Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2012; 8(8):1085-1096. doi:10.7150/ijbs.4406 This issue Cite
Research Paper
Molecular and Biochemical Characterization of a Novel β-N-Acetyl-D-Hexosaminidase with Broad Substrate-Spectrum from the Aisan Corn Borer, Ostrinia Furnacalis
School of Life Science and Biotechnology, Dalian University of Technology, Dalian, China.
* These authors contributed equally to this work.
Received 2012-3-27; Accepted 2012-8-19; Published 2012-8-31
Abstract
Insect β-N-acetyl-D-hexosaminidases with broad substrate-spectrum (IBS-Hex) are the homologues of human β-N-acetyl-D-hexosaminidase A/B (HsHex A/ B). These enzymes are distributed in most insect species and vary in physiological roles. In this study, the gene encoding an IBS-Hex, OfHEX2, was cloned from the Asian corn borer, Ostrinia furnacalis. Recombinant OfHex2 was expressed in Pichia pastoris and purified to homogeneity. By structure-based sequence alignment, three sequence segments with high diversity among IBS-Hexs were firstly concluded. Furthermore, the residue pair N423-R424/ D452-L453 important for the specificity of human β-N-acetyl-D-hexosaminidase subunits α/β toward charged/ non-charged substrates was not conserved in OfHex2 and other IBS-Hexs. Unlike HsHex A, OfHex2 could not degrade charged substrates such as 4-methylumbelliferyl-6-sulfo-N-acetyl-β-D-glucosaminide, ganglioside GM2 and peptidoglycan. OfHex2 showed a broad substrate-spectrum by hydrolyzing β1-2 linked N-acetyl-D-glucosamines from both α3 and α6 branches of biantennary N-glycan and β1-4 linked GlcNAc from chitooligosaccharides as well as β1-3 linked or β1-4 linked N-acetyl-D-galactosamine from oligosaccharides of glycolipids. Real-time PCR analysis demonstrated that the expression of OfHEX2 was up-regulated in the intermolt stages (both larva and pupa), and mainly occurred in the carcass rather than in the midgut during the feeding stage of fifth (final) instar larva. This study reported a novel IBS-Hex with specific biochemical properties, suggesting biodiversity of this class of enzymes.
Keywords: β-N-acetyl-D-hexosaminidase, insect, glycoside hydrolase, N-glycan, Ostrinia furnacalis.
INTRODUCTION
β-N-acetyl-D-hexosaminidases (EC 3.2.1.52) belong to the glycoside hydrolase family 20 and catalyze the removal of N-acetyl-D-glucosamine (GlcNAc) or N-acetyl-D-galactosamine (GalNAc) from the non-reducing ends of a variety of physiological substrates, such as oligosaccharides, glycoproteins and glycolipids. These enzymes are present in numerous species of diverse organisms, in which they play different physiological roles [1].
In insects, two β-N-acetyl-D-hexosaminidase isoforms with strict substrate spectra have been extensively studied. One is chitinolytic β-N-acetyl-D-hexosaminidase, which is specifically involved in chitin degradation during insect metamorphosis [2-6]. This enzyme degrades linear chitooligosaccharides with high efficiency but cannot degrade the sugar parts derived from glycoconjugates [such as biantennary N-glycan (GnGn)]. The second is the N-glycan processing β-N-acetyl-D-hexosaminidase, which is a membrane-bound enzyme that is involved in the degradation of N-glycans [7-9]. It selectively removes β1-2 linked GlcNAc from the α3 branch of GnGn, but cannot remove β1-2 linked GlcNAc from the α6 branch of GnGn or β1-4 linked GlcNAc from chitooligosaccharides.
In addition to the above-mentioned β-N-acetyl-D-hexosaminidases, another, broad substrate-spectrum β-N-acetyl-D-hexosaminidase (IBS-Hex) can be found in insects. IBS-Hexs are distributed widely in insects with the exception of the fruitfly, Drosophila melanogaster [10] and the medfly, Ceratitis capitata [11]. IBS-Hexs are found to have high sequence similarity to human β-N-acetyl-D-hexosaminidases but low sequence similarity with other insect Hex enzymes. The physiological importance of human β-N-acetyl-D-hexosaminidase A (HsHex A, αβ heterdimer) and B (HsHexB, ββ homodimer) are linked to the degradation of glycoconjugates (such as GM2 ganglioside) [12,13]. However, little is known about IBS-Hexs. IBS-Hexs from several insect species have been isolated and briefly characterized. The crude preparations of recombinant IBS-Hexs from Spodoptera frugiperda (SfGlcNAcase1 and SfGlcNAcase3) [14] and Bombyx mori (BmGlcNAcase2 and BmGlcNAcase3) [15] were found to have an acidic pH-optimum and could remove the terminal GlcNAc from both chitooligosaccharides and α3 and α6 branches of GnGn. SfHex, another IBS-Hex purified from the culture broth of S. frugiperda cells, had 11-fold preference for GnGn-PA than (GlcNAc)3-PA and was postulated to act as a N-glycan processing enzyme [16]. However, SfHex and SfGlcNAcase3 were actually encoded by the same gene [9,14,16]. Thus, to clarify the physiological role of IBS-Hexs, the substrate specificity of these enzymes toward various substrates with different glycan structures needs to be assessed. Moreover, the gene expression pattern of IBS-Hex was only partially determined for BmGlcNAcase2 [17], and thus more experimental data is needed to clarify their physiological importance.
In this study, we cloned, expressed and characterized one IBS-Hex, named OfHex2, from the Asian corn borer, Ostrinia furnacalis (Guenée), a lepidopteran pest that severely affects corn production. Sequence characteristics, enzymatic property and the expression patterns of its gene were studied to reveal the biochemical and physiological aspects of OfHex2.
MATERIALS AND METHODS
Insect culture
The Asian corn borer, O. furnacalis, was kindly provided by Prof. Kanglai He from the Institute of Plant Protection, Chinese Academy of Agricultural Sciences. The larvae were reared using an artificial diet at 26-28 °C under a relative humidity of 70-90 % and a photoperiod of 16 h light and 8 h darkness. Insects at different developmental stages were frozen in liquid nitrogen and stored at -80 °C.
