the human and mouse (Fig. 1 and Fig. 2).
The exon ⁄ intron organizations of porcine OLFML3 gene.
| No. | Exon size (bp) | Intron size (bp) | 5' splice donor | 3' Splice acceptor |
|---|---|---|---|---|
| 1 | 114 | 704 | TTAGAGgtaagt | |
| 2 | 289 | 324 | cttcagGAACGG | TGACAGgtaagt |
| 3 | 821 | caccagACTGTG |
Coding sequences are shown in uppercase letters; noncoding regions are in lowercase letters. The conserved GT ⁄ AG junctions are shown in italic font.
Deduced amino acid sequence of porcine OLFML3 compared with mouse OLFML3 (GenBank accession no. NP_598620.2) and human OLFML3 (GenBank accession no. NP_064575.1). Shading shows identical amino acid residues among the three species. Common structural domains are indicated by boxes including coiled coil region and Olfactomedin-like domains.
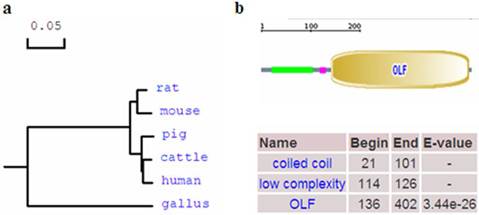
Phylogenetic tree and domain of porcine OLFML3 protein. (a) Phylogenetic tree of OLFML3 in different species. The GenBank accession numbers of those sequences are as follows: human: NP_064575.1, cattle: NP_001068665.1, rat: NP_001101178.1, mouse: NP_598620.2, Gallus: NP_001035107.1. The porcine OLFML3 amino acid sequence was deduced from the full cDNA sequence in this study. (b) Domain of porcine OLFML3 protein. The porcine OLFML3 precursor protein contains coiled coil region and Olfactomedin-like domain.
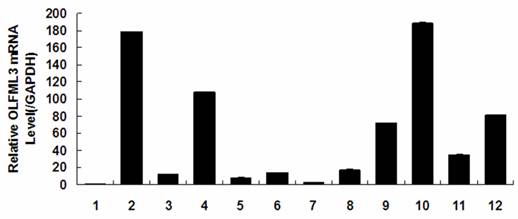
Tissue distribution and dynamical expression of OLFML3 gene
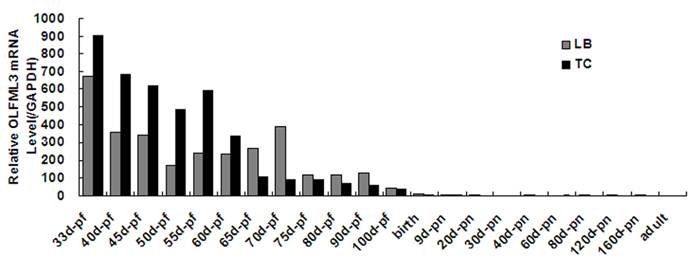
Quantitative real-time PCR analysis showed that porcine OLFML3 mRNA expression varied greatly in diverse tissues, it was highly expressed in liver and pancreas, moderately in lung, small intestines and placenta, and weakly in other tissues (Fig. 3). As shown in Fig. 4, the OLFML3 gene exhibited different dynamical expression patterns in prenatal skeletal muscle and a lower level in post-natal stages between Landrace and Tongcheng pigs. In the prenatal skeletal muscle development of Landrace pigs, the OLFML3 was down-regulated during 33-50 days, subsequently up-regulated during 50-70 days, and then down-regulated during 70-100 days. While in Tongcheng pigs, the OLFML3 was down-regulated at 33-50 dpc, subsequently up-regulated at 50-55 dpc and then down-regulated at 55-100 dpc. Moreover, there was higher expression in prenatal muscle of 33-60 dpc in Tongcheng pigs than in Landrace, while this expression pattern is reversed at 65-100 dpc.
OLFML3 expression in different tissues of the adult Tongcheng pig. Tissues were (1) heart, (2) liver, (3) spleen, (4) lung, (5) kidney, (6) stomach, (7) fat, (8) longissimus dorsi, (9) small intestines, (10) pancreas, (11) uterus, and (12) placenta. The values are the average (±SE) levels of OLFML3 normalized to GAPDH from three independent experiments. The value of OLFML3 in heart was arbitrarily set to 1.0.
