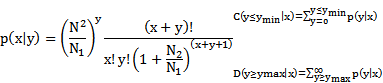10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2012; 8(7):1013-1022. doi:10.7150/ijbs.3836 This issue Cite
Research Paper
MicroRNA Expression Profile in RAW264.7 cells in Response to Brucella melitensis Infection
1. State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan 430070, China;
2. Department of Preventive Veterinary Medicine, College of Animal Science & Technology, Shihezi University, Shihezi city, China
* Contributed equally to this work.
Received 2011-11-19; Accepted 2012-7-25; Published 2012-8-3
Abstract
MicroRNA (miRNA) is small non-coding RNA with approximate 22 nt in length. Recent studies indicate that miRNAs play significant roles in pathogen-host interactions. Brucella organisms are Gram-negative facultative intracellular bacteria that cause Brucellosis. Brucella strains infect macrophages and establish chronic infection by altering host life activities including apoptosis and autophagy. Here, we report a comprehensive analysis of miRNA expression profiles in mock- and Brucella-infected RAW264.7 cells using high-throughput sequencing approach. In total, 344 unique miRNAs were co-expressed in the two libraries, in which 57 miRNAs were differentially expressed. Eight differentially expressed miRNAs with high abundance were subjected to further analysis. The GO enrichment analysis suggests that the putative target genes of these differentially expressed miRNAs are involved in apoptosis, autophagy and immune response. In particular, a total of 25 target genes are involved in regulating apoptosis and autophagy, indicating that these miRNAs may play important regulatory roles in the Brucella-host interactions. Furthermore, the interactions of miR-1981 and its target genes, Bcl-2 and Bid, were validated by luciferase assay. The results show that miR-1981 mimic up-regulated the luciferase activity of psiCHECK-2 Bcl-2 3′ UTR, but the luciferase activity of psiCHECK-2 Bid 3′ UTR was not changed significantly. Taken together, these data provide valuable framework on Brucella induced miRNA expression in RAW264.7 cells, and suggest that Brucella may establish chronic infection by regulating miRNA expression profile.
Keywords: Brucella, RAW264.7, microRNA, high-throughput sequencing, apoptosis, autophagy
Introduction
Brucellae are Gram-negative, facultative intracellular, coccobacillus bacteria that cause Brucellosis, a zoonotic disease affecting humans and animals worldwide [1]. Brucellosis is characterized by undulant fever, arthritis, endocarditis, and meningitis in humans, as well as abortion and infertility in animals, which develops to chronic infection if not treated [2]. Brucella strains express uncanonical virulence factors including lipopolysaccharides (LPS), virulence regulator proteins and phosphatidylcholine, but lack most of classical virulence factors such as invasive protease, extoxin, or virulence plasmids [3]. The genus Brucella consists of at least six species: B.abortus, B.melitensis, B.suis, B.ovis, B.canis, and B.neotomae, of which B.melitensis is the most widely distributed [4].
Macrophage, a kind of early defense cell against pathogenic bacteria, is primary target of Brucella [2, 5]. Upon invasion, Brucella utilizes vesicles formed by macrophage endocytosis to transport and block the fusion between Brucella-containing vesicles and lysosomes [6, 7]. Besides, apoptosis of macrophages is inhibited, facilitating Brucella′s latent infection [8, 9]. Previous studies revealed the mRNA expression profiles of macrophages infected by Brucella based on microarray [10, 11]. These studies provide important clues to the mechanism how Brucella strains establish chronic infection.
MicroRNA (miRNA) is endogenous small non-coding RNA molecule that functions in post-transcriptional regulation [12, 13]. MiRNAs regulate target genes expression by directly binding to complementary sites within the 3′ untranslated region (UTR) to either block or enhance translation or increase degradation of mRNA [14]. Diverse biological activities including programmed cell death are regulated by miRNAs [15]. Recent studies indicate that host miRNA expression can be regulated by pathogens, which suggest that pathogens may establish infection niche through miRNA pathway [16-21].
In this study, miRNA expression profiles of mock- and Brucella-infected RAW264.7 cells were identified by high-throughput sequencing, the differentially expressed miRNAs, target genes, and biological activities of infected macrophage were further analyzed and validated, and possible mechanism how Brucella establish chronic infection is discussed.
Materials and methods
Bacterial and cell lines
Brucella melitensis strain 027 [22] was grown either on tryptic soy agar (TSA) plates or in tryptic soy broth (TSB), or in cells as described [10, 11]. Experiments involving handling bacteria operation were performed in biological safety level 3 (BSL-3) laboratory.
The RAW264.7 and Hela cell lines (obtained from China Center For Type Culture Collection, Wuhan) were cultured in Dulbecco′s modified Eagle′s medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Gibco) at 37 oC with 5% CO2 (vol/vol).
RAW264.7 infection
RAW264.7 cells were infected as described with slight modification [10]. Briefly, RAW264.7 cells were cultured in T25 flasks (Corning) containing 5 ml DMEM (10% FBS). Each T25 flask contains ~2×106 cells. Prior to infection, B.melitensis strain 027 was cultured in TSB for 40 h, and RAW264.7 cells were infected at a multiplicity of infection (MOI) of 200. After infection for 30 min at 37 oC, the cells were washed for three times with phosphate buffered saline (PBS), and incubated for 1 h in fresh DMEM (10% FBS) with 50 μg/ml gentamicin to kill extracellular bacteria. Then the cells were incubated in fresh DMEM (10% FBS). Mock-infected cells were treated with the same procedure except for the bacteria infection.
