10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2012; 8(7):1055-1061. doi:10.7150/ijbs.4460 This issue Cite
Short Research Communication
Protein Interactions on Telomeric Retrotransposons in Drosophila
1. Department of Genetics, University of Szeged, H-6701 Szeged, Hungary
2. Laboratory of Molecular Genetics, National Institute of Environmental Health Sciences, Research Triangle Park, North Carolina 27709 USA
† Developmental Biology Center, University of California, Irvine CA 92697 USA.
Received 2012-4-11; Accepted 2012-7-4; Published 2012-8-15
Abstract
Telomere length in Drosophila is maintained by targeted transposition of three non-LTR retrotransposons: HeT-A, TART and TAHRE (HTT), but understanding the regulation of this process is hindered by our poor knowledge of HTT associated proteins. We have identified new protein components of the HTT array: Chromator (Chro), the TRF2/DREF complex and the sumoylation machinery. Chro was localized on telomeric HTT arrays by immunostaining, where it may interact with Prod directly, as indicated by yeast two-hybrid interaction, co-IP, and colocalization on polytene chromosomes. The TRF2/DREF complex may promote the open structure of HTT chromatin. The protein interactions controlling HTT chromatin structure and telomere length may be modulated by sumoylation.
Keywords: chromatin, Chromator, prod, putzig, Z4, sumoylation, telomere, Drosophila, retrotransposon
Introduction
Drosophilids lack telomerase; their chromosome length is maintained by targeted transposition of HTT elements and homologous recombination between these elements (1;2). To understand the mechanisms that regulate telomere length, it is important to unravel the protein components of HTT-associated chromatin, however these are difficult to identify genetically because of the absence of a predictable mutant phenotype. Three components of HTT chromatin have been identified by immunostaining: Z4, JIL-1 and Prod (3;4), and HP1 was detected on HTT by immunoprecipitation (5). The potential roles of HP1 and Prod on the HTT array has been partly elucidated, but those of Z4 and JIL-1 are not known.
HP1 is encoded by the Su(var)205 gene. Increased levels of HeT-A transcript observed in Su(var)205 mutants is attributed to reduced HP1 on the HTT array (5), while a defect in the capping function in mutants for these genes may lead to increased retroelement transposition, terminal gene conversion, and elongated telomeres (2;4).
We have shown previously that Prod binds upstream of the HeT-A promoter and acts as a repressor of HeT-A transcription, however we did not detect elongated telomeres in prod heterozygous mutants (4).
Z4, encoded by pzg, is a component of two protein complexes that generate open chromatin: TRF2/DREF (6) and NURF (7). pzg mutants, however, are haplo-suppressors and triplo-enhancers of position effect variegation (PEV), indicative of an opposite role in chromatin condensation, and accordingly polytene chromosomes in pzg mutants exhibit decondensation (8).
JIL-1 phosphorylates histone H3 at Ser 10 (9). Mutations that reduce JIL-1 function increase condensation of euchromatic regions in polytene chromosomes and are enhancers of PEV (10), consistent with a role of JIL-1 in maintaining an “open” chromatin configuration.
We searched for novel protein components of HTT chromatin based on an interaction with Prod in a protein interaction screen, and tested the presence of Prod-interacting proteins on telomeres. One interaction could be verified by co-immunoprecipitation.
Materials and methods
Yeast two-hybrid screen
The Clontech Matchmaker cDNA library was screened (Cat. no: IL4003AH) according to the manufacturers instructions. The bait was full length prod cDNA in the pBTM116 vector. The library makes use of the pACT2 vector, thus the interaction rescues Leu auxotrophy and activates the Gal4 gene for blue selection. Blue positives were transferred to new plates and restained for β-gal activity three times, to minimize background. Approximately 120,000 cDNA clones were tested, and the best 100 potential interactors were sequenced.