Relative OLFML3 mRNA expression level in porcine skeletal muscle from Landrace and Tongcheng pigs at different developmental stages. Expression was normalized to GAPDH and expression levels are the average (±SE) of three independent experiments during all stages. The value of OLFML3 in adult of Tongcheng pigs was arbitrarily set to 1.0. pf, post- fecundation; pn, post-natal.
OLFML3 was a target of microRNA-155
Using TargetScan and PicTar target prediction programs, OLFML3 was identified as a putative miR-155 target in human and mouse. The target site between OLFML3 and miR-155 is highly conserved among mammals (Fig. 5). To validate whether OLFML3 is directly targeted by miR-155 in pig, a dual luciferase assay was performed in PK15 and PIEC cells. The result showed that the luciferase activity of psiCHECK-2, which contains the OLFML3 3'UTR sequence, was significantly decreased (65.1% of control in PK15 and 62.9% of control in PIEC cell line). To further validate the specific target site, we mutated the binding sites of the OLFML3 3'UTR and the results indicated that the luciferase activity of psiCHECK-2, which contains the mutant, was not affected by miR-155 (Fig. 6).
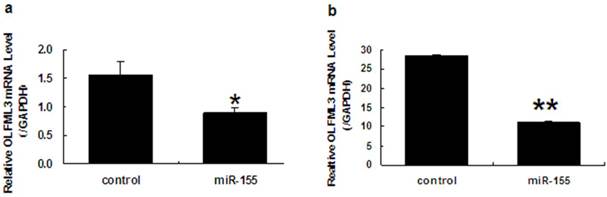
Since OLFML3 is a target of miR-155, we investigated whether endogenous mRNA levels of OLFML3 were affected by the overexpression of miR-155. PK15 cells were transfected with control and miR-155 overexpression vectors, and total cellular RNA was harvested 48 h after transfection. Mature miR-155 and OLFML3 mRNA levels were tested by RT-PCR. The result showed that overexpression of miR-155 down-regulated OLFML3 mRNA to 60% of the level in control cells (Fig. 7). To validate this result, we cotransfected the miR-155 overexpression vector and psiCHECK-2, which contains the OLFML3 mRNA sequence, in the PK15 cells, real-time PCR results suggested that the OLFML3 mRNA level was downregulated 61% of the level in control cells by overexpression of miR-155 (Fig. 7). These results documented that the OLFML3 gene was regulated by miR-155 in pigs.
OLFML3 3'UTR has conserved miR-155 target sites. (a) OLFML3 was predicted as a target of miR-155 using the PicTar. (b) OLFML3 was predicted as a target of miR-155 using TargetScan program (Version 5.1). (c) The complementary site for OLFML3 and miR-155 was mapped using DNAMAN. The upper part represents OLFML3 and the lower part represents miR-155. The corresponding plot shows conservation across species.

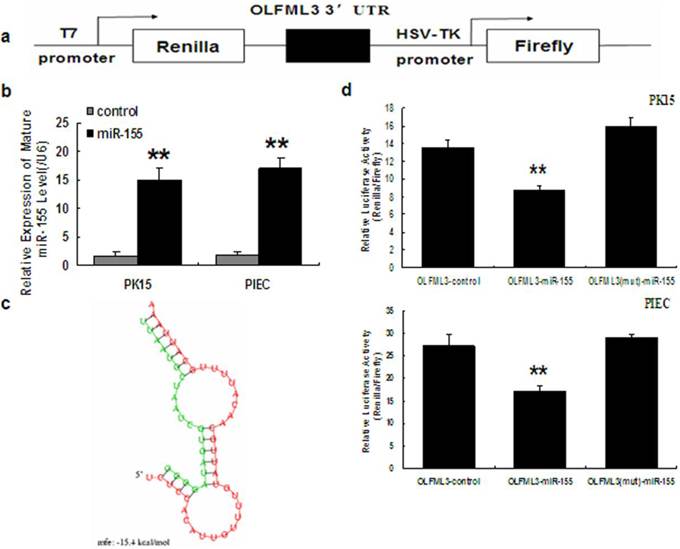
OLFML3 is a primary target for miR-155. a. Schematic representation of the construct used in the luciferase assay. A 485-bp region of the porcine OLFML3 3'UTR containing one putative miR-155 target site was cloned into the psiCHECK-2 vector. b. Overexpression of porcine miR-155 in PK15 and PIEC cells. Histogram indicates overexpression of miR-155 after transfection for 48 h. Mature miR-155 was analyzed by real-time PCR. c. miR-155:OLFML3 3'UTR duplex. The sequence marked in red is OLFML3 3'UTR, and the sequence marked in green is the seed sequence of miR-155 in human. d. Cotransfection of porcine pre-miR-155 or control and porcine OLFML3 3'UTR-derived psiCHECK-2 construct or mutant in PK15 and PIEC cells. Renilla activity at 48 h post-transfection shows a significant decrease in normalized values than in control and mutant in both cell types. Five replicates were performed for each group and the experiment was repeated at least three times. ** indicates a p-value of less than 0.01 in a Student's t-test.
