10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2012; 8(10):1291-1309. doi:10.7150/ijbs.4966 This issue Cite
Research Paper
Multiple Sodium Channel Variants in the Mosquito Culex quinquefasciatus
1. Department of Entomology and Plant Pathology, Auburn University, Auburn AL 36849, USA;
2. Current address: College of Plant Protection, Southwest University, Chongqing, China;
3. Genomics Laboratory, Auburn University, Auburn, AL 36849, USA.
* These authors contributed equally to this work.
Received 2012-7-31; Accepted 2012-9-27; Published 2012-10-25
Abstract
Voltage-gated sodium channels are the target sites of both DDT and pyrethroid insecticides. The importance of alternative splicing as a key mechanism governing the structural and functional diversity of sodium channels and the resulting development of insecticide and acaricide resistance is widely recognized, as shown by the extensive research on characterizing alternative splicing and variants of sodium channels in medically and agriculturally important insect species. Here we present the first comparative study of multiple variants of the sodium channel transcripts in the mosquito Culex quinquefasciatus. The variants were classified into two categories, CxNa-L and CxNa-S based on their distinguishing sequence sizes of ~6.5 kb and ~4.0 kb, respectively, and generated via major extensive alternative splicing with minor small deletions/ insertions in susceptible S-Lab, low resistant HAmCqG0, and highly resistant HAmCqG8 Culex strains. Four alternative Cx-Na-L splice variants were identified, including three full length variants with three optional exons (2, 5, and 21i) and one with in-frame-stop codons. Large, multi-exon-alternative splices were identified in the CxNa-S category. All CxNa-S splicing variants in the S-Lab and HAmCqG0 strains contained in-frame stop codons, suggesting that any resulting proteins would be truncated. The ~1000 to ~3000-fold lower expression of these splice variants with stop codons compared with the CxNa-L splicing variants may support the lower importance of these variants in S-Lab and HAmCqG0. Interestingly, two alternative splicing variants of CxNa-S in HAmCqG8 included entire ORFs but lacked exons 5 to18 and these two variants had much higher expression levels in HAmCqG8 than in S-Lab and HAmCqG0. These results provide a functional basis for further characterizing how alternative splicing of a voltage-gated sodium channel contributes to diversity in neuronal signaling in mosquitoes in response to pyrethroids, and possibly indicates the role of these variants in the development of pyrethroid resistance.
Keywords: Sodium channel, transcript variants, alternative splicing, insecticide resistance, Culex quinquefasciatus.
INTRODUCTION
The insects' voltage-gated sodium channels are the targets for insecticides, such as DDT and pyrethroids [1-4], which are responsible for the rising phase of action potentials in the membranes of neurons and most electrically excitable cells [5]. Pyrethroids and DDT deliver their toxic, insecticidal effects primarily by binding to the sodium channel, thus altering its gating properties and keeping the sodium channel open for unusual long time, thereby causing a prolonged flow of sodium current that initiates repetitive discharges and prevents the repolarization phase of action potentials [5-7]. The common feature found in sodium channels is that relatively small changes, such as point mutations or substitutions [3, 8-14], short sequence insertions or deletions, or alternative splicing [15-23] in the structure of these channels significantly affect their behavior and are sufficient to change neuronal firing, resulting in different phenotypes. Modifications of the insect sodium channel structure can cause insensitivity of the channels to DDT and pyrethroids via a reduction in or an elimination of the binding affinity of the insecticides to proteins [6-7], and hence result in the development of insecticide resistance.
In mammalian systems, molecular characterization of voltage-gated sodium channel genes has revealed the existence of multiple genes [5, 24-26]: ten paralogous voltage-gated sodium channel genes have been identified in humans [25]; 8 in zebra fish [27]; and 6 in electric fish [28]. Several invertebrate species have also been found to include multiple sodium channel genes in their genome; for example, 4 sodium channel genes have been characterized in Hirudo medicinalis (leech) [29] and 2 in Halocynthia roretzi (ascidia) [30-31]. Compared to the fairly well defined multiple vertebrate sodium channel genes, it appears that a single sodium channel gene that has been well characterized in many insect species, homologous to para (currently DmNaV) of Drosophila melanogaster [32-33] encodes the equivalent of the α-subunit of the mammalian sodium channelsLoughney et al. 1989). While mammals rely on the selective expression of at least ten different sodium channel genes in various tissues to achieve sodium channel diversity [25], insects may produce a range of diverse sodium channels with different functional and pharmacological properties from a single sodium channel by extensive alternative splicing [16-23, 34].