Immunoprecipitation
107 S2 cells were centrifuged for 1 min. at 6000 rpm, washed in 1xPBS and resuspended in 1ml lysis buffer (50mM Tris 7.5, 50mM NaCl, 0.2% NP-40, 0.5mM EDTA, 1mM PMSF, Roche complete mini protease inhibitor, 1mM DTT). After 1h on ice, the lysate was centrifuged for 2 min. at 4°C, 10,000 rpm, then the supernatant was recentrifuged at 13,000 rpm for 10 min. For preclearing 25μl bead volume protein-A sepharose was added to the supernatant and rotated for 1h at 4°C, then centrifuged at 2000 rpm for 1 min. Beads were washed 3 times in lysis buffer, then taken up in 1x Laemmli and run in control lanes.
Precomplex was made by incubating 50μl bead-vol protein-A sepharose with 6μl anti-PROD-AB (or 10μl anti-CHRO-AB) in 1 ml lysis buffer for 1h at 4°C, then washed 3 times for 5 min in lysis buffer. Half of the precomplex was run as a control in AB lane, the other half was added to the precleared lysate and incubated overnight at 4°C. Beads were centrifuged for 1 min at 2000 rpm. 1/50th of supernatant was run in lane REST to monitor the IP efficiency. Beads were washed 3 times for 5 min in lysis buffer, then resuspended in 40μl 1x Laemmli and loaded on gels.
Immunofluorescence
Polytene chromosomes and immunostainings were prepared as described previously (4). Rabbit anti-Prod antibodies have been described (11). Rabbit anti-Chro (12) and mouse anti-Z4 antibodies were a gift of Dr. Harald Saumweber. Rabbit anti-Smt3 antibodies (13) were provided by Dr. Jacob Seeler. Rabbit anti-DREF antibodies (14) were from Imre Boros. Antibodies were used in the following dilutions: Prod 1:2000, Z4 1:2, Chro 1:1000, Smt3 1:50, DREF 1:500. The appropriate Alexa Fluor 546- and 488-conjugated secondary antibodies (Invitrogen) were used to visualize primary labeling.
Fly stocks
prodU is null allele with a stop codon after aa 94. prodE is a hypomorphic allele (G61E). smt304493 is a P-induced null; lwr5 is a weak hypomorph; lwr05486 is null; pzg1.3 is a null. chroΔKG12 resulted from an imprecise excision that deleted most of the coding region (12). JIL-13 is a gain of function mutation, while JIL-1z2 is a loss of function null allele (9). Other mutants are described at FlyBase.org.
DNA extraction and qPCR
HeT-A primers and qPCR are described in (4). The mutants tested were all heterozygous (except Tel1). The y w67c23; Sco/SM1 and y w67c23; Sb/TM6 stocks were produced through a series of crosses to substitute unbalanced chromosomes with those from our y w67c23; Oregon-R control. Before the qPCR assessment of relative genomic HeT-A copy number, mutants were crossed repeatedly to appropriate balancer stocks to generate similar genetic backgrounds, then maintained in stock for two years to allow the retrotransposon array to grow.
Results and Discussion
Yeast two-hybrid screening
Interactors that were identified more than once, gave strong staining, or could be related to telomeres are listed on Table 1.
Prod was found four times, suggesting that Prod interacts with itself, as suggested earlier (15). Smt3 activating enzyme 2 (Uba2) was identified six times, and Ubc9 (encoded by lwr), the SUMO-specific E2 conjugating enzyme, was identified as one of the strongest interacting peptides. Thus, two major members of the sumoylation pathway interact with Prod, suggesting that either Prod is sumoylated, or Prod recruits the SUMO machinery to modify Prod interacting proteins.
Among other interacting peptides Z4 and Chro are of particular significance. Z4 is one of the few known proteins associated with the HTT domain (3). Chro a chromodomain protein, (synonymous name Chriz), was identified by its interaction with Z4 (8) and was shown to localize on XL and 2L telomeres (12). Chro also interacts directly with JIL-1 (16), although the latter was not found in our screen, and Chro, JIL-1 and Z4 colocalize in the interbands of polytene chromosomes (12). It is reassuring that both Z4 and Chro showed up in the screen, suggesting that these proteins can be part of a common complex, and Chro might also colocalize with Z4 and JIL-1 on telomeres.