Molecular cloning of OfHEX2
Gene-specific primers for OfHEX2, named Of2F1, Of2F2, Of2R1, Of2R2 and Of2R3 were designed and synthesized based on the conserved regions reported for insect β-N-acetyl-D-hexosaminidases (Supplementary Material: Table S1). Total RNA was extracted from day-3 fifth instar O.furnacalis larvae (5L3) using TaKaRa RNAiso Reagent (TaKaRa, Dalian, China). One mg of total RNA was used as template for the synthesis of cDNA by PrimeScriptTM 1st Strand cDNA Synthesis Kit (TaKaRa) according to the manufacturer's instruction. Using the cDNA as template, two fragments of OfHEX2 (Of2-1, Of2-2) were obtained by PCR using Of2F1/Of2R1 and Of2F2/Of2R2 as primers. The PCR products were purified with the TaKaRa Agarose Gel DNA purification Kit (TaKaRa) and sequenced with an ABI377 DNA sequencer (Applied Biosystems, Foster City, USA). Primers Of2F3 and Of2R3 were designed on the basis of the PCR product sequences of OfHEX2. Using the cDNA as template, the third fragment of OfHEX2 (Of2-3) was obtained by PCR using Of2F3/Of2R3 as primers. The PCR products were purified and sequenced as described above. To obtain the full length OfHEX2, 3′ and 5′-RACE were performed using TaKaRa 3′-Full RACE Core Set (TaKaRa) and TaKaRa 5′-Full RACE kit (TaKaRa). The cloning strategies and the sequences of the primers are summarized in Supplementary Material: Table S1.
Sequence and phylogenetic analysis
Structure-based multiple sequence alignments were performed with PROMALS3D [18] and the results were demonstrated with ESPript [19]. The phylogenetic tree was constructed with MEGA 4 using the neighbor-joining method with a bootstrap evaluation of 1,000 replications [20]. The amino acid sequence from the protozoa, Entamoeba histolytica, was used as an outgroup (GenBank ID: XP_650273).
Gene expression of OfHEX2
To detect the developmental expression pattern of OfHEX2 during insect development, insect samples were collected from fifth instar larvae from day-1 to day-5 (5L1 to 5L5), prepupae (PP), white pupae (WP, pupa day-0), day-1 to day-7 pupae (P1 to P7) and adults (A). To examine gene expression in specific tissues, larvae (5L3) were dissected to obtain integument, midgut and carcass (which included whole body minus integument and midgut). Total RNAs were isolated respectively and used as templates for cDNA synthesis using TaKaRa RNAiso Reagent (TaKaRa). The expression abundance of OfHEX2 was detected by real-time PCR using SYBR PremeScript RT-PCR kit (TaKaRa) on a Rotor-Gene 3000 System (Corbett Research, Sydney, Australia). Gene-specific primers 5′-TAAAGGCAACCAACCACACA-3′ and 5′- TCGGGAGCCTATGACGAGA-3′ were designed according to the most conserved region of OfHEX2. Real-time PCRs were performed in triplicate under the following cycling conditions: 30 s at 95 °C, followed by 35 cycles at 95 °C for 5 s, 62 °C for 30 s and 72 °C for 40 s. RT-PCRs of O. furnacalis ribosomal protein 3 (OfRpS3, GenBank ID: EU275206) transcripts were performed with the same cDNA templates using gene-specific primers 5′-agcgtttcaacatccctgaac-3′ and 5′-CACACCATAGCAGGCACGA-3′ and served as an internal control to normalize differences in template levels between samples. For developmental expression analysis, the expression level of OfHEX2 in PP was given the value of 1, and then the expression levels for other sample relative to the level of expression in PP were calculated. For tissue-specific expression analysis, the expression level of OfHEX2 in the midgut was given the value of 1, and then the expression levels in the integument and carcass relative to the level of expression in the midgut were calculated.
Expression and purification of recombinant OfHex2
The OfHEX2 cDNA was cloned into the plasmid pPIC9 (Invitrogen, Carlsbad, CA) using the same method as we described previously [21]. Plasmid pPIC9-OfHex2 was linearized with Pme I (New England Biolabs, Beverly, MA) and transformed into Pichia pastoris strain GS115 (Invitrogen) by electroporation. The selection of His+ and Mut+ transformant was performed according to the manufacturer's instructions. The recombinant P. pastoris was cultured in BMGY/ BMMY media in 30 °C, 220 rpm. The culture was supplemented with 1 % methanol every 24 h. The culture supernatant was harvested after 144 h by centrifugation at 8,000 rpm for 15 min using Eppendorf centrifuge 5810R (Eppendorf, Hamburg, Germany).
Then solid ammonium sulfate was added to culture supernatant to 65 % saturation. After incubation at 4 °C for 2 h, the sample was recentrifuged at 12,000 rpm for 30 min. Then the pellet was dissolved in buffer A (20 mM sodium phosphate, 0.5 M NaCl, pH 7.4) and loaded onto an IMAC Sepharose High Performance column (20 ml, GE Healthcare, Uppsala, Sweden). After washing with buffer B (20 mM sodium phosphate, 0.5 M NaCl, 100 mM imidazole, pH 7.4), recombinant OfHex2 was eluted with buffer C (20 mM sodium phosphate, 0.5 M NaCl, 250 mM imidazole, pH 7.4). The purity of the recombinant OfHex2 was verified by SDS-PAGE and the enzymatic activity was measured using p-nitrophenyl-N-acetyl-β-D-glucosaminide (pNP-β-GlcNAc, Sigma-Aldrich, St. Louis, MO) as substrate [4].
Molecular mass measurement and dimer determination
The molecular mass of OfHex2 was measured under denatured and native conditions. Under denatured condition, OfHex2 was separated by 10 % SDS-PAGE together with low molecular mass standard proteins (Bio-Rad, Hercules, CA). Under native condition, OfHex2 was analyzed by size exclusion chromatography on a Superdex 200 10/ 30 GL column (GE Healthcare) by using buffer D (20 mM sodium phosphate, 0.15 M NaCl, pH 6.8) at a flow rate of 0.4 ml/ min. For comparison, bovine serum albumin (BSA) (Sigma-Aldrich) containing monomer (66 kDa), dimer (132 kDa) and minor trimer (198 kDa) was also separated using the same chromatographic condition.
Enzymatic activity assay
The optimal pH was determined by using Britton-Robinson wide range pH buffers (pH 2-12) [22]. Enzymatic reactions were performed in 60-μl reaction mixtures containing 6 μl of 1 mM pNP-β-GlcNAc, 4 μl of 0.05 μM OfHex2 solution and 50 μl of buffer with different pH at 30 °C. The reactions were terminated by adding 60 μl of 0.5 M Na2CO3 and the absorbances at 405 nm were measured using a microplate reader (TECAN, Männedorf, Switzerland).
The kinetic parameters of OfHex2 for pNP-β-GlcNAc were determined by endpoint colorimetric assay. The 60-μl reaction mixtures contain 30 μl of pNP-β-GlcNAc at different concentrations (1, 2, 3, 4, 5 mM), 2 μl of 0.05 μM OfHex2 and 28 μl of buffer at pH 5.5, 30 °C. The reactions were terminated by adding 60 μl of 0.5 M Na2CO3. The amount of p-nitrophenol (pNP) product was quantified by standard curve of pNP with known concentrations. The substrate consumption was limited to less than 10 % and the experiments were performed in triplicate. The Km and kcat values were calculated by linear regression of data in Lineweaver-Burk plots.