Isolation of total RNA
At 4 h, 12 h, 24 h and 36 h postinfection, cells were washed three times with PBS, and 4 ml TRIZOL reagent (Invitrogen) was added to each flask in both groups. The homogenized samples were incubated for 5 min at room temperature, and total RNA was isolated according to the manufacturer's protocols.
Solexa sequencing
Total RNA was electrophoresed in formaldehyde-denatured agarose gel, and quantified with spectrophotometer (NanoDrop 2000). Ten μg RNA was mixed and then subjected to BGI (Huada Genomics Institute Co. Ltd, China) for solexa sequencing of small RNAs less than 30 nt.
Bioinformatic analysis of small RNA sequences
After Solexa sequencing, the sequence reads went through the data cleaning procedule as described earlier [23]: (1) filter and remove the low quality reads, (2) get rid of three primer adaptor sequences, (3) trim adaptor contaminations formed by adaptor ligation, and (4) retain only trimmed reads of sizes from 18 to 30 nt. Then, overview of small RNA libraries was obtained, including length distribution, common and specific sequences between samples, genome mapping and annotation. Filtered sequences were mapped to mouse (Mus musculus) genome or Brucella genome using SOAP algorithm [24]. The clean tags that cannot be annotated to any category were taken to predict novel miRNA using MIEREAP (http://cloud.genomics.cn/).
Brucella melitensis 16M genome (GenBank accession no. NC_003317, NC_003318) was retrieved from the NCBI Genome database. Mouse (Mus musculus) genome sequences were retrieved from the UCSC Genome Browser (ftp://hgdownload.cse.ucsc.edu/goldenPath/mm9/chromosomes/).
Differential expression analysis
The miRNAs between the mock- and Brucella-infected cells were compared as described previously [25]: (i) Normalize the expression of two samples to obtain the expression in per million, (ii) Calculate fold-change and P-value from the normalized expression.
Fold-change formula: Fold_change=log2 (treatment/control).
P-value:
MiRNA target prediction and GO analysis
The miRanda algorithm was used to identify miRNA target genes with parameters: -score 140, -en -20 [26].
The GO functional analysis of the putative target genes was performed by WEGO program [27].
Gene enrichment analysis
The target genes were subjected to GO enrichment analysis using GO::TermFinder [28]. The significantly enriched GO terms of the target gene sets (4997 genes in total) were detected when compared to the genome-wide background. The default parameters of Hypergeometric test and Benjamini & Hochberg false discovery rate (FDR) were utilized to adjust the P-value (P<0.05).
Constructs
The 3′ UTRs of Bcl-2 and Bid were amplified by polymerase chain reaction (PCR) from genome DNA derived from RAW264.7 cells, then the PCR products were cloned to pMD18-T vector. After digestion by Xho I and Not I, each of the fragment containing 3′ UTR of Bcl-2 or Bid was cloned into psiCHECK-2 vector (Promega). The primers used are: Bcl-2 forward primer 5′-CTCGAGATCAATCAAAGCCAAGCAGAC-3′ (Xho I), and Bcl-2 reverse primer 5′-GCGGCCGCGCCACCCATTATCTACATTCA-3′ (Not I). Bid forward primer 5′- CTCGAGAGACAACAGTATGGGAAAGGG-3′(Xho I), and Bid reverse primer 5′- GCGGCCGCGTGGTTCTGGTATCTTGTCCC-3′(Not I).
Luciferase assay
The miRNA mimics were synthesized from GenePharma (Shanghai, China). The sequences of mimics are as follows: miR-1981 mimic, 5′-GUAAAGGCUGGGCUUAGACGUGGC-3′(sense)/5′-CACGUCUAAGCCCAGCCUUUACUU-3′(antisense); mutant miR-1981 mimic, 5′-GUGGAGGCUGGGCUUAGACGUGGC-3′ (sense)/ 5′-CACGUCUAAGCCCAGCCUCCACUU-3′ (antisense); negative control, 5′-UUCUCCGAACGUGUCACGUTT-3′(sense)/ 5′-ACGUGACACGUUCGGAGAATT-3′(antisense). Hela cells were cultured in 24-well plates and co-transfected with 200 ng psiCHECK-2 vector containing 3′ UTR of Bcl-2 or 3′ UTR of Bid and 40 nM miRNA mimics per well. Transfections were performed using Lipofectamine 2000 (Invitrogen). The luciferase analysis was performed 24 h later using Dual-Luciferase Reporter Assay (Promega, Cat. no. E1910) according to the manufacturer's protocol. The relative firefly luciferase activity was obtained after normalizing to renilla luciferase activity.