Proteins interacting with Prod identified in yeast two-hybrid screen.
| gene name | protein function | no. of hits | strength of interaction |
|---|---|---|---|
| Uba2 | Smt3 activating enzyme | 6 | strong |
| prod | DNA binding protein | 4 | strong |
| Nc73EF | oxoglutarate dehydrogenase | 3 | strong |
| Cys | cysteine protease inhibitor | 2 | strong |
| CG4847 | cysteine peptidase | 2 | strong |
| windei | cofactor of the H3K9 methyl transferase | 2 | strong |
| bip2 | TAFII155, DNA binding protein | 2 | strong |
| CG5001 | chaperone | 2 | strong |
| NaCP60E | sodium ion transport | 2 | strong |
| dhd | nuclear electron transport | 2 | moderate |
| lwr | ubiquitin conjugating enzyme | 1 | strong |
| CG9797 | Zn finger transcription factor | 1 | strong |
| phol | Zn finger transcription factor | 1 | strong |
| CG4557 | transcription factor | 1 | strong |
| CG31365 | Zn finger transcription factor | 1 | strong |
| Chro | chromatin binding (chromo-domain) | 1 | strong |
| pzg (Z4) | DNA-binding interband protein | 1 | strong |
| E(z) | histone methylation | 1 | weak |
Prod binds Chro in vivo directly
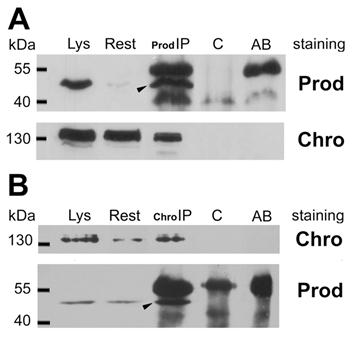
To confirm the in vivo relevance of the possible Prod-Z4 and Prod-Chro interactions we performed co-immunoprecipitation experiments from Drosophila S2 cells. Figure 1 demonstrates that Chro can be co-immunoprecipitated with Prod, and a small fraction of these proteins seem to be associated in vivo. Together with the yeast two-hybrid result this suggests that Prod binds Chro directly, and these proteins can be part of a common complex, at least at those sites where they colocalize. The Prod-Z4 interaction could not be confirmed the same way.
Coimmunoprecipitation of Chro and Prod from S2 cells. (A) Arrowhead indicates that Prod was immunoprecipitated with anti-Prod antibody from the cell lysate (Lys) with minor left-over (Rest). The secondary anti-rabbit antibody used in the Western blot also detects the rabbit-anti-Prod antibodies in lane Prod-IP as major bands above and below Prod, as indicated in lane AB, where anti-Prod AB was run alone. The lower filter, stained for Chro, indicates that Chromator is also present in the Prod-IP lane, although most of it remained in the supernatant (Rest). The Prod-IP lane stained for Prod contains 1/20th of total IP, while Prod-IP lane stained for Chro contains all the rest (19/20th). As both lanes Lys and Rest contain 1/50th of total extract, and the Prod-IP lane is roughly estimated to contain half Chro compared to Rest, it follows that less than 1% of Chro is associated with Prod in vivo. (B) In the reciprocal experiment Chro was immunoprecipitated from an S2 extract and a small fraction of total Prod was co-immunoprecipitated with it (arrowhead). Control lanes (C) show that the same immunoprecipitates made in the absence of Prod or Chro antibodies contain neither Prod nor Chro.
Immunolocalization on telomeres
To investigate whether Chro and Z4 colocalize with Prod at telomeres, we performed immunofluorescence on polytene chromosomes of Tel/Oregon-R hybrids. In contrast to Oregon-R chromosomes, the chromosomes maintained in the presence of Tel1 have greatly extended HTT arrays. This facilitates the identification of proteins binding to the asymmetric HTT arrays of the hybrids (3;4). As both the available Prod and Chro antibodies were derived from rabbit, we could not use them together in these experiments.