Degradation of chitooligosaccharides was assayed in 60-μl reaction mixture containing 28 μl of 5 mM phosphate buffer (pH 5.5), 5 μl of 0.05 μM OfHex2 solution and 30 μl of at different concentrations (0.5, 1, 2, 3, 4 mM). The experiments were performed in triplicate at 30 °C and the substrate consumption was limited to less than 10 %, then the samples were boiled and filtered through 0.22-μm Millipore filters. The amounts of the chitooligosaccharide degradation products and residual substrates were quantified using Koga's method [23], and the hydrolysis rates were calculated by the consumptions of the initial substrates. The Km and kcat values were also calculated by linear regression of data in Lineweaver-Burk plots.
Substrate specificity
The enzymatic activities for substrates 4-methy-lumbelliferyl-N-acetyl-β-D-glucosaminide (MU-β-GlcNAc) (Sigma-Aldrich), 4-methylumbelliferyl-N-acetyl-β-D-galactosaminide (MU-β-GalNAc) (Sigma-Aldrich) and 4-methylumbelliferyl-6-sulfo-N-acetyl-β-D-glucosaminide (MU-β-GlcNAc-SO4-) (Merck, San Diego, CA) were measured at 30 °C. The 100-μl reaction mixture contained 92.5 μl of 20 mM phosphate buffer (pH 5.5), 2.5 μl of 0.02 μM OfHex2 solution and 5 μl of 4 mM substrate solution. The experiments were performed in triplicate at 30 °C and the substrate consumption was limited to less than 10 %. The reaction was terminated by adding 100 μl of 1 M Glycine/NaOH (pH 10.6). The 4-methylumbelliferone released was detected by fluorescence using excitation/ emission wavelengths of 360/ 450 nm on a fluorescence microplate reader (Thermo, Waltham, USA).
Degradation of chitooligosaccharides was assayed in 60-μl reaction mixture containing 28 μl of 5 mM phosphate buffer (pH 5.5), 5 μl of 0.05 μM OfHex2 solution and 30 μl of 0.4 mM substrate solution. The substrate consumption was limited to less than 10 % and the experiments were performed in triplicate at 30 °C, then the samples were boiled and filtered through 0.22-μm Millipore filters.
Degradation of pyridylaminated oligosaccharides (TaKaRa), including the pyridylaminated globotetraose of globoside (Gb4-PA), the pyridylaminated trisaccharide of ganglioside GA2 (GA2-PA) and the pyridylaminated tetrasaccharide of ganglioside GM2 (GM2-PA), were assessed respectively in a 50-μl reaction mixture containing 20 μl of 20 mM phosphate buffer (pH 5.5), 5 μl of 0.5 μM OfHex2 solution and 25 μl of 10 μM substrate solution. The experiments were performed in triplicate at 30 °C and the substrate consumption was limited to less than 10 %, then the samples were boiled and filtered through 0.22-μm Millipore filters.
The peptidoglycan degradation experiment was performed by using the EnzChek Lysozyme Assay Kit (Invitrogen) according to the manufacturer's instruction. Three copies of 25 μl of 50 μg/ ml substrates were incubated with 25 μl of human β-N-acetyl-D-hexosaminidase (Sigma-Aldrich), 25 μl of purified OfHex2 (with same activity toward pNP-β-GlcNAc) and 25 μl of 10 U/ ml lysozyme (Invitrogen) at pH 4.5, 5.5 and 7.5, respectively, for 2 h 30 °C. Fluorescence was then measured using excitation/ emission wavelengths of 494/ 518 nm.
HPLC analysis of oligosaccharides hydrolytic products
Chitooligosaccharides (Toronto Research Chemicals, North York, Canada) were analyzed using a TSK-Gel Amide-80 column (0.46 × 25 cm) (Tosoh, Tokyo, Japan) on a HPLC system (Agilent Technologies, Santa Clara, USA) [23]. A 10-μl portion of the reaction mixture was injected into the column, and then eluted with 70 % acetonitrile at 0.7 ml/min at 25 °C. The chitooligosaccharides were monitored at 210 nm.
Pyridylaminated oligosaccharides were analyzed using a Hypersil ODS2 column (0.46 × 25 cm) (Thermo) and monitored by fluorescence using excitation/ emission wavelengths of 320/ 400 nm. HPLC analyses for GnGn-PA and GA2-PA (TaKaRa) were carried out using modified Altmann's methods [24]. A 40-μl portion of the GnGn-PA reaction mixture was loaded on the column and eluted with a linear gradient of 0-6 % (v/v) methanol in 0.1 M ammonium acetate (pH 4.0) in 50 min at a flow rate of 0.8 ml/min at 20 °C. A 40-μl portion of GA2-PA reaction mixture was loaded onto the column and eluted with a linear gradient of 0-1.8 % (v/v) methanol in 0.1 M ammonium acetate (pH 8.0) in 50 min at a flow rate of 0.8 ml/min at 25 °C. HPLC analyses for Gb4-PA and GM2-PA (TaKaRa) were performed in the same condition. A 40-μl portion of reaction mixture was loaded onto the column and eluted with a linear gradient of 0-0.1 % (v/ v) butyl alcohol in 50 mM acetic acid-triethylamine (pH 5.0) in 20 min at a flow rate of 1.0 ml/ min at 40 °C. The amounts of the pyridylaminated residual substrate and degradation products were quantified by standard curve with known concentrations, and the hydrolysis rates were calculated by the consumptions of the initial substrates.
RESULTS
cDNA cloning of OfHEX2
To isolate a cDNA encoding IBS-Hex from O. furnacalis, four conserved sequence segments from known lepidopteran IBS-Hexs were selected. The segments IWGILRGLE, FHWHIVDDQ, IPEFDVPGH, and RVWPRASAVA were used to design specific primers for RT-PCR amplification of the gene of interest (Supplementary Material: Table S1). 5′ and 3′-RACE reactions were performed to obtain a full-length cDNA. The nucleotide sequence of a putative β-N-acetyl-D-hexosaminidase, named OfHEX2 (GenBank ID: EF469203) was obtained from the fifth instar larvae of O. furnacalis. The full-length cDNA of OfHEX2 contains a predicted 1734-bp ORF encoding a polypeptide of 557 amino acids (OfHex2), a 43-bp untranslated region at the 5′-end and a 628-bp untranslated region at the 3′ end. A pairwise sequence alignment using BLASTP revealed that OfHex2 shares amino acid sequence identity with SfGlcNAcase1 (56 % identity) [14], BmGlcNAcase2 (55 %, GenBank ID: BAF52532) [15], BmGlcNAcase2 (55 %, GenBank ID: AAT99455. Note: This is a different enzyme from that in reference 15, but authors used the same abbreviation) [17], SfGlcNAcase3 (53 %) [14] and SfHex (53 %) [16]. OfHex2 also displays approximately 40 % identity with Homo sapiens or other vertebrate β-N-acetyl-D-hexosaminidases. However, OfHex2 had unexpectedly low identities with other insect β-N-acetyl-D-hexosaminidases that possess strict substrate specificity, such as chitinolytic β-N-acetyl-D-hexosaminidase OfHex1 (26 %) and N-glycan modifying β-N-acetyl-D-hexosaminidase DmFDL (23 %).