Because of the importance of alternative splicing as a key mechanism for generating structural and functional diversity in sodium channels [19], following the first discovery of the existence of alternative splicing of the para sodium channel gene from Drosophila melanogaster [32], alternative splicing events were subsequently characterized in many medically or agricultural important insect and arachnid pest species [17, 19-23, 34-36]. As yet, however, there have been no reports of alternative splicing in Culex quinquefasciatus, an important mosquito vector of human pathogens such as St. Louis encephalitis virus (SLEV), West Nile virus (WNV), and the parasitic Wuchereria bancrofti nematode in many urban settings throughout the tropical and temperate regions of the world [37-39]. Here we present the first comparative study of full length sequences of the para-orthologue sodium channel transcripts from the Culex quinquefasciatus mosquito and examine multiple variants obtained through the mechanism of alternative splicing.
MATERIALS AND METHODS
Mosquito strains
Three strains of mosquito Cx. quinquefasciatus were studied. S-Lab is an insecticide susceptible strain provided by Dr. Laura Harrington (Cornell University). HAmCqG0 is a low insecticide resistant strain with a 10-fold level of resistance to permethrin compared with the laboratory susceptible S-Lab strain [40]. It was originally collected from Huntsville, Alabama, in 2002 and established in our laboratory without further exposure to insecticides [41]. The HAmCqG8 strain is the 8th generation of permethrin-selected HAmCqG0 offspring and has a 2,700-fold level of resistance [40]. All mosquitoes were reared at 25±2oC under a photoperiod of 12:12 (L:D) h.
Amplification of the full length of sodium channel transcripts in Cx. quinquefasciatus
For each of the three mosquito populations, total RNA was extracted from the 4th instar larvae, and different tissues (head + thorax, and abdomen) from 2-3 day-old adult females before blood feeding using the acidic guanidine thiocyanate-phenol-chloroform method [42]. Messenger RNAs (mRNAs) were isolated using Oligotex-dT suspension (QIAGEN). The full length cDNA of the Cx. quinquefasciatus sodium channel gene was subsequently isolated from each of the mosquitoes populations by RT-PCR using the Expand Long Range, dNTPack kit (Roche) with a specific primer pair, KDR S16 (TGTTGGCCATATAGACAATGACCGA) /KDR AS09 (GCTTCTGAATCTGAATCAGAGGGAG), synthesized based on the respective 5' and 3' end sequences of the putative sodium channel cDNAs [43], accession numbers: JN695777, JN695778, and JN695779]. The PCR reaction was conducted following a PCR cycle of 92°C for 2 min, 10 cycles of 92°C for 10 s, 55°C for 15 s, and 68°C for 6 min, and 35 cycles of 92°C for 10 s, 55°C for 15 s, and 68°C for 8 min, with a final extension of 68°C for 10 min. All PCR products were cloned into PCRTM 2.1 Original TA cloning vector (Invitrogen) and sequenced. Cloning and sequence analyses of sodium channel cDNA fragments were repeated at least two times for each mosquito strain with different preparations of RNAs and mRNAs. The inserts of Culex sodium channel clones were sequenced and analyzed.