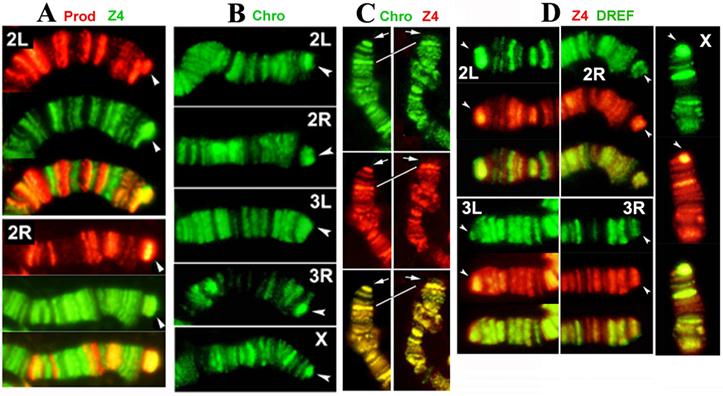
Our results show that Z4 colocalizes with Prod at the HTT arrays of all chromosomes (Figure 2A), confirming previous immunocytochemistry observations with single antibody staining, which showed that Z4 binds to HTT (3). In contrast to a previous report (12), we found that Chro is present on all telomeres (Figure 2B). In addition, our immunostaining results demonstrate that Chro colocalizes with Z4 not only in interbands but also on telomeres (Figure 2C), in the same pattern that has been demonstrated for Prod (4). Large terminal deficiencies removing both telomere-associated sequence (TAS) repeats and HTT seem to abolish Z4/Chro/Prod telomeric binding, while removing TAS alone does not affect telomere binding of either protein, indicating that HTT is a predominant target for a potential Z4/Chro/Prod protein complex.
The Prod and Z4/Chro staining pattern of chromosome arms is generally complementary with few overlaps (Figure 2A); their HTT colocalization seems to be an exception rather than the rule. This makes it likely that the Prod-Chro interaction is restricted to these overlapping sites, including HTT, and can explain why only a small fraction of Prod and Chro are associated with each other in vivo. Chro is also part of the spindle matrix, where it interacts with Skeletor and EAST (19). As Chro also directly interacts with Z4 (8), JIL-1 (16) and Prod (present study), Chro appears to be an adaptor protein, which can connect different chromatin components in a context dependent manner.
As Z4 is part of the TRF2/DREF complex (7;8), we also tested for the presence of DREF on HTT arrays with immunostaining. DREF was found to be abundant on all HTT arrays (Figure 2D), suggesting that the entire TRF2/DREF complex is present, which may contribute to the euchromatic structure of HTT reported previously (20). As HTT elements lack the DREF target sequence TATCGATA (21), the complex must be anchored differently, possibly through Z4.
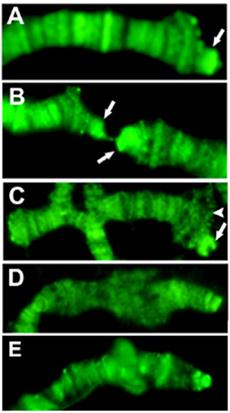
The single Drosophila SUMO homologue is Smt3 (22), so sumoylated proteins can be identified by anti-Smt3 antibodies. We tested the presence and distribution of this modification on telomeres with the same method as above. Telomeric ditribution of Smt3 indicates that the HTT array is heavily sumoylated in a pattern that coincides with Prod (Figure 3).