Results
Sequencing and overview of small RNAs
In order to investigate whether miRNAs were differentially expressed in RAW264.7 cells during Brucella infection, small RNA (sRNA) libraries were constructed and submitted to Illumina/solexa sequencing. After deep sequencing, 20,052,540 and 18,971,297 raw reads were obtained from the Brucella- and mock-infected cells sRNA library, respectively. After data cleaning, 17,689,706 and 15,911,556 clean reads were generated from the two sRNA libraries, accounting for 89.26% and 84.88% of total reads of Brucella- and mock-infected cells, repectively. Small RNAs ranging between 18 nt and 30 nt in length were subjected to further analysis. The distributions of selected reads were analyzed and are shown in Figure 1 A and C. The majority of reads accounting for approximate 70% in the sRNA libraries were in range of 20 to 24 nt in length, most of which contain 5′ A or 5′ U. The two sRNA libraries share 96.88% common sequences, while 0.49% and 2.63% represent the mock-infected specific and Brucella-infected specific sRNAs, respectively (Additional file 1: Figure S1). Aligned against the Mus and Brucella genomes utilizing SOAP program [24], about 40% reads were mapped to the Mus and Brcuella genomes, while sequences mapped to Brucella genome were in quite low abundance (Additional file 2: Table S1). Distributions of small RNAs among different categories are shown in Figure 1 B and D. The 2,591,363 and 33,562,043 reads belong to miRNA in mock- and Brucella-infected libraries, which account for 17.96% and 20.14%, respectively.
Overview of total small RNA libraries. (A) Length distribution of mock-infected RAW264.7 cells library (6,443,327 mapped reads, corresponding to 15,813,792 total reads). Small RNAs with length in a range of 20 nt to 24 nt are highlighted by light yellow background, of which first nucleotide distribution is presented in pie chart. (B) Distribution of mock-infected small RNAs among different categories. Some small RNAs can be mapped to more than one category. To make sure that every unique small RNA was mapped to only one category, we follow the following priority rule: rRNAetc (in which Genbank > Rfam) > known miRNA > repeat > exon > intron. (C) Length distribution of Brucella-infected RAW264.7 cells library (7,975,853 mapped reads, corresponding to 17,654,028 total reads). Small RNAs between 20 nt and 24 nt in length and their first nucleotide distribution is presented as (A). (D) Distribution of Brucella-infected small RNAs among different categories. The priority rule is as same as described in (B).
Analysis of miRNA expression in mock- and Brucella-infected libraries
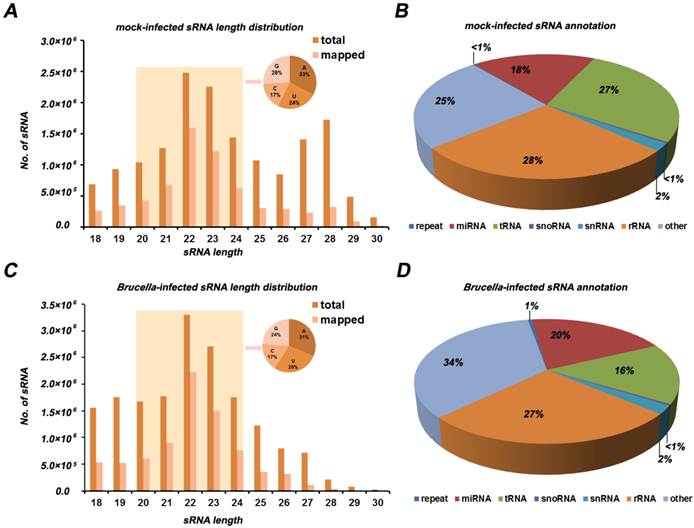
The Illumina/solexa sequencing method provides an insight into identification of miRNA expression profile. The miRNA frequency in the mock- and Brucella-infected cells is presented in Figure 2 and Additional file 2: Table S2. In order to identify known miRNA in the two libraries, read sequences were aligned against the miRBase (Release version 16.0). We obtained 254 miRNA families including 368 unique miRNAs, among which 344 unique miRNAs were co-expressed in the two libraries. The analysis of miRNA family and counts of known miRNA are shown in Additional file 2: Table S2. In mock-infected libraries, mmu-miR-21, mmu-let-7c, mmu-miR-146b, mmu-let-7f, mmu-miR-378, mmu-let-7a, mmu-miR-146a, mmu-miR-182 were the dominantly expressed miRNAs, with more than 100,000 reads for each miRNA. However, the dominantly expressed miRNAs in Brucella-infected cells were mmu-miR-21, mmu-let-7c, mmu-miR-146b, mmu-let-7f, mmu-miR-146a, mmu-let-7a, mmu-miR-378 and mmu-let-7b, with more than 100,000 reads for each one. This result suggests that the mmu-miR-21, mmu-let-7c, mmu-miR-146b, mmu-let-7f, mmu-miR-146a, mmu-let-7a and mmu-miR-378 are most abundant in both libraries. Interestingly, mmu-miR-140* and mmu-miR-221* are quite different from other miRNAs. The expression level of mmu-miR-140* is much higher than its corresponding miRNA, while mmu-miR-221* has the almost same expression level compared to mmu-miR-221. This results indicate that these miRNA*s have potential function as well as miRNAs.