Phylogenetic analysis of OfHex2 and other IBS-Hexs
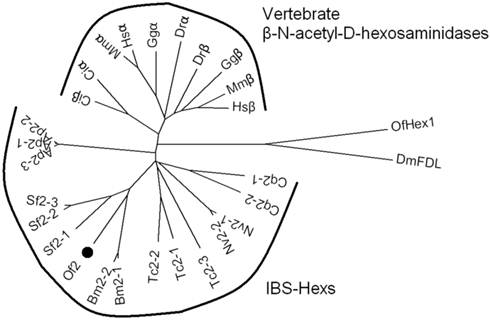
As IBS-Hexs shared high sequence similarities with vertebrate β-N-acetyl-D-hexosaminidases, a phylogenetic analysis of these enzymes was performed. The sequences of IBS-Hexs used for the analysis were selected from insect species with completely sequenced genomes, including Acyrthosiphon pisum (Ap1-Ap3), B. mori (Bm2-1 and Bm2-2), Culex quinauefasciatus (Cq2-1 and Cq2-2), Nasonia vitripennis (Nv2-1 and Nv2-2) and Tribolium castaneum (Tc2-1, Tc2-2 and Tc2-3). In addition, we included the three IBS-Hexs from S. frugiperda (Sf2-1 [14], Sf2-2 and Sf2-3 [14,16]), two of which have been characterized. The sequences of vertebrate β-N-acetyl-D-hexosaminidases were selected from representative species including the sea squirt, Ciona intestinalis (Ciα and Ciβ), the zebrafish Danio rerio (Drα and Drβ), the chicken Gallus gallus (Ggα and Ggβ), the mouse Mus musculus (Mmα and Mmβ), and Homo sapiens (Hsα and Hsβ).
As shown in Fig. 1, IBS-Hexs and vertebrate β-N-acetyl-D-hexosaminidases fell into two clusters while chitinolytic OfHex1 and N-glycan processing DmFDL were out-branched. Vertebrate β-N-acetyl-D-hexosaminidases were further divided into two subclusters which contained the sequences encoding subunits α and β respectively. However, the Ciα and Ciβ from the lower species C. intestinalis were undivided and located between the IBS-Hex cluster and vertebrate cluster. This result demonstrated the differentiation of vertebrate β-N-acetyl-D-hexosaminidase subunits α and β was likely to occur after the species differentiation of insects and vertebrates. OfHex2 was positioned in the subcluster containing other lepidopteran IBS-Hexs. It is interesting to note that the IBS-Hexs from the same insect species differ in phylogenetic distance. Bm2-1 (BmGlcNAcase2 [15]) and Bm2-2 (BmGlcNAcase2 [17]) from B. mori, Ap2-1, Ap2-2 and Ap2-3 from A. pisum and Nv2-1 and Nv2-2 from N. vitripennis were nearly identical to each other. However, Cq2-1 and Cq2-2 from C. quinquefasciatus, and Tc2-1, Tc2-2 and Tc2-3 from T. castaneum, for example, were more divergent from each other. Sf2-1(SfGlcNAcase1 [14]) from S. frugiperda is more divergent from Sf2-2 and Sf2-3(SfGlcNAcase2 [14] and SfHex [16]).
Structure-based sequence alignment of OfHex2 with other IBS-Hexs
Structure-based sequence comparisons of OfHex2 and other IBS-Hexs were performed according to the structure-known human β-N-acetyl-D-hexosaminidase α subunit (Hsα, PDB ID: 2GJX) and β subunit (Hsβ, PDB ID: 1O7A). Most of the amino acid residues comprising the active pocket of Hsβ are conserved in OfHex2 [25-27], including the catalytic residues (H294, D354 and E355); residues for making hydrogen bonds with the non-reducing end GlcNAc of the substrate (R211 and E491) and residues that comprise the hydrophobic wall of the active pockets (W405, W424, Y450 and W489, Supplementary Material: Fig. S1).
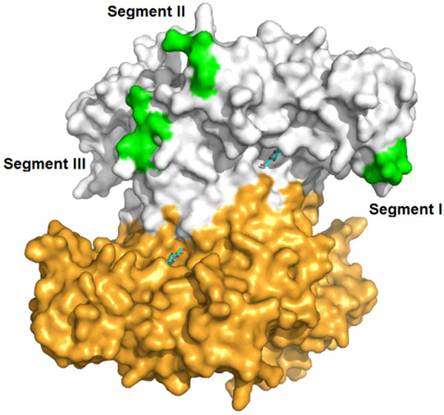
Three segments (Segment I, II and III) displaying sequence diversity were firstly discovered when aligning IBS-Hexs and human β-N-acetyl-D-hexosaminidase subunits (Fig. 2A). Based on the crystal structure of HsHexB, the three segments were all located away from both the active pocket and the dimer interface. Segment I is located between strand β3 and helix α3; Segment II is located between strands β7 and β8, and finally Segment III follows helix α8 (Fig. 3).
In addition, structure-based sequence alignment revealed that the residues determining the specificity of Hsα/ Hsβ toward charged/ non-charged substrate (N423-R424 in Hsα and D452-L453 in Hsβ) were not conserved in IBS-Hexs (Fig. 2B) [26-28]. R424 in Hsα responsible for binding both the 6-sulfate group in MU-β-GlcNAc-SO4- and the sialic acid moiety in GM2 was absent in IBS-Hexs [28]. D452 in Hsβ was instead occupied by N396, N417 and N324 in Ap2-1, Ap2-2 and Ap2-3, S392 and S430 in Nv2-2 and Tc2-2 and H492 in Sf2-2.
Phylogenetic analysis of insect β-N-acetyl-D-hexosaminidases and vertebrate β-N-acetyl-D-hexosaminidases. The GenBank ID of the β-N-acetyl-D-hexosaminidase protein sequences are: A. pisum Ap2-1(XP_003248256), Ap2-1(XP_001945979) and Ap2-1(XP_001943356); B. mori Bm2-1 (BAF52532) and Bm2-2(AAT99455); C. intestinalis Ciα (XP_002121203) and Ciβ (XP_002126592); C. quinauefasciatus Cq2-1 (EDS44384) and Cq2 (EDS44385); D. melanogaster DmFDL (AAL55992); D. rerio Drα (AAH93192) and Drβ (CAM16012); G. gallus Ggα (CAG32597) and Ggβ (XP_424791); H. sapiens Hsα (AAB00965) and Hsβ (AAA52645); M. musculus Mmα (AAC53246) and Mmβ (AAA18776); N. vitripennis Nv2-1 (XP_001600369) and Nv2-2(XP_001600369); O. furnacalis Of2 (ABO65045) and OfHex1 (ABI81756); S. frugiperda Sf2-1 (ABB76924), Sf2-2 (ABY57947) and Sf2-3 (ABB76925); T. castaneum Tc2-1 (EFA05960), Tc2-2 (EFA07069) and Tc2-3 (EFA05959).