Prod, Z4, Chro and DREF bind to the HTT array. (A) Double-staining of Tel/Oregon R polytene chromosomes with anti-Prod (red) and anti-Z4 (green) antibodies. 2L and 2R telomeres are shown, but other telomeres stain similarly. Prod and Z4 staining patterns of chromosome arms are different, but both proteins localize to the elongated HTT array (arrowhead), as indicated by the strong yellow color in the merged images. (B) Staining of Tel/Oregon R polytene chromosomes with anti-Chro antibodies show localization at the extended HTT array of the Tel homologue (arrowhead), similar to Prod and Z4. (C) The localization of Z4 (red) and Chro (green) completely overlap not only in interbands but also on telomeres (arrows). The left panel shows the y1 w X chromosome, which lacks TAS (17), nevertheless Z4 and Chro telomeric localization remains the same as in wild type (not shown). The right panel shows an X chromosome with terminal deficiency, Df(1)RT858 (18), which removes both TAS and HTT (white line connects identical bands in 1C) but retains a functional capping complex. Strong telomeric Z4 and Chro staining disappears from this telomere indicating that they are associated with HTT and not with the cap. (D) Double-staining of Tel/Oregon R polytene chromosomes with anti-Z4 (red) and anti-DREF (green) antibodies. The extended HTT homologues (arrowheads) stain with both antibodies.
SUMO staining of Tel/OreR telomeres indicates that proteins on HTT are intensely sumoylated, while the sumoylation level on the cap and TAS is not prominent. Only chromosome tips are shown, and arrows point at the elongated HTT array on 3R (A), 2R and 2L (B). The terminal 2L deficiency Df(2L)GB26/Cy (23) (C) shows significant sumoylation only on the balancer chromosome (arrow), not at the terminal deficiency chromosome (arrowhead), although it also has a capping complex. The sumoylation level of the X chromosome telomere in wild type (E) is comparable to that of the y1 w chromosome (D), which lacks X TAS, indicating that TAS associated proteins may not be sumoylated significantly.
Sumoylation is often associated with transcriptional repression (24), thus sumoylation may contribute to HTT transcriptional inactivation together with Prod and HP1, or it may act as a switch between chromatin states. We do not know which HTT binding proteins are sumoylated. Prod, Chro and JIL-1 each have 2-3 very high probability sumoylation sites, while Z4 does not. It is notable that we identified Windei, a cofactor of the H3K9 methyl transferase dSETDB1/Eggless (25), as a strong interactor of Prod, although this interaction remains to be confirmed. Eggless is the only essential H3K9 methyltransferase in Drosophila required for H3K9 trimethylation in the female germ line (26), where HTT retrotransposons are active, and it was shown to silence HeT-A transcription (27). Binding of the telomeric component HP1 depends on H3K9 modification made by Eggless, which has been shown to be strongly sumoylated (25).
Telomere length in heterozygous mutants
The chromatin state of HTT may influence telomere length through modulating retrotransposition or gene conversion (1). In heterozygous mutants the reduced protein concentration can lead to phenotypic change if the threshold value required for wild type exceeds 50% gene activity. For example, longer telomeres are detected in Su(var)205 heterozygotes (2). We tested whether mutants of known HTT components have an effect on telomere length by comparing the HeT-A content of mutant heterozygotes to wild type using quantitative PCR (Figure 4).
Telomere length comparison of heterozygous mutants. Graph represents relative copy number of HeT-A (average 2∆ct values ), normalized to the copy number of ribosomal protein S17 (RpS17). Error bars represent the standard deviation of threshold values from the average of triplicate reactions run on different days, each in triplicate. Blue columns represent negative controls; the green column shows the positive Tel1 control (28). All strains except Tel carry y w67c23.