Novel miRNA prediction
The unannotated sRNAs were taken to predict novel miRNA. Mireap software was utilized to predict novel miRNAs by exploring the secondary structure, the Dicer cleavage site and the minimum free energy of the unannotated small RNA tags which could be mapped to genome. A total of 34 novel miRNAs were identified in mock-infected RAW264.7 cells, while 81 novel miRNAs in Brucella-infected cells. There are 16 same novel miRNAs between the two samples, and 24 novel miRNAs are mapped to Brucella genome. However, all of these novel miRNAs are in low expression level (less than 400 reads) (Additional file 2: Table S3 and S4).
Putative target genes of the differentially expressed miRNAs
Differentially expressed miRNAs were summarized in Additional file 2: Table S5. Eight differentially expressed miRNAs with high abundance (Table 1) were taken to predict putative target genes using miRanda algorithm [26]. Then 13,241 transcription products were obtained as described in Material and Methods. These predicted targets were classified according to KEGG annotation [29] and subjected to GO functional analysis utilizing WEGO program [27]. The WEGO output of target genes is presented in Figure 3. The results reveal that these target genes are involved in cell death, translation regulation, and immune response. Furthermore, GO enrichment analysis reveals that target genes related to apoptosis, autophagy and immune response are significantly enriched in comparison to genes in the mouse genome as a whole (Additional file 2: Table S6).
The regulatory impacts of host genes by differentially expressed miRNAs
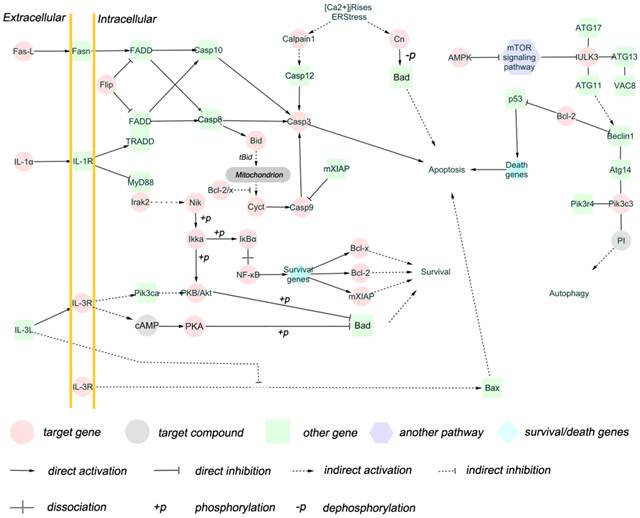
Brucella is capable to avoid activation of the innate immune system and utilize autophagy process to transport, survive and replicate, and inhibit apoptosis of host cells [6-9]. Therefore, we selected target genes related to apoptosis and/or autophagy for impact analysis. A total of 25 target genes were obtained (Additional file 2: Table S7), and the functions of these target genes in biological pathway were summerized in Figure 4. The result shows that these target genes are involved extrinsic pathway and intrinsic pathways of apoptosis, as well as autophagy regulation pathway (Figure 5). Fas-l, Bcl-2, IL-1, IL-3R, Cn and AMPK are involved in apoptosis and autophagy pathway [30-32], and these genes are regulated by miRNAs.
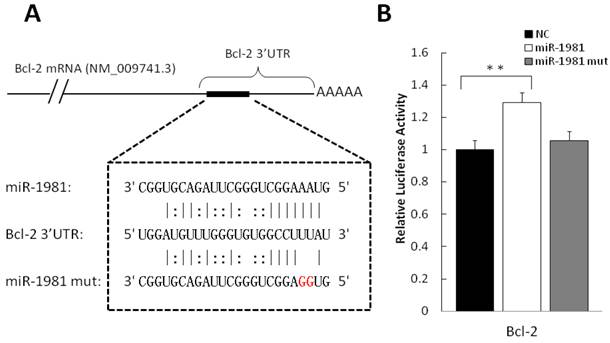
Validation of putative target
In order to validate the putative targets of differentially expressed miRNAs, luciferase assay report system was employed. Among candidate interactions between miRNA and mRNA, we analysed miR-1981-Bcl-2 and miR-1981-Bid. The 3′ UTR of the two genes were cloned to 3′ of Renilla luciferase gene in the dual-reporter vector psiCHECK-2. As shown in Figure 6A, the miR-1981 mimic and mutant miR-1981 mimic were employed to confirm the miR-1981 binding site in Bcl-2 mRNA 3′ UTR. After 24 h post transfection, the miR-1981 mimic enhanced the luciferase activity of psiCHECK-2 Bcl-2 3′ UTR in Hela cells, whereas mutant miR-1981 mimic had no significant effect (Fig. 6B). This luciferase assay was repeated three times. When treated with miR-1981, the luciferase activity of psiCHECK-2 Bcl-2 3′ UTR was enhanced approximate 30%, indicating that the expression of Bcl-2 was significantly up-regulated (P<0.01). Meanwhile, the luciferase activity of psiCHECK-2 Bid 3′ UTR was not changed significantly in the presence of the miR-1981 mimic or the mutant miR-1981 mimic (data not shown).