Quantitative real-time PCR (qRT-PCR)
The total RNA (0.5 µg/sample) from each mosquito sample was reverse-transcribed using SuperScript II reverse transcriptase (Stratagene) in a total volume of 20 μl. The quantity of cDNAs was measured using a spectrophotometer prior to qRT-PCR, which was performed with the SYBR Green master mix Kit and ABI 7500 Real Time PCR system (Applied Biosystems). Each qRT-PCR reaction (25 μl final volume) contained 1x SYBR Green master mix, 1 μl of cDNA, and a sodium channel transcript specific primer pair designed according to each of the sodium transcript or allele sequences (Table 1 shows the accession number for each of the sodium channel transcripts or alleles) at a final concentration of 3-5 μM. All samples, including the A 'no-template' negative control, were performed in triplicate. The reaction cycle consisted of a melting step of 50ºC for 2 min then 95°C for 10 min, followed by 40 cycles of 95ºC for 15 sec and 60ºC for 1 min. Specificity of the PCR reactions was assessed via a melting curve analysis for each PCR reaction using Dissociation Curves software [44]. Relative expression levels for the sodium channel transcripts were calculated by the 2-ΔΔCT method using SDS RQ software [45]. The 18S ribosome RNA, an endogenous control, was used to normalize the expression of targets [46-49]. Preliminary qRT-PCR experiments with a primer pair (Table 1) designed according to the sequences of the 18S ribosome RNA had revealed that the expression of this gene remained constant among all 3 mosquito strains, so the 18S ribosome RNA was used for internal normalization in the qRT-PCR assays. Each experiment was repeated three to four times with different preparations of RNA samples. The statistical significance of the gene expressions was calculated using a Student's t-test for all 2-sample comparisons and a one-way analysis of variance (ANOVA) for multiple sample comparisons (SAS v9.1 software); a value of P≤0.05 was considered statistically significant. Significant overexpression was determined using a cut-off value of a ≥2-fold change in expression [50].
Oligonucleotide primers used in qRT-PCR reactions for amplifying sodium channel variants.
| Mosquito population | Variants | Forward Primer | Reverse Primer |
|---|---|---|---|
| S-Lab | CxNav-Lv1 | 5' AATCAGCTGTAAAAGTGATGGCGC 3' | 5' AGCTCGTAGTACCCTGAATGTTCT 3' |
| CxNav-Lv2 | 5' CACTGCAAAAGCCCGTAAAGTGAG 3' | 5' ATAGTATACTGGAACGATGATTGCA 3' | |
| CxNav-Lv3 | 5' ACAAGGGCAAGAAGAACAAGCAGC 3' | 5' CTTTATACTGGCAGTGTCATCGTC 3' | |
| CxNav-Sv1 | 5' ATCGATATCTGAGAGAACGTAGTT 3' | 5' TTCATCCTCGTCCTCATCGTCGTA 3' | |
| HAmCqG0 | CxNav-Lv4 | 5' GGTCGGAAGAAAAAGAAAAGAGA 3' | 5' TATCCTTTCCTTTACTAACTACTA 3' |
| CxNav-Lv5 | 5' GCCAAAAAAAGTACTACAACGCAA 3' | 5' TCCCGTCTGCTTGTAGTGAT 3' | |
| CxNav-Lv6 | 5' AGCACAACCATCTCAGTTGGATAT 3' | 5' TCGTCGTCGAGTTCTTCGTCAATT 3' | |
| CxNav-Sv2 | 5' CAAAAGTTCGACATGATCATCATG 3' | 5' TGAAGAACGACATCCCGAAGATG 3' | |
| CxNav-Sv3 | 5' TACTACATGGACAGGATATTCAC 3' | 5' CAGGTTTATGAGCGAGAGCATCA 3' | |
| HAmCqG8 | CxNav-Lv7 | 5' TCGAGTGTTCAAGCTAGCAAA 3' | 5' AATGCAGAGCACAAACGTCAG 3' |
| CxNav-Lv8 | 5' TTCCAGTATACTATGCTAATTTAG 3' | 5' TTGGTGTCGACGTAGGACATGTT 3' | |