Structure-based sequence alignment of OfHex2 with other IBS-Hexs and human β-N-acetyl-D-hexosaminidases. Only partial results are shown here and complete results are shown in Supplementary Material: Fig. S1. Numbers before the amino acid indicate nucleotide position. The identical residues are with red background blocks. (A) Three sequence segments with low similarities among IBS-Hexs and human β-N-acetyl-D-hexosaminidase subunits. (B) The two residues that determine the substrate specificity of HsHexA/ HsHexB for charged/ non-charged substrates are indicated by arrows. The alignment was generated by PROMALS3D and the picture was generated by ESPript.

Spatial location of segments I, II and II in the crystal structure of human β-N-acetyl-D-hexosaminidase B. The PDB ID of HsHexB is 1O7A. The two subunits of HsHexB were coloured in white and gold, respectively. The positions of the three segments were coloured in green. The positions of the active pocket are marked by the binding inhibitor, 2-acetamido-2-deoxy-D-glucono-1,5-lactone. The picture was prepared by PyMOL (DeLano Scientific, http://www.pymol.org).
Expression, purification and characterization of the recombinant OfHex2
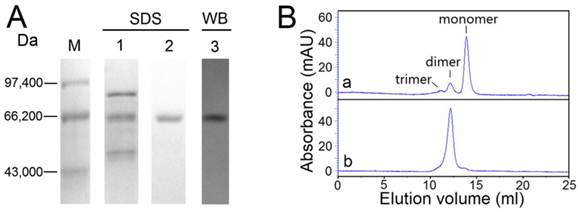
Approximately 1.75 mg of purified recombinant OfHex2 was obtained from 1 L of P. pastoris culture supernatant. The efficiency of the protein purification procedure is summarized in Supplementary Material: Table S2. The purified protein was resolved by SDS-PAGE and a single band with a molecular mass of 65 kDa, and identified by western blotting using anti-His tag antibody (Fig. 4A). Furthermore, the retention volume of OfHex2 (12.18 ml) in size exclusion chromatography was similar to that of dimeric BSA (12.14 ml) (Fig. 4B), the molecular mass of which is 132 kDa. Thus, we conclude that OfHex2 is a homodimer.
The pH optimum of OfHex2 enzymatic activity was 5.5 when using pNP-β-GlcNAc as substrate. The Km and Kcat values of OfHex2 for pNP-β-GlcNAc were 0.463±0.027 mM and 151.05±6.21 s-1, respectively (Table 1).
Substrate spectrum and enzymatic kinetics of OfHex2
Synthesized monosaccharide substrates
To study the substrate spectrum of OfHex2, mono N-acetyl-β-D-hexosamine substrates including MU-β-GlcNAc, MU-β-GalNAc and MU-β-GlcNAc-SO4- were used for enzymatic assays. -OfHex2 hydrolyzed MU-β-GlcNAc and MU-β-GalNAc at the rates of 22.74±0.38 nmol∙min-1∙μg-1 and 25.59±0.34 nmol∙min-1∙μg-1, respectively (Table 2). This higher relative catalytic activity for MU-β-GalNAc (113 %) was different from the reported IBS-Hex BmGlcNAcase2 [15] and SfHex [16]. OfHex2 degraded negatively charged substrate MU-β-GlcNAc-SO4- only at a low rate, approximately 250 times lower than the rate for MU-β-GlcNAc (Table 2).
Chitooligosaccharides
The activities of OfHex2 toward chitooligosaccharides were determined using 0.2 mM (GlcNAc)n (n=2,3,4,6) as substrates (Fig. 5, Table 2) OfHex2 could efficiently degrade chitooligosaccharides. The hydrolytic rate decreased as the “n” increased from 3 to 6. However, the hydrolytic rate for (GlcNAc)2 (6.970±0.010 nmol∙min-1∙μg-1) was lower than that for (GlcNAc)3 (8.355±0.011 nmol∙min-1∙μg-1). This differed OfHex2 from many of other reported insect β-N-acetyl-D-hexosaminidases, which hydrolyze (GlcNAc)2 faster than (GlcNAc)3 [4].
To investigate the mechanism behind this result, the kinetic parameters of OfHex2 for (GlcNAc)n (n=2,3,4,6) were determined (Table 1). The Km values of OfHex2 for (GlcNAc)n increased with the increase of n from 2 to 6, whereas the kcat values of OfHex2 fluctuated. It is worthy to note that the kcat value of OfHex2 for (GlcNAc)3 was 1.5-fold higher than that for (GlcNAc)2, suggesting OfHex2 prefers (GlcNAc)3 to (GlcNAc)2 as substrate.
N-glycan
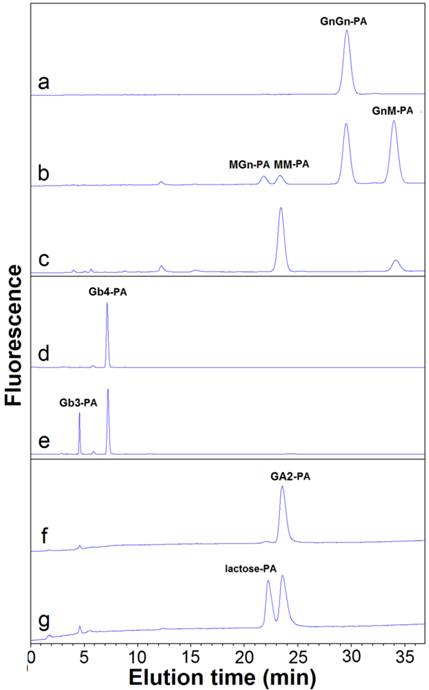
The activity of OfHex2 toward N-glycan was determined using GnGn-PA as substrate (Fig. 5 and Fig. 6). OfHex2 could release β1-2 linked GlcNAc residue from either the α3 branch or the α6 branch of GnGn-PA, because both intermediates GnM-PA and MGn-PA were produced (Fig. 6B), which were further degraded to MM-PA during the hydrolytic processes (Fig. 6C). Furthermore, during GnGn-PA degradation, the resulted amount of GnM-PA was higher than that of MGn-PA, indicating a configurational preference of OfHex2 to the GlcNAc residue on the α3 branch. The quantative analysis showed that OfHex2 hydrolyzed GnGn-PA approximately 24-fold slower than MU-β-GlcNAc and exhibited a lower relative activity for GnGn-PA than for other neutral oligosaccharides (Table 2).