Summary of the top 8 differentially expressed miRNAs with high abundance.
| miR-name | count in mock-infection | count in Brucella-infection | fold-change(log2 mix/CK-mix) | p-value | Significant label | Sequence (5'-3') |
|---|---|---|---|---|---|---|
| mmu-let-7b | 43093 | 113171 | 1.0986932 | 0 | ** | UGAGGUAGUAGGUUGUGUGGUU |
| mmu-miR-93 | 37823 | 15754 | -1.5578313 | 0 | ** | CAAAGUGCUGUUCGUGCAGGUAG |
| mmu-miR-151-3p | 20457 | 12520 | -1.00264556 | 0 | ** | CUAGACUGAGGCUCCUUGAGG |
| mmu-miR-92a | 3795 | 10584 | 1.18542781 | 0 | ** | UAUUGCACUUGUCCCGGCCUG |
| mmu-miR-142-5p | 2202 | 6455 | 1.2573173 | 1.86E-307 | ** | CAUAAAGUAGAAAGCACUACU |
| mmu-miR-99a | 1517 | 4062 | 1.12668333 | 1.02E-163 | ** | AACCCGUAGAUCCGAUCUUGUG |
| mmu-miR-181b | 1192 | 4159 | 1.50856643 | 3.63E-261 | ** | AACAUUCAUUGCUGUCGGUGGGU |
| mmu-miR-1981 | 486 | 1367 | 1.19769834 | 7.14E-62 | ** | GUAAAGGCUGGGCUUAGACGUGGC |
** denotes extremely significant.
Profiles of mock- and Brucella-infected sRNA libraries. (A) Each known miRNA reads for mock and Brucella-infected libraries. Several miRNAs (i.e., miR-21, let-7a, let-7b, let-7c, let-7f, miR-146b, miR-378, miR-146a, miR-182) with high abundance (more than 100,000 reads) are indicated. (B) Differential expression of known miRNA. Each point in the figure represents a miRNA. The X and Y axis represent expression level of miRNAs in mock- and Brucella-infected samples, respectively. Red points represent miRNAs with ratio>2; Blue points represent miRNAs with 1/2<ratio<2; Green points represent miRNAs with ratio<1/2. Ratio =normalized expression of the Brucella-infected sample / normalized expression of the mock-infected sample.
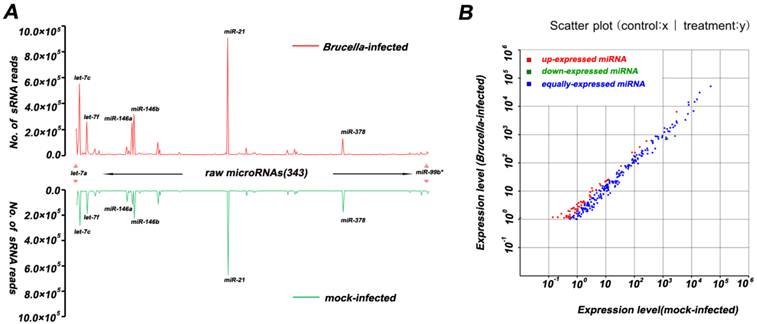
GO analysis on Mus targets of top 8 differentially expressed miRNAs. GO functional analysis shows that 4997 target genes are involved in several biological processes including cell dreath, transcription regulator, translation regulator, immune response, immune system process, and response to stress.
Gene regulatory network of several miRNAs and their targets in Mus genome. The network was formed by the six miRNAs (round rectangles) and their targets (circles). Eight differentially expressed miRNAs with high abundance were taken into predicting target genes, of which 6 miRNAs are involved in regulating apoptosis and autophagy.
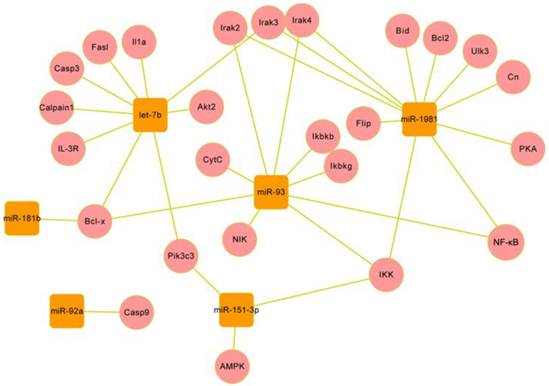
Network of 25 target genes and their interactive genes involved in regulating apoptosis and autophagy. The target genes (cycles) are coloured with red, the genes (round rectangles ) interacting with target genes are coloured with green, the survival/death genes (diamond) are coloured with blue, the another pathways (hexagon) are coloured with purple, and the target compounds are coloured with gray. The solid line with delta tip represents direct activation, the solid line with “T” tip represents direct inhibition, the dash line with delta tip represents indirect activation, the dash line with “T” tip represents indirect inhibition, the cross-shape line represent dissociation, and “+p”, “-p” represent phosphorylation and dephosphorylation, respectively. The interactions between target genes and other genes are involved in apoptosis extrinsic (including fas,IL-1,IL-3 pathway) and intrinsic (mitochondria-associated pathway) pathways, and autophagy induction and vesicle nucleation processes.