| CxNav-Sv4 | 5' TCCAAGGTGATAGGCAATTCTATT 3' | 5' TCAATTCCTAGGTCCTCCTTGCT 3' | |
| CxNav-Sv5 | 5' ACTACTACGAATGTCTTAATGTTT 3' | 5' TGTACTAAAATATAAATAGCTACG 3' |
Results
Generation of sodium channel transcripts in Cx. quinquefasciatus
To examine the number of transcripts of the para-type sodium channel gene in the genome of Culex mosquitoes, RNAs isolated from S-Lab, HAmCqG0 and HAmCqG8 were subjected to PCR amplification using a primer pair: KDR S16 (TGTTGGCCATATAGACAATGACCGA)/KDR AS09 (GCTTCTGAATCTGAATCAGAGGGAG), synthesized based on the respective 5' and 3' end sequences of the putative sodium channel cDNAs (Table 1, 43). Two distinct molecular sizes of sodium channel cDNAs with ~6.5 and ~4 kb were generated by PCR amplification from each of the three mosquito strains, namely susceptible (S-Lab), intermediate (HAmCqG0), and highly resistant (HAmCqG8), when only a single primer pair, KDR S16/ KDR AS09 was used (Fig. 1). The PCR products of both the ~6.5 and ~4.0 kb fragments from each strain were then cloned and sequenced. Sequence analysis of insertions of clones (3, 3, and 2 clones for S-lab, HAmCqG0, and HAmCqG8, respectively) with ~6.5 kb and clones (2, 7, and 2 clones for S-lab, HAmCqG0, and HAmCqG8, respectively ) with~4.0 kb PCR amplified products from each strain indicated that all the cDNA clones were indeed the sodium channel transcripts. Interestingly, nucleotide sequence analysis of these sodium channel transcripts revealed the existence of multiple variants in each of ~6.5 and ~4.0 kb PCR amplification products in each mosquito strains. These variants were then assigned to two categories, CxNa-L and CxNa-S, based on their sizes of ~6.5 and ~4.0 kb, respectively.
Polymerase chain reaction (PCR) amplification of para-type sodium channel transcripts from genomic RNAs of Culex mosquitoes. Sodium channel cDNA transcripts amplified from RNAs isolated from S-Lab, HAmCqG0 and HAmCqG8 mosquito strains were subjected to PCR amplification using a primer pair: KDR S16 (TGTTGGCCATATAGACAATGACCGA)/KDR AS09 (GCTTCTGAATCTGAATCAGAGGGAG), synthesized based on the respective 5' and 3' end sequences of the putative sodium channel genes [43].
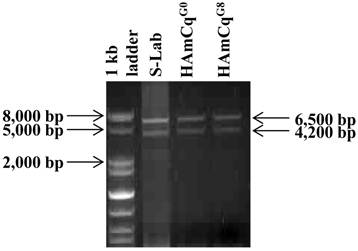
The CxNa-L PCR products included three cDNA sequences of the sodium channel, CxNa-Lv1, CxNa-Lv2, and CxNa-Lv3, in the S-Lab strain, with molecular sequence sizes of 6246, 6273, and 6234 bps, respectively (Fig. 2); three sodium channel cDNA sequences, CxNa-Lv4, and CxNa-Lv5 and CxNa-Lv6, in the HAmCqG0 strain, with 6276, 6285, and 6063 bps, respectively (Fig. 3); and two cDNA sequences, CxNa-Lv7 and CxNa-Lv8, in the HAmCqG8 strain, with 6267 and 6273 bps, respectively (Fig. 4). In contrast, the CxNa-S PCR products contained only one cDNA sequence of the sodium channel in S-Lab, CxNa-Sv1, with a molecular size of 3891 bps (Fig. 2); two cDNA sequences in HAmCqG0, CxNaSv2 and CxNa-Sv3, with molecular sizes of 3615 and 3417 bps, respectively (Fig. 3); and two sodium channel cDNA sequences in HAmCqG8, CxNa-Sv4 and CxNa-Sv5, with 4068 and 3987 bps, respectively, (Fig. 4). This discovery provides strong evidence supporting the existence of multiple transcripts of the sodium channel gene in the mosquito Cx. quinquefasciatus, as we previously suggested [51].