Glycolipids
The activities of OfHex2 toward glycolipids were determined using glycosphingolipids as substrates. Gb4-PA, GA2-PA and GM2-PA are the pyridylaminated oligosaccharides of globoside, ganglioside GA2 and ganglioside GM2, respectively (Fig. 5). OfHex2 could release the terminal β1-3 linked GalNAc residue from Gb4-PA (Fig. 6E) and the β1-4 linked GalNAc residue from GA2-PA (Fig. 6G), but could not release GalNAc residue from GM2-PA (data not shown), suggesting that OfHex2 is capable of degrading neutral sugar parts of glycolipids but is inactive toward charged substrates. Table 3 showed the hydrolysis rates measured with 5 μM substrates. OfHex2 hydrolyzed GA2-PA at the rate of 0.1387±0.0017 nmol∙min-1∙μg-1, approximately 2.2-fold faster than Gb4-PA and 5-fold faster than GnGn-PA.
Peptidoglycan
Human β-N-acetyl-D-hexosaminidase was recently found to be secreted by macrophages and to display a bactericidal role by hydrolyzing the peptidoglycan of bacterial cell wall (Fig. 5) [29]. To determine whether OfHex2 has a similar role in insect, it was incubated with the fluorescein-labeled Micrococcus lysodeikticus cell wall for 2 h. Human β-N-acetyl-D-hexosaminidase (with the same pNP-β-GlcNAc activity as OfHex2) and lysozyme from chicken egg white were used as positive controls. Human β-N-acetyl-D-hexosaminidase exhibited 22 % of the lysozyme activity, but OfHex2 did not show any activity toward peptidoglycan substrate (data not shown), suggesting that its role is not relevant in insect defense.
To summarize, OfHex2 can remove the β1-4 linked GlcNAc from synthesized monosaccharide substrates and chitooligosaccharides, β1-2 linked GlcNAc from both branches of GnGn-PA, β1-3 linked GalNAc from Gb4-PA and β1-4 linked GalNAc from GA2-PA. These results demonstrate that OfHex2 is a “broad-spectrum” enzyme that hydrolyzes glycans of different structures and from various sources including oligosaccharides, glycoproteins and glycolipids.
Purification efficiency and analysis of the molecular mass of OfHex2. (A) SDS-PAGE analysis of the recombinant OfHex2 obtained at each purification step. Lane M, low molecular mass protein markers; lane 1, ammonium sulfate precipitation; lane 2, metal chelate affinity chromatography; lane 3, western blotting of purified OfHex2 using anti-Histag antibody. (B) Size exclusion chromatography of native bovine serum albumin (BSA) (a) and OfHex2 (b). The three compositions of BSA mixture presented different retention volumes based on molecular mass, which was BSA trimer (198 kDa, 11.08 ml), BSA dimer (132 kDa,12.14 ml) and BSA monomer(66 kDa, 13.93 ml). Under the same condition, OfHex2 presented a retention volume of 12.18 ml, which was approximately the same as BSA dimer.
Schematic structures of the substrates used in this study.

HPLC analysis of hydrolyzed products of glycans by OfHex2. Pyridylaminated substrates (10 μM) were incubated with purified OfHex2 at pH 5.5, 30 °C. Recombinant OfHex1 which exhibites no hydrolytic activity toward GnGn-PA, Gb4-PA and GA2-PA was used as negative control (with same activity toward pNP-β-GlcNAc). For GnGn-PA degradation analysis, OfHex1 exhibited no hydrolytic activity after incubation for 8 h (A), while OfHex2 degraded GnGn-PA to GnM-PA, MGn-PA and MM-PA after incubated for 30 min (B), and continued to degrade the intermediates GnM-PA and MGn-PA to MM-PA after a 4h-incubation (C). For Gb4-PA degradation analysis, OfHex1 exhibited no hydrolytic activity after incubation for 8 h (D), while OfHex2 degraded Gb4-PA to pyridylaminated globotriose (Gb3-PA) after 2 h-incubation (E). For GA2-PA degradation analysis, OfHex1 exhibited no drolytic activity after incubation for 8 h (F), while OfHex2 degraded GA2-PA to lactose-PA after 2 h-incubation (G).
Kinetic parameters of OfHex2
| Substrates | Km (mM)a | kcat (s-1)a | kcat/ Km (s-1 mM-1) |
|---|---|---|---|
| pNP-β-GlcNAc | 0.463±0.027 | 151.05±6.21 | 326.2 |
| (GlcNAc)2 | 0.169±0.008 | 11.64±0.30 | 68.91 |
| (GlcNAc)3 | 0.224±0.010 | 17.98±0.11 | 80.27 |
| (GlcNAc)4 | 0.304±0.005 | 11.08±0.05 | 36.45 |
| (GlcNAc)6 | 0.529±0.012 | 8.65±0.15 | 16.35 |
aData are the mean ±S.D. of three independent experiments.
Substrate specificity of OfHex2 for monosaccharide substrates and chitooligosaccharide
| Substrates | Hydrolysis rate a (nmol∙min-1∙μg-1) | Relative activity (%) |
|---|---|---|
| MU-β-GlcNAc | 22.74±0.38 | 100 |
| MU-β-GalNAc | 25.59±0.34 | 112.6 |
| MU-β-GlcNAc-SO4- | 0.09101±0.002 | 0.4002 |
| (GlcNAc)2 | 6.970±0.010 | 30.65 |
| (GlcNAc)3 | 8.355±0.011 | 36.74 |
| (GlcNAc)4 | 3.469±0.018 | 17.21 |
| (GlcNAc)6 | 2.371±0.019 | 10.43 |
a Enzyme activity was measured at pH 5.5 with 0.2 mM substrate. Data are the mean ±S.D. of three independent experiments.
Substrate specificity of OfHex2 for N-glycan and sugar parts of glycosphingolipids with terminal GlcNAc/ GalNAc residues.
| Substrates | Hydrolysis rate a (nmol∙min-1∙μg-1) | Relative activity (%) |
|---|---|---|
| MU-β-GlcNAc | 0.7349±0.0043 | 100 |
| GnGn-PA | 0.03037±0.0008 | 4.132 |
| Gb4-PA | 0.06218±0.0014 | 8.461 |
| GA2-PA | 0.1387±0.0017 | 18.87 |
| GM2-PA | N.D.b | N.D. |
a Enzyme activity was measured at pH 5.5 with 5 μM substrate. Data are the mean ±S.D. of three independent experiments.
b. N.D. indicates not detectable.