Validation of puptative target by luciferase assay. (A) Shown is the predicted interaction between miR-1981 and its target gene Bcl-2 3′ UTR. Red letters indicate the mutation sites of miR-1981. (B) miR-1981 mimic, mutant miR-1981 mimic and negative control were co-transfected with psiCHECK-2 Bcl-2 3′ UTR report plasmid into Hela cells. At 24 h post-transfection, luciferase activity was measured with Dual-Luciferase Reporter Assay System (Promega). The luciferase activity of negative control was regarded as 1. This result is the representive data of four experiments.
Discussion
High-throughput sequencing provides a convenient and sensitive approach to study small RNA. In this study, the expression profiles of miRNA in mock- and Brucella-infected RAW264.7 cells were determined, and the detailed biological processes involved in apoptosis and autophagy were comprehensively analyzed. In addition, two of the predicted targets of miR-1981 were validated by luciferase assay.
MiRNAs are able to affect protein expression via regulating mRNA degradation or translation [14]. The differential expression of miRNA in cells could be induced by pathogen and may have the potential to regulate the host response to pathogen. In heat-killed Candida albicans-infected macrophage, miR-155, miR-455, miR-148b, miR-135a*, miR125-5p, miR-146a, miR-125a-3p, miR-192, miR-139-5p and miR-99b were differentially expressed, and miR-155, miR-125-5p, miR-146a, miR-125a-3p and miR-99b were up-regulated by LPS induction. MiR-155 was also upregulated by F. novicida, Helicobacter pylori and Salmonella [16, 19]. In our study, 57 miRNAs were regulated during Brucella infection, of which let-7b, miR-93, miR-151-3p, miR-92a, miR-142-5p, miR-99a, miR-181b and miR-1981 were differentially expressed (with more than 1000 reads). However, there is no significant difference of miR-155 expression between Brucella- and mock-infected RAW264.7 cells.
Brucella noncanonical LPS could account for the different miRNA induction. Several studies proposed that lipopolysacharide (LPS) of Brucella was one of the major virulence factors [33, 34]. LPS plays pivotal roles both in structural and functional integrities. LPS also serves as the primary target of innate immune system [35]. Compared with classic LPS of Escherichia coli, LPS in Brucella is non-classical [34]. CD14 and toll-like receptor (TLR) family proteins, which activate NF-κB signaling pathway, are involved in recognition of bacterial LPS [36, 37]. Since the NF-κB signaling pathway regulates gene transcription, LPS may induce miRNA expression, and classical and non-classical LPS regulate different miRNA expression via different pathways. In our study, miR-155 expression was not changed obviously, which is consistent to this possibility.
Macrophages are Brucella target host cells, in which Brucella is capable of surviving and multiplying [38]. RAW264.7 cell line is a good model for macrophage infection. Apoptosis and autophagy play important roles in host defense mechanism [39, 40]. However, Brucella strains use guileful strategy to regulate life activities with the aim to avoid activation of the innate immune system and to transform the environment for survival and replication [41]. Brucella strains inhibit apoptosis of monocytes, utilize autophage vesicles to transport, and inhibit the fusion of BCV and lysosome [42, 43]. He et al. described the mRNA profiles of Brucella-infected RAW264.7 cells, revealing that some of genes regulating apoptosis such as Bcl-2, Bax, and Bad, were up- or down-expressed [10]. Similar description was reported by Linda et al [11]. In this study, we validated some putative targets involved in apoptosis. The expression of miR-1981 was up-regulated in respond to Brucella infection in RAW264.7 cells. Thus the interaction between miR-1981 mimic and 3′ UTR of its target gene Bcl-2 was validated using the luciferase assay. As expected, the luciferase activity of psiCHECK-2 Bcl-2 3′ UTR was enhanced extremely significantly. Strikingly, no significant change of the luciferase activity of psiCHECK-2 Bid 3′ UTR was observed.
There are two major pathways of apoptosis, namely the extrinsic (Fas and other TNFR superfamily members and ligands) and the intrinsic (mitochondria-associated) pathways [30] , both of which have been found to be the differentially expressed miRNAs target genes in our study. Fas-L and IL-1, the putative target genes of miRNA let-7b, are death ligands interacting with Fas and IL-1R, respectively [44]. Fas and IL-1 pathway activate caspase 8 via FADD or TRADD FADD [45]. Apoptosis regulator Flip, the target gene of mmu-miR-1981, inhibits FADD or TRADD FADD. Caspase molecules are categorized into initiator, effectors or executioners and inflammatory. The target gene caspase 9 belongs to initiator that can be activated by FADD or TRADD FADD and cytochrome C. Caspase 3, activated by caspase 8 and caspase 9 and serving as effector or executioner in caspase pathway, was also predicted as a target gene in our study. The IL-3 pathway, which promotes cell survival, was regulated by let-7b, miR-1981, and miR-151-3p. Members of NF-κB signaling pathway such as IκBα, Ikka, and p53 signaling pathway were also regulated by miRNAs in our study. These transcription factors are associated with the expression of survival genes and/or death genes including Bcl-x and Bcl-2. Bid is able to promote cytochrome C release after truncation, antagonising Bcl-2/x. The inducer and inhibitor for apoptosis pathway such as cytochrome C were also regulated by miRNAs.