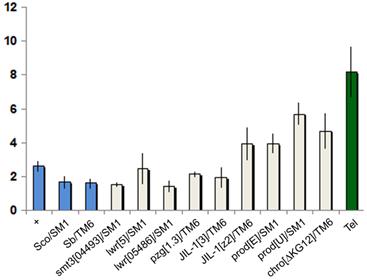
We found that reduced levels of Prod, Chro, JIL-1, Z4, SUMO, or Ubc9 have little or no influence on telomere length. Previously we found a slight (1.1-1.6x) but insignificant difference between the HeT-A copy number of prod mutants and Oregon-R control (4), while in the present study we found slightly larger increases (1.5-2.2x), possibly because more effort was made to control the genetic background. However after making a Bonferroni correction, with a stringent p-value to exclude possible false positives, only prodU shows a significant increase. Altogether the four prod alleles tested in two different labs show a slight increase in HeT-A copy number, suggesting that the increase may be real, although weak. Again, after making the Bonferroni correction, none of the other mutants tested showed an indication of significantly longer telomeres, or the observable effect may be very weak, as suggested for prod mutants. So in the tested heterozygous mutants the gene dosage changes do not sufficiently alter chromatin structure or sumoylation of HTT proteins to increase either transposition frequency or gene conversion at telomeres.
Conclusions
Chro seems to be a central mediator of protein interactions on the HTT domain, where it can interact directly with Prod, JIL-1, and Z4 (part of the DREF complex). Prod binds as a multimer upstream of HeT-A promoters and may recruit the SUMO machinery, which may in turn modify the binding affinity of different components. It is doubtful, however, that any of these activities influence telomere length.
Acknowledgements
We thank Harald Saumweber, Pavel Georgiev and the Bloomington Drosophila Stock Center for fly stocks, and Harald Saumweber, Jacob Seeler and Imre Boros for antibodies. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, OTKA T/F 037422. The publication is supported by the European Union and co-funded by the European Social Fund. Project title: “Broadening the knowledge base and supporting the long term professional sustainability of the Research University Centre of Excellence at the University of Szeged by ensuring the rising generation of excellent scientists.” Project number: TÁMOP-4.2.2/B-10/1-2010-0012.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Capkova Frydrychova R, Biessmann H, Mason JM. Regulation of telomere length in Drosophila. Cytogenet Genome Res. 2008;122(3-4):356-64
2. Savitsky M, Kravchuk O, Melnikova L, Georgiev P. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol Cell Biol. 2002;22(9):3204-18
3. Andreyeva EN, Belyaeva ES, Semeshin VF, Pokholkova GV, Zhimulev IF. Three distinct chromatin domains in telomere ends of polytene chromosomes in Drosophila melanogaster Tel mutants. J Cell Sci. 2005;118(Pt 23):5465-77
4. Török T, Benitez C, Takacs S, Biessmann H. The protein encoded by the gene proliferation disrupter (prod) is associated with the telomeric retrotransposon array in Drosophila melanogaster. Chromosoma. 2007;116(2):185-95
5. Capkova Frydrychova R, Mason JM, Archer TK. HP1 is distributed within distinct chromatin domains at Drosophila telomeres. Genetics. 2008;180(1):121-31
6. Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420(6914):439-45
7. Kugler SJ, Nagel AC. A Novel Pzg-NURF Complex Regulates Notch Target Gene Activity. Mol Biol Cell. 2010;21(19):3443-3448
8. Eggert H, Gortchakov A, Saumweber H. Identification of the Drosophila interband-specific protein Z4 as a DNA-binding zinc-finger protein determining chromosomal structure. J Cell Sci. 2004;117(Pt 18):4253-64
9. Zhang W, Deng H, Bao X, Lerach S, Girton J, Johansen J, Johansen KM. The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development. 2006;133(2):229-35