Structural analysis of deduced sodium channel protein sequences within each strain and/or among different strains of Culex mosquitoes
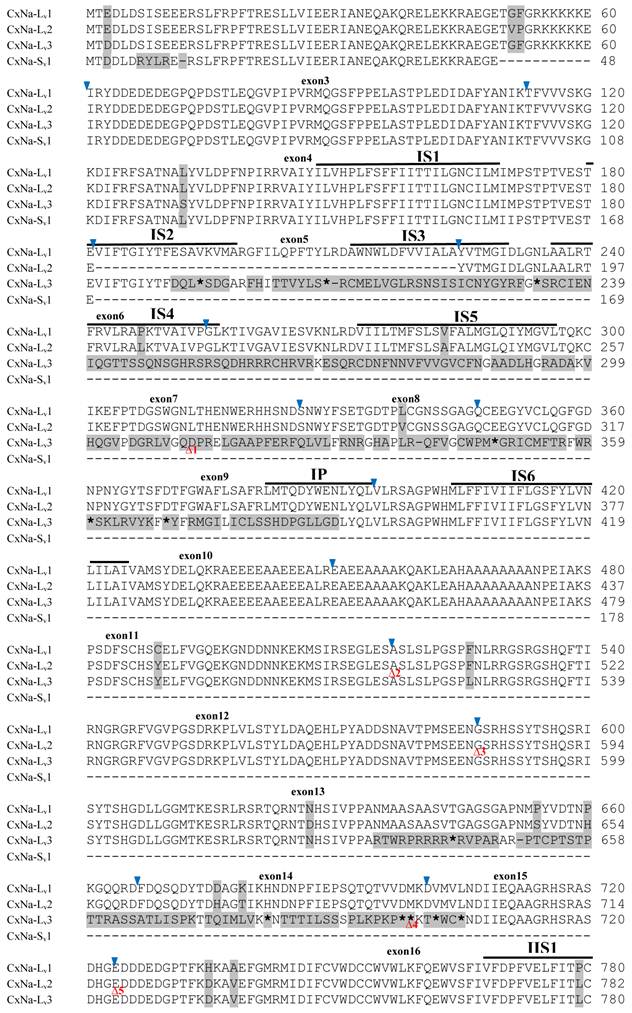
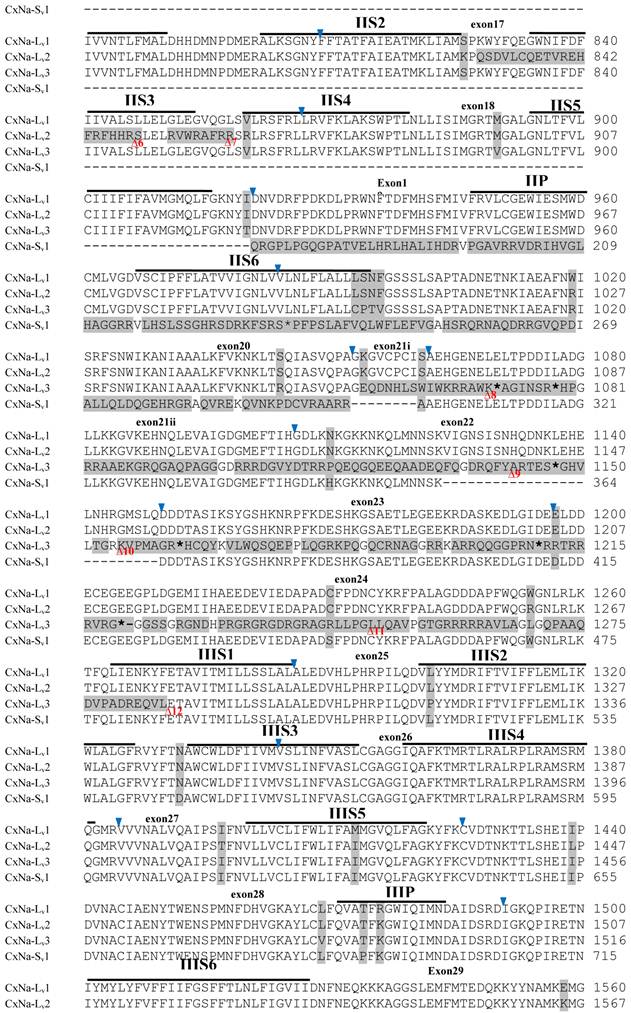
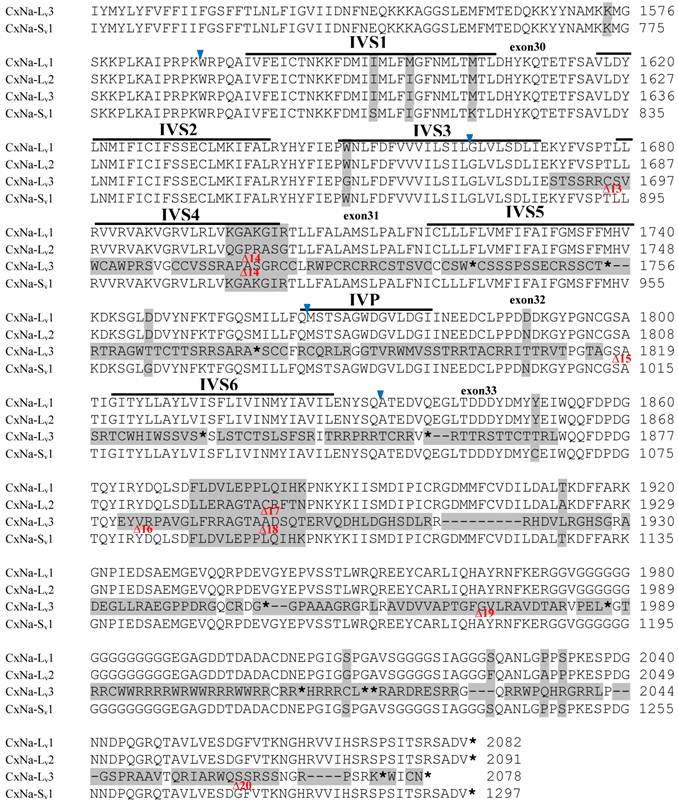
The putative amino acid sequences for the CxNa-L and the CxNa-S transcript sequences were compared for each of the three mosquito strains studied. In the S-Lab strain, of the three transcripts identified in the CxNa-L category, two, CxNa-Lv1 and CxNa-Lv2 (Accession numbers: JN695777 and JX424546), consisted of full length sodium channel sequences encoding the entire ORFs of the sodium channel proteins with 2082 and 2091 amino acid residues, respectively. These exons were numbered 1 through 33 (Fig. 5), based on the silkworm Bombyx mori sodium channel BmNav [35] and house fly sodium channel sequences [11, 52-53]. However, the Culex mosquito sodium channel lacked the exon 12 present in both the BmNav and DmNaV sodium channel sequences. CxNa-Lv1 and CxNa-Lv2 shared very high sequence similarity (96%), except for a missing exon 5 as a result of the alternative splicing (Figs. 2 and 5) and several short insertions identified in the CxNa-Lv2 sequence (Figs. 2 and 5). The remaining transcript, CxNa-Lv3, incorporated several