Autophagy process consists of autophagy induction, vesicle nucleation, vesicle expansion and completion, and docking and fusion [46]. The mTOR signaling pathway, which inhibits ULK3, is inhibited by AMPK [47-49]. Pik3c3 interacts with ATG14, PIK3R4, and Beclin1 to form complex to serve as regulator [50], while Beclin1 is inhibited by Bcl-2 [51, 52]. In our study, AMPK, ULK3, and Pik3c3 are putative target genes, indicating that autophagy pathway might be utilized by Brucella for intracellular survival.
In our study, miR-1981 is a regulator for Bcl-2, but not for Bid. Bcl-2 is able to inhibit apoptosis by blocking the release of cytochrome C, while Bid can induce the release of cytochrome C after truncation [53]. This result may account for the phenomenon that apoptosis is inhibited in Brucella-infected RAW264.7 cells [54]. The possible reason for the up-regulation of Bcl-2 3′ UTR is that miR-1981 might reduce the mRNA complex to make it convenient for translation. This hypothesis remains to be confirmed by future experiments.
Taken together, our study unveiled the miRNA expression profile in Brucella-infected RAW264.7 cells and confirmed one pair of interations between miRNA and target genes. We consider that the differently expressed miRNAs in RAW264.7 cells in response to Brucella infection fulfil important but as yet to be discovered functions in Brucella life cycle. We believe these data provide important clues as to and accelerate the understanding of the pathogenesis of Brucella and other intracellular bacteria, as well as the host biological pathway process.
Supplementary Material
Additional File 1Figure S1.
Table S1-S7.
Acknowledgements
We are grateful to Di-Jun Chen (College of Life Science, Huazhong Agricultural University) for suggestive discussions. We thank Zhen Fang Fu (College of Veterinary Medicine, Georgia University) and Wei Zhang (College of Veterinary Medicine, Huazhong Agricultural University) for critical reading and suggestions of this manuscript. This work was supported by 973 plan (2010CB530203) to Z.-F. Liu and C.-F. Chen, and autonomous research grant from State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, to H. -C. Chen and Z.- F. Liu.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Schurig GG, Sriranganathan N, Corbel MJ. Brucellosis vaccines: past, present and future. Vet Microbiol. 2002;90:479-96
2. Liautard JP, Gross A, Dornand J, Kohler S. Interactions between professional phagocytes and Brucella spp. Microbiologia. 1996;12:197-206
3. Roop RM 2nd, Gee JM, Robertson GT, Richardson JM, Ng WL, Winkler ME. Brucella stationary-phase gene expression and virulence. Annu Rev Microbiol. 2003;57:57-76
4. Benkirane A. Ovine and caprine brucellosis: World distribution and control/eradication strategies in West Asia/North Africa region. Small Ruminant Res. 2006;62:19-25
5. Baldwin CL, Winter AJ. Macrophages and Brucella. Immunol Ser. 1994;60:363-80
6. Jones SM, Winter AJ. Survival of virulent and attenuated strains of Brucella abortus in normal and gamma interferon-activated murine peritoneal macrophages. Infect Immun. 1992;60:3011-4
7. Harmon BG, Adams LG, Frey M. Survival of rough and smooth strains of Brucella abortus in bovine mammary gland macrophages. Am J Vet Res. 1988;49:1092-7
8. Gross A, Terraza A, Ouahrani-Bettache S, Liautard JP, Dornand J. In vitro Brucella suis infection prevents the programmed cell death of human monocytic cells. Infect Immun. 2000;68:342-51
9. Tolomeo M, Di Carlo P, Abbadessa V, Titone L, Miceli S, Barbusca E. et al. Monocyte and lymphocyte apoptosis resistance in acute and chronic brucellosis and its possible implications in clinical management. Clin Infect Dis. 2003;36:1533-8
10. He Y, Reichow S, Ramamoorthy S, Ding X, Lathigra R, Craig JC. et al. Brucella melitensis triggers time-dependent modulation of apoptosis and down-regulation of mitochondrion-associated gene expression in mouse macrophages. Infect Immun. 2006;74:5035-46
11. Eskra L, Mathison A, Splitter G. Microarray analysis of mRNA levels from RAW264.7 macrophages infected with Brucella abortus. Infect Immun. 2003;71:1125-33
12. Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165-73
13. Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85
14. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
15. Melino G, Knight RA. MicroRNAs meet cell death. Cell Death Differ. 2010;17:189-90
16. Cremer TJ, Ravneberg DH, Clay CD, Piper-Hunter MG, Marsh CB, Elton TS. et al. MiR-155 induction by F. novicida but not the virulent F. tularensis results in SHIP down-regulation and enhanced pro-inflammatory cytokine response. PLoS ONE. 2009;4:e8508
17. Cameron JE, Yin Q, Fewell C, Lacey M, McBride J, Wang X. et al. Epstein-Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol. 2008;82:1946-58
18. Monk CE, Hutvagner G, Arthur JS. Regulation of miRNA transcription in macrophages in response to Candida albicans. PLoS ONE. 2010;5:e13669
19. Xiao B, Liu Z, Li BS, Tang B, Li W, Guo G. et al. Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J Infect Dis. 2009;200:916-25
20. Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026-33
21. Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z. et al. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol. 2008;82:5295-306
22. Audic S, Lescot M, Claverie J-M, Scholz HC. Brucella microti: the genome sequence of an emerging pathogen. BMC Genomics. 2009;10:352
23. Wu YQ, Chen DJ, He HB, Chen DS, Chen LL, Chen HC. et al. Pseudorabies virus infected porcine epithelial cell line generates a diverse set of host microRNAs and a special cluster of viral microRNAs. PLoS ONE. 2012;7:e30988
24. Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713-4
25. Huang J, Ju Z, Li Q, Hou Q, Wang C, Li J. et al. Solexa sequencing of novel and differentially expressed microRNAs in testicular and ovarian tissues in Holstein cattle. Int J Biol Sci. 2011;7:1016-26
26. Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1
27. Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z. et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293-7
28. Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM. et al. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710-5
29. Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277-80
30. Elmore S. Apoptosis: A Review of Programmed Cell Death. Toxicol Pathol. 2007;35:495-516
31. Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 2008;16:70-8
32. Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr Opin Cell Biol. 2004;16:663-9
33. Cardoso P, Macedo G, Azevedo V, Oliveira S. Brucella spp noncanonical LPS. Microb Cell Fact. 2006;5:13
34. Lapaque N, Moriyon I, Moreno E, Gorvel JP. Brucella lipopolysaccharide acts as a virulence factor. Curr Opin Microbiol. 2005;8:60-6
35. Erridge C, Bennett-Guerrero E, Poxton IR. Structure and function of lipopolysaccharides. Microbes Infect. 2002;4:837-51
36. Wang ZJ, Zhang FM, Wang LS, Yao YW, Zhao Q, Gao X. Lipopolysaccharides can protect mesenchymal stem cells (MSCs) from oxidative stress-induced apoptosis and enhance proliferation of MSCs via Toll-like receptor(TLR)-4 and PI3K/Akt. Cell Biol Int. 2009;33:665-74
37. Arimilli S, Johnson JB, Alexander-Miller MA, Parks GD. TLR-4 and -6 agonists reverse apoptosis and promote maturation of simian virus 5-infected human dendritic cells through NFkB-dependent pathways. Virology. 2007;365:144-56
38. Porte F, Liautard JP, Kohler S. Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect Immun. 1999;67:4041-7
39. Kobayashi SD, Braughton KR, Whitney AR, Voyich JM, Schwan TG, Musser JM. et al. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc Natl Acad Sci U S A. 2003;100:10948-53
40. Grassme H, Jendrossek V, Gulbins E. Molecular mechanisms of bacteria induced apoptosis. Apoptosis. 2001;6:441-5
41. Barquero-Calvo E, Chaves-Olarte E, Weiss DS, Guzman-Verri C, Chacon-Diaz C, Rucavado A. et al. Brucella abortus uses a stealthy strategy to avoid activation of the innate immune system during the onset of infection. PLoS ONE. 2007;2:e631
42. Arenas GN, Staskevich AS, Aballay A, Mayorga LS. Intracellular trafficking of Brucella abortus in J774 macrophages. Infect Immun. 2000;68:4255-63
43. Pizarro-Cerda J, Meresse S, Parton RG, van der Goot G, Sola-Landa A, Lopez-Goni I. et al. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect Immun. 1998;66:5711-24
44. van Eijk M, Medema JP, de Groot C. Cutting edge: cellular Fas-associated death domain-like IL-1-converting enzyme-inhibitory protein protects germinal center B cells from apoptosis during germinal center reactions. J Immunol. 2001;166:6473-6
45. Chen L, Zhuang G, Li W, Liu Y, Zhang J, Tian X. RGD-FasL induces apoptosis of pituitary adenoma cells. Cell Mol Immunol. 2008;5:61-8
46. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42
47. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132-41
48. Silver N, Proctor GB, Arno M, Carpenter GH. Activation of mTOR coincides with autophagy during ligation-induced atrophy in the rat submandibular gland. Cell Death Dis. 2010;1:e14
49. Shang L, Wang X. AMPK and mTOR coordinate the regulation of Ulk1 and mammalian autophagy initiation. Autophagy. 2011;7:924-6
50. Itakura E, Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy. 2009;5:534-6
51. Marzo I, Naval J. Bcl-2 family members as molecular targets in cancer therapy. Biochem Pharmacol. 2008;76:939-46
52. Rong Y, Distelhorst CW. Bcl-2 protein family members: versatile regulators of calcium signaling in cell survival and apoptosis. Annu Rev Physiol. 2008;70:73-91
53. Tait SW, de Vries E, Maas C, Keller AM, D'Santos CS, Borst J. Apoptosis induction by Bid requires unconventional ubiquitination and degradation of its N-terminal fragment. J Cell Biol. 2007;179:1453-66
54. Chen F, He Y. Caspase-2 mediated apoptotic and necrotic murine macrophage cell death induced by rough Brucella abortus. PLoS ONE. 2009;4:e6830
Author contact
![]() Corresponding author: Zheng-Fei Liu Tel: +86-27-87282608; Fax: +86-27-87282608; E-mail: lzf6789hzau.edu.cn
Corresponding author: Zheng-Fei Liu Tel: +86-27-87282608; Fax: +86-27-87282608; E-mail: lzf6789hzau.edu.cn

 Global reach, higher impact
Global reach, higher impact