Gene expression pattern of OfHEX2
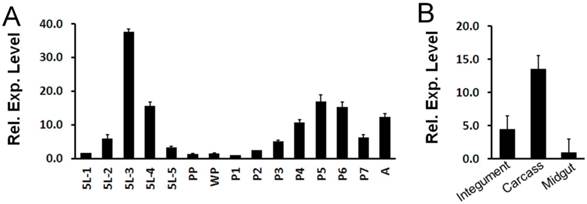
Expression pattern of OfHEX2 at different developmental stages of O. furnacalis was determined by real-time PCR. As shown in Fig 7A, the expression level of OfHEX2 was significantly up-regulated in the mid-fifth instar (5L2, 5L3 and 5L4), subsequently down-regulated to background levels during larval-pupal molt (PP and WP), and then up-regulated in the pupa (P4, P5 and P6). This result indicates that OfHex2 may function during the larval intermolt stage and pupal stage, but not during the larval-pupal molt.
The expression levels of OfHEX2 in different tissues (integument, midgut and carcass) at the fifth (final) feeding stage (5L3) were also determined by real-time PCR. The result indicated that the expression levels of OfHEX2 in the carcass and integument were 13.9 and 4.7 times higher than that in the midgut (Fig 7B), suggesting OfHex2 functions mainly in the carcass rather than in the intestine.
DISCUSSION
Insect β-N-acetyl-D-hexosaminidases with broad substrate-spectrum (IBS-Hexs) are widely distributed among different insect species, but their physiological roles remain obscure [14-17]. They have an interesting evolutionary position as they share a closer phylogenetic relationship with vertebrate β-N-acetyl-D-hexosaminidases than with other insect β-N-acetyl-D-hexosaminidases (Fig. 1). In this study, OfHex2, a novel IBS-Hex from O. furnacalis, was characterized by sequence analysis, gene expression pattern and substrate spectrum.
By phylogenetic analysis, IBS-Hexs and vertebrate β-N-acetyl-D-hexosaminidases fell into two individual clusters. Though most of the key residues of vertebrate β-N-acetyl-D-hexosaminidases are conserved in IBS-Hexs [25-27], several unique sequence characteristics are found in OfHex2 and other IBS-Hexs. Three sequence segments with low similarity with Hsα and Hsβ or other IBS-Hexs were noted for the first time (Fig. 2A). These segments locate neither in the active pocket, nor in the dimer interface when compared to the crystal structure of HsHexB [25,26] (Fig. 3). Currently, we could not infer the function of all these segments. So far, the function of the corresponding Segment I in human β-N-acetyl-D-hexosaminidases has been deduced. The R312-Q313-N314-K315 in Segment I in pro-Hsβ is removed by specific proteolytic processing after its delivery to lysosome [26]. However in Hsα, the G280-S281-E282-P283 in Segment I constitutes a flexible loop that binds a specific lipid binding protein named GM2 activator protein, which significantly stimulates the hydrolysis rate of HsHexA toward the negatively-charged ganglioside GM2 [27, 30]. Moreover, the residue pair N423-R424 (Hsα) / D452-L453 (Hsβ), which determines the substrate specificity for charged/ non-charged substrates of HsHexA/ HsHexB [25-28], are not conserved in OfHex2 and other IBS-Hexs (Fig. 2B). Correspondingly, OfHex2 showed very low activity toward MU-β-GlcNAc-SO4- and could not hydrolyze GM2-PA and peptidoglycan. We speculate that inactivity toward charged substrates maybe a common property of IBS-Hexs.
Though IBS-Hexs have a broad substrate spectrum, they still display relative specificities among different types of substrates. Most previous studies on IBS-Hexs provided non-quantitative results of the substrate specificity. Only SfHex was examined in any quantitative detail [16]. OfHex2 and SfHex showed different preferences toward chitooligosaccharides and GnGn-PA substrates. SfHex exhibited 11-fold higher activity toward GnGn-PA than toward (GlcNAc)3-PA. On the contrary, the hydrolysis activity of OfHex2 toward GnGn-PA was 5-fold and 2-fold lower than those toward GA2 and Gb4, respectively (Table 3). The relative activity toward (GlcNAc)3 was approximately 9-fold higher than that toward GnGn-PA (Table 2 and 3). Additionally, OfHex2 hydrolyzed MU-β-GalNAc faster than MU-β-GlcNAc, which also differed OfHex2 from SfHex as well as BmGlcNAcase2. These differences in substrate preference suggest that different IBS-Hexs may play distinct physiological roles.
The expression level of OfHEX2 is up-regulated in intermolt stages but dropped to a negligible level during molting (Fig. 7A). However, BmGLcNAcase2, a reported IBS-Hex gene, expresses in embryo and larval stages but not in pupal stages [17]. Furthermore, the spatial expression of OfHEX2 and BmGLcNAcase2 are also different. OfHEX2 expresses at a higher level in the carcass than in the integument and midgut (Fig. 7B), whereas BmGLcNAcase2 expresses at the same levels in these tissues [17]. Such differences in the developmental patterns and tissue specificities of expression of IBS-Hexs implied that they have different physiological functions.
To investigate the physiological role, we performed RNA interference, which showed that the gene silencing of OfHEX2 was non-lethal but severely affected the development of larval abdomen, pupal wing and adult appendages (unpublished data).Interestingly, a similar phenotype was observed in T. castaneum by the RNAi-mediated knockdown of the chitinase TcCHT7 [31,32]. It will be interesting to study the function of OfHex2 further, and if it plays a role comparable to TcCHT7-like endoglycosidase in glycan degradation during appendage development.
Temporal and tissue-specific OfHEX2 gene expression patterns. RT-PCRs of OfRpS3 transcripts were performed with the same cDNA templates as an internal control to normalize differences in template levels between samples. Data are the mean ±S.D. of three independent experiments. (A) Expression pattern of OfHEX2 at different developmental stages. Insect samples were collected from the fifth instar larvae to adults. The expression levels of OfHEX2 relative to the level of expression in prepupa are shown. 5L1-5L5: samples from day-1 to day-5 fifth instar larvae; PP: prepupa; WP: white pupa; P1-P7: samples from day-1 to day-7 pupae; A: adult. (B) Expression pattern of OfHEX2 in different tissues of fifth instar larvae. The expression levels of OfHEX2 relative to the level of expression in midgut are shown.
In conclusion, a novel member of IBS-Hex, OfHex2, was identified in this study. The sequence characteristics, the substrate spectrum as well as the temporal and spatial gene expression patterns revealed biodiversity of this class of enzymes.
Supplementary Material
Table S1-S2, and Fig.S1.
Acknowledgements
This work was supported by the National Key Project for Basic Research (2010CB126100), the National Natural Science Foundation of China (31070715, 31101671), the National High Technology Research and Development Program of China (2011AA10A204), the National Key Technology R&D Program (2011BAE06B05), the Fundamental Research Funds for the Central Universities (DUT11ZD113, DUT11RC(3)73). Also thanks to Dr. Alan K. Chang at Dalian University of Technology for reviewing the language of this manuscript.
CONFLICT OF INTERESTS
The authors have declared that no conflict of interest exists.