10. Bao X, Deng H, Johansen J, Girton J, Johansen KM. Loss-of-function alleles of the JIL-1 histone H3S10 kinase enhance position-effect variegation at pericentric sites in Drosophila heterochromatin. Genetics. 2007;176(2):1355-8
11. Török T, Harvie PD, Buratovich M, Bryant PJ. The product of proliferation disrupter is concentrated at centromeres and required for mitotic chromosome condensation and cell proliferation in Drosophila. Genes Dev. 1997;11(2):213-25
12. Gortchakov AA, Eggert H, Gan M, Mattow J, Zhimulev IF, Saumweber H. Chriz, a chromodomain protein specific for the interbands of Drosophila melanogaster polytene chromosomes. Chromosoma. 2005;114(1):54-66
13. Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem. 2000;275(18):13321-9
14. Papai G, Komonyi O, Toth Z, Pankotai T, Muratoglu S, Udvardy A, Boros I. Intimate relationship between the genes of two transcriptional coactivators, ADA2a and PIMT, of Drosophila. Gene. 2005;348:13-23
15. Török T, Gorjanacz M, Bryant PJ, Kiss I. Prod is a novel DNA-binding protein that binds to the 1.686 g/cm(3) 10 bp satellite repeat of Drosophila melanogaster. Nucleic Acids Res. 2000;28(18):3551-7
16. Rath U, Ding Y, Deng H, Qi H, Bao X, Zhang W, Girton J, Johansen J, Johansen KM. The chromodomain protein, Chromator, interacts with JIL-1 kinase and regulates the structure of Drosophila polytene chromosomes. J Cell Sci. 2006;119(Pt 11):2332-41
17. Abad JP, de PB, Osoegawa K, De Jong PJ, Martin-Gallardo A, Villasante A. Genomic analysis of Drosophila melanogaster telomeres: full-length copies of HeT-A and TART elements at telomeres. Mol Biol Evol. 2004;21(9):1613-9
18. Biessmann H, Carter SB, Mason JM. Chromosome ends in Drosophila without telomeric DNA sequences. Proc Natl Acad Sci U S A. 1990;87(5):1758-61
19. Johansen J, Johansen KM. The spindle matrix through the cell cycle in Drosophila. Fly (Austin ). 2009;3(3):213-20
20. Biessmann H, Prasad S, Walter MF, Mason JM. Euchromatic and heterochromatic domains at Drosophila telomeres. Biochem Cell Biol. 2005;83(4):477-85
21. Matsukage A, Hirose F, Yoo MA, Yamaguchi M. The DRE/DREF transcriptional regulatory system: a master key for cell proliferation. Biochim Biophys Acta. 2008;1779(2):81-9
22. Talamillo A, Sanchez J, Cantera R, Perez C, Martin D, Caminero E, Barrio R. Smt3 is required for Drosophila melanogaster metamorphosis. Development. 2008;135(9):1659-68
23. Walter MF, Jang C, Kasravi B, Donath J, Mechler BM, Mason JM, Biessmann H. DNA organization and polymorphism of a wild-type Drosophila telomere region. Chromosoma. 1995Dec;104(4):229-41
24. Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci U S A. 2003;100(23):13225-30
25. Koch CM, Honemann-Capito M, Egger-Adam D, Wodarz A. Windei, the Drosophila homolog of mAM/MCAF1, is an essential cofactor of the H3K9 methyl transferase dSETDB1/Eggless in germ line development. PLoS Genet. 2009;5(9):e1000644
26. Yoon J, Lee KS, Park JS, Yu K, Paik SG, Kang YK. dSETDB1 and SU(VAR)3-9 sequentially function during germline-stem cell differentiation in Drosophila melanogaster. PLoS One. 2008;3(5):e2234
27. Gou D, Rubalcava M, Sauer S, Mora-Bermudez F, Erdjument-Bromage H, Tempst P, Kremmer E, Sauer F. SETDB1 is involved in postembryonic DNA methylation and gene silencing in Drosophila. PLoS One. 2010;5(5):e10581
28. Siriaco GM, Cenci G, Haoudi A, Champion LE, Zhou C, Gatti M, Mason JM. Telomere elongation (Tel), a new mutation in Drosophila melanogaster that produces long telomeres. Genetics. 2002;160(1):235-45
Author contact
![]() Corresponding author: Tibor Török, Department of Genetics, University of Szeged, H-6701 Szeged, Hungary.
Corresponding author: Tibor Török, Department of Genetics, University of Szeged, H-6701 Szeged, Hungary.

 Global reach, higher impact
Global reach, higher impact