MiR-155 regulates OLFML3 at the mRNA level. a. Transfection of porcine pre-miR-155 in PK15 cells shows a significant decrease in the normalized endogenous OLFML3 mRNA level. b. Cotransfection of porcine pre-miR-155 and porcine OLFML3 -CDS-3′UTR- psiCHECK-2 construct in PK15 cells shows a significant decrease in the normalized OLFML3 mRNA level. Three replicates were performed for each group and the experiment repeated at least three times. * indicates a p-value of less than 0.05 in a Student's t-test and ** indicates a p-value of less than 0.01 in a Student's t-test.
Discussion
The results of our study provide the complete fundamental structure of the porcine OLFML3 gene. Comparison of its genome structure with that human OLFML3, demonstrated remarkably high similarity between the two species; they both contained three exons and two introns. The coding sequence (CDS) region and the deduced amino acid sequence of porcine OLFML3 shared 92% and 94% identity to that of human respectively. The Olfactomedin-like domain, may be involving intracellular accumulation of folded proteins [36], is highly conserved among human, mouse and pig.
Comparing our results with human and mouse OLFML3 microarray expression data (http://genome.ucsc.edu/), we found that the expression pattern of porcine OLFML3 gene was partial coincided with the expression of the corresponding gene in human and mouse, it was expressed at considerable levels in liver and lung. Our results were also in agreement with the northern blot analysis results in human tissues [11].
OLFML3 gene play an important roles in embryonic development [3], our study documented that it exhibits different expression patterns during prenatal skeletal muscle development from Landrace to Tongcheng pigs, indicating that OLFML3 may affect prenatal skeletal muscle development in porcine. The previous studies showed that the establishment of the total number of fibers involves two major waves of fiber generation: primary muscle fiber formation at 35-60 dpc and secondary muscle fibers assembled at 54-90 dpc [37-39], the expression patterns of OLFML3 gene during prenatal skeletal muscle development in our study indicating that it mainly involved in the formation of primary muscle fibers. The differential expression patterns from Landrace to Tongcheng pigs also showed that there are developmental synchronizations in prenatal skeletal muscle development between the two pig breeds, the result was consistent with our previous report [31, 40], which concluded Landrace pigs may have a quicker muscle growth rate than Tongcheng pigs. Prenatal muscle development programs postnatal performance. The total number of fibers was fixed by the number of primary muscle fibers before birth in pig [37]. The facts documented that OLFML3 may affect the phenotype of postnatal muscle in different pig breeds.
Previous studies have reported that miRNA play important roles in gene expression regulation. However, the expression and regulation mechanism of OLFML3 gene in pig have not been reported. Our study documented that OLFML3 expression was regulated by miR-155. The overexpression of miR-155 decreased the luciferase activity of psiCHECK-2, which contains the OLFML3 3'UTR sequence, and the suppression level was similar to that observed in other studies [41]. Moreover, the miR-155 overexpression vector did not decrease the luciferase activity of psiCHECK-2 with the OLFML3 3'UTR mutant sequence, indicating that the target site is specific and unique in porcine. However, we do not exclude the possibility that additional miR-155-independent effects (e.g., mRNA stability, alteration of RNA secondary structure, or disruption/addition of a binding site for an unknown regulatory factor) may be attributed to specific point mutations introduced in the OLFML3 3'UTR[42]. Furthermore, the overexpression of miR-155 significantly decreased the OLFML3 mRNA level in PK15 cells, indicating that the downregulation of OLFML3 by miR-155 may be due to the mRNA degradation. The results are similar to those of previous reports of miRNA-dependent mRNA degradation in other systems [43]. MiR-155-mediated OLFML3 mRNA degradation is probably performed by mRNA degradation machinery, because the miRNA-mediated mRNA degradation in mammals is not thought to involve endonucleolytic cleavage [44]. Previous studies have proved that miR-155 may accelerate cell proliferation and differentiation [28, 29], and therefore, we speculated that OLFML3 was regulated by miR-155 and involved in proliferation of muscle cells and further affect prenatal skeletal muscle development. Moreover, we have tried to explore the interaction between OLFML3 and miR-155 at the protein level, however, without the antibody in pigs, we have not obtained the target strip with Olfactomedin-like antibody in human (GTX120065, GeneTex, CA, USA). The sequences of the 3'UTR of OLFML3 gene around the miR-155 target sites and the seed sequence of mature miR-155 are well conserved in mammals, suggesting that the target region is probably important in OLFML3 regulation, and the regulation of OLFML3 by miR-155 may exist in other species.