References
1. Slámová K, Bojarová P, Petrásková L. et al. β-N-Acetylhexosaminidase: What's in a name…? Biotechnol Adv. 2010;28:682-93
2. Merzendorfer H, Zimoch L. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J Exp Biol. 2003;206:4393-412
3. Nagamatsu Y, Yanagisawa I, Kimoto M. et al. Purification of a chitooligosaccharidolytic β-N-acetylglucosaminidase from Bombyx mori larvae during metamorphosis and the nucleotide sequence of its cDNA. Biosci Biotechnol Biochem. 1995;59:219-25
4. Yang Q, Liu T, Liu F. et al. A novel β-N-acetyl-D-hexosaminidase from the insect Ostrinia furnacalis (Guenée). FEBS J. 2008;275:5690-702
5. Liu T, Zhang H, Liu F. et al. Structural determinants of an insect β-N-acetyl-D-hexosaminidase specialized as a chitinolytic enzyme. J Biol Chem. 2011;286:4049-58
6. Zheng YP, Krell PJ, Doucet D. et al. Cloning, expression, and localization of a molt-related β-N-acetylglucosaminidase in the Spruce budworm, Choristoneura fumiferana. Arch. Insect Biochem Physiol. 2008;68:49-59
7. Altmann F, Schwihla H, Staudacher E. et al. Insect cells contain an unusual, membrane-bound β-N-acetylglucosaminidase probably involved in the processing of protein N-glycans. J Biol Chem. 1995;270:17344-9
8. Léonard R, Rendić D, Rabouille C. et al. The Drosophila fused lobes gene encodes and N-acetylglucosaminidase involved in N-glycan processing. J Biol Chem. 2006;281:4867-75
9. Geisler C, Aumiller JJ, Jarvis DL. A fused lobes gene encodes the processing β-N-acetylgl ucosaminidase in Sf9 cells. J Biol Chem. 2008;283:11330-9
10. Cattaneo F, Pasini ME, Intra J. et al. Identification and expression analysis of Drosophila melanogaster genes encoding β-hexosaminidases of the sperm plasma membrane. Glycobiology. 2006;16:786-800
11. Pasini ME, Intra J, Gomulski LM. et al. Identification and expression profiling of Ceratitis capitata genes coding for β-hexosaminidases. Gene. 2011;473:44-56
12. Mahuran DJ. Biochemical consequences of mutations causing the GM2 gangliosidoses. Biochim Biophys Acta. 1999;1455:105-38
13. Hepbildikler ST, Sandhoff R, Kölzer M. et al. Physiological substrates for human lysosomal β-hexosaminidase S. J Biol Chem. 2002;277:2562-72
14. Aumiller JJ, Hollister JR, Jarvis DL. Molecular cloning and functional characterization of β-N-acetylglucosaminidase genes from Sf9 cells. Protein Expr Purif. 2006;47:571-90
15. Okada T, Ishiyama S, Sezutsu H. et al. Molecular cloning and expression of two novel β-N-acetylglucosaminidases from silkworm Bombyx mori. Biosci Biotechnol Biochem. 2007;71:1626-35
16. Tomiya N, Narang S, Park J. et al. Purification, characterization, and cloning of a Spodoptera frugiperda Sf9 β-N-acetylhexosaminidase that hydrolyzes terminal N-acetylglucosamine on the N-glycan core. J Biol Chem. 2006;281:19545-60
17. Kokuho T, Yasukochi Y, Watanabe S. et al. Molecular cloning and expression profile analysis of a novel β-D-N-acetylhexosaminidase of domestic silkworm (Bombyx mori). Genes Cells. 2010;15:525-36
18. Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295-300
19. Gouet P, Robert X, Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320-3
20. Tamura K, Dudley J, Nei M. et al. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol. 2007;24:1596-9
21. Liu T, Liu F, Yang J. et al. Expression, purification and characterization of the chitinolytic β-N-acetyl-D-hexosaminidase from the insect Ostrinia furnacalis. Protein Expr Purif. 2009;68:99-103
22. Britton HTS, Robinson RA. Universal buffer solutions and the dissociation constant of veronal. J Chem Soc. 1931:1456-62
23. Koga D, Yoshioka T, Arakane Y. HPLC analysis of anomeric formation and cleavage pattern by chitinolytic enzyme. Biosci Biotechnol Biochem. 1998;62:1643-6
24. Altmann F, Kornfeld G, Dalik T. et al. Processing of asparagine-linked oligosaccharides in insect cells. N-acetylglucosaminyltransferase I and II activities in cultured lepidopteran cells. Glycobiology. 1993;3:619-25
25. Maier T, Strater N, Schuette C. et al. The X-ray crystal structure of human β-hexosaminidase B provides new insights into Sandhoff disease. J Mol Biol. 2003;328:669-81
26. Mark BL, Mahuran DJ, Cherney MM. et al. Crystal structure of human β-Hexosaminidase B: understanding the molecular basis of Sandhoff and Tay-Sachs disease. J Mol Biol. 2003;327:1093-109
27. Lemieux MJ, Mark BL, Cherney MM. et al. Crystallographic structure of human β-hexosaminidase A: interpretation of Tay-Sachs mutations and loss of GM2 ganglioside hydrolysis. J Mol Biol. 2006;359:913-29
28. Sharma R, Bukovac S, Callahan J. et al. A single site in human β-hexosaminidase A binds both 6-sulfate-groups on hexosamines and the sialic acid moiety of GM2 ganglioside. Biochim Biophys Acta. 2003;1637:113-8
29. Koo IC, Ohol YM, Wu P. et al. Role for lysosomal enzyme β-hexosaminidase in the control of mycobacteria infection. Proc Natl Acad Sci USA. 2008;105:710-5
30. Werth N, Schuette CG, Wilkening G. et al. Degradation of Membrane-bound Ganglioside GM2 by β-Hexosaminidase A STIMULATION BY GM2 ACTIVATOR PROTEIN AND LYSOSOMAL LIPIDS. J Biol Chem. 2001;276:12685-90
31. Zhu Q, Arakane Y, Beeman RW. et al. Functional specialization among insect chitinase family genes revealed by RNA interference. Proc Nat Acad Sci USA. 2008;105:6650-5
32. Arakane Y, Muthukrishnan S. Insect chitinase and chitinase-like proteins. Cell Mol Life Sci. 2010;67:201-16
Author contact
![]() Corresponding author: Prof. Qing Yang, School of Life Science and Biotechnology, Dalian University of Technology, Dalian 116024, China. Tel.: 86-411-84707245; Fax: 86-411-84707245; E-mail: qingyangedu.cn.
Corresponding author: Prof. Qing Yang, School of Life Science and Biotechnology, Dalian University of Technology, Dalian 116024, China. Tel.: 86-411-84707245; Fax: 86-411-84707245; E-mail: qingyangedu.cn.

 Global reach, higher impact
Global reach, higher impact