in-frame premature stop codons, with the first occurring at domain I segment 2 (IS2) (Figs. 2 and 5). Short deletions and insertions were also identified in CxNa-Lv3 compared with CxNa-Lv1 and CxNa-Lv2.
Similar length transcripts/variants were identified in the CxNa-L transcripts of the HAmCqG0 mosquitoes (Figs. 3 and 5), in which two of the three transcripts identified, CxNa-Lv4 and CxNa-Lv5 (Accession numbers: JN695778 and JX424547), were entire sodium channel ORFs, encoding 2092 and 2095 amino acid residues, respectively, and sharing 99% sequence similarity. Comparing the transcript sequences of CxNa-Lv4 and CxNa-Lv5 revealed that an alternative splicing exon 2, a short in-frame insertion in exon 12, and a short in-frame deletion in exon 21ii were present in CxNa-Lv5 (Figs. 3 and 5). The third CxNa-L transcript, CxNa-Lv6, in the HAmCqG0 mosquitoes was again the exception, incorporating several in-frame premature stop codons, with the first occurred in the linker between IIS6 and IIIS1, in addition to short insertions and deletions (Figs. 4 and 5). CxNa-Lv6 also exhibited an alternative splicing of exon 2 compared to that identified in CxNa-Lv5. The two CxNa-L transcripts, CxNa-Lv7 and CxNa-Lv8 (Accession numbers: JN695779 and JX424548), identified in HAmCqG8 were