In conclusion, we reported the molecular characterization of the porcine OLFML3 gene and found that OLFML3 exhibits different expression patterns during prenatal skeletal muscle development between Landrace and Tongcheng pigs, indicating that OLFML3 gene may affect prenatal skeletal muscle development. These facts also suggested that OLFML3 may contribute to muscle phenotype with different genotype breed pigs. Finally, we have verified that porcine OLFML3 mRNA expression was regulated by miR-155. Overall, our work provides fundamental data regarding OLFML3 and miR-155 and suggests that OLFML3 gene may affect muscle development of pig.
Acknowledgements
The authors thank Dr. Jianhua Cao in Huazhong Agricultural University for providing psiCHECK-2 vector. This work was supported by National Natural Science Foundation of China (31171192, 30830080), National Key Project (2011ZX08009-001) and National Basic Research Program of China (No. 2009CB941604).
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Hillier BJ, Vacquier VD. Amassin, an olfactomedin protein, mediates the massive intercellular adhesion of sea urchin coelomocytes. J Cell Biol. 2003;160:597-604
2. Snyder DA, Rivers AM, Yokoe H. et al. Olfactomedin: purification, characterization, and localization of a novel olfactory glycoprotein. Biochemistry. 1991;30:9143-9153
3. Zeng LC, Han ZG, Ma WJ. Elucidation of subfamily segregation and intramolecular coevolution of the olfactomedin-like proteins by comprehensive phylogenetic analysis and gene expression pattern assessment. FEBS Lett. 2005;579:5443-5453
4. Stone EM, Fingert JH, Alward WL. Identification of a Gene That Causes Primary Open Angle Glaucoma. Science. 1997;275:668-670
5. Polansky JR. Nguyen TD. The TIGR gene, pathogenic mechanisms, and other recent advances in glaucoma genetics. Curr Opin Ophthalmol. 1998;9:15-23
6. Morissette J, Clepet C, Moisan S. et al. Homozygotes carrying an autosomal dominant TIGR mutation do not manifest glaucoma. Nat Genet. 1998;19:319-321
7. Paper W, Kroeber M, Heersink S. et al. Elevated amounts of myocilin in the aqueous humor of transgenic mice cause significant changes in ocular gene expression. Exp Eye Res. 2008;87:257-267
8. Adam MF, Belmouden A, Binisti P. et al. Recurrent mutations in a single exon encoding the evolutionarily conserved olfactomedin-homology domain of TIGR in familial open-angle glaucoma. Hum Mol Genet. 1997;6:2091-2097
9. Barembaum M, Moreno TA, LaBonne C. et al. Noelin-1 is a secreted glycoprotein involved in generation of the neural crest. Nat Cell Biol. 2000;2:219-225
10. Moreno TA, Bronner-Fraser M. The secreted glycoprotein Noelin-1 promotes neurogenesis in Xenopus. Dev Biol. 2001;240:340-360
11. Zeng LC, Liu F, Zhang X. et al. hOLF44, a secreted glycoprotein with distinct expression pattern, belongs to an uncharacterized olfactomedin-like subfamily newly identified by phylogenetic analysis. FEBS Lett. 2004;571:74-80
12. Ikeya M, Kawada M, Nakazawa Y. et al. Gene disruption/knock-in analysis of mONT3: vector construction by employing both in vivo and in vitro recombinations. Int J Dev Biol. 2005;49:807-823
13. Sakuragi M, Sasai N, Ikeya M. et al. Functional analysis of chick ONT1 reveals distinguishable activities among olfactomedin-related signaling factors. Mechanisms of Development. 2006;123:114-123
14. Inomata H, Haraguchi T, Sasai Y. Robust stability of the embryonic axial pattern requires a secreted scaffold for chordin degradation. Cell. 2008;134:854-865
15. Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5'UTR as in the 3'UTR. Proceedings of the National Academy of Sciences. 2007;104:9667-9672
16. Kulkarni S, Savan R, Qi Y. et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495-498
17. Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-355
18. Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281-297
19. Griffiths-Jones S, Grocock RJ, van Dongen S. et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140-144
20. Taganov KD, Boldin MP, Baltimore D. MicroRNAs and Immunity: Tiny Players in a Big Field. Immunity. 2007;26:133-137
21. Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131-140
22. Chen JF, Mandel EM, Thomson JM. et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228-233
23. Kim HK, Lee YS, Sivaprasad U. et al. Muscle-specific microRNA miR-206 promotes muscle differentiation. The Journal of Cell Biology. 2006;174:677-687
24. Naguibneva I, Ameyar-Zazoua M, Polesskaya A. et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278-284
25. Winbanks CE, Wang B, Beyer C. et al. TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J Biol Chem. 2011;286:13805-13814
26. Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol. 2011;192:69-81
27. Forrest AR, Kanamori-Katayama M, Tomaru Y. et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia. 2009;24:460-466
28. Xie Q, Chen X, Lu F. et al. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer. 2011 doi: 10.1002/cncr.26566
29. Seok HY, Tatsuguchi M, Callis TE. et al. miR-155 Inhibits Expression of the MEF2A Protein to Repress Skeletal Muscle Differentiation. J Biol Chem. 2011;286:35339-35346
30. Tang ZL, Li Y, Zhao SH. et al. A modified GLGI method for identification of novel porcine genes from long serial analysis of gene expression tags. Chinese Journal of Agricultural Biotechnology. 2007;4:111-115
31. Tang ZL, Li Y, Wan P. et al. LongSAGE analysis of skeletal muscle at three prenatal stages in Tongcheng and Landrace pigs. Genome Biol. 2007;8:R115
32. Friedman RC, Farh KK, Burge CB. Most Mammalian mRNAs Are Conserved Targets of MicroRNAs. Genome Research. 2009;19:92-105
33. Sharbati S, Friedlander MR, Sharbati J. et al. Deciphering the porcine intestinal microRNA transcriptome. BMC Genomics. 2010;11:275
34. Chen C, Ridzon DA, Broomer AJ. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179
35. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408
36. Caballero M, Borras T. Inefficient processing of an olfactomedin-deficient myocilin mutant: potential physiological relevance to glaucoma. Biochem Biophys Res Commun. 2001;282:662-670
37. Wigmore PM, Stickland NC. Muscle development in large and small pig fetuses. J Anat. 1983;137(Pt 2):235-245
38. Ashmore CR, Addis PB, Doerr L. Development of muscle fibers in the fetal pig. J Anim Sci. 1973;36:1088-1093
39. Davoli R, Braglia S, Russo V. et al. Expression profiling of functional genes in prenatal skeletal muscle tissue in Duroc and Pietrain pigs. J Anim Breed Genet. 2011;128:15-27
40. Tang ZL, Zhang XJ, Yang SL. et al. The chromosomal localization, expression pattern and polymorphism analysis of porcine FSCN1 gene differently expressed from LongSAGE library. Mol Biol Rep. 2009;37:2361-2367
41. Trakooljul N, Hicks JA, Liu HC. Identification of target genes and pathways associated with chicken microRNA miR-143. Anim Genet. 2010;41:357-364
42. Teng G, Hakimpour P, Landgraf P. et al. MicroRNA-155 Is a Negative Regulator of Activation-Induced Cytidine Deaminase. Immunity. 2008;28:621-629
43. Bagga S, Bracht J, Hunter S. et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553-563
44. Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459-469
Author contact
![]() Corresponding author: Zhonglin Tang, 1. State Key Laboratory for Animal Nutrition, Beijing 100193, P. R. China; 2.The Key Laboratory for Domestic Animal Genetic Resources and Breeding of Ministry of Agriculture of China, Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing 100193, P. R. China. E-mail: zhonglinqy_99com.
Corresponding author: Zhonglin Tang, 1. State Key Laboratory for Animal Nutrition, Beijing 100193, P. R. China; 2.The Key Laboratory for Domestic Animal Genetic Resources and Breeding of Ministry of Agriculture of China, Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing 100193, P. R. China. E-mail: zhonglinqy_99com.

 Global reach, higher impact
Global reach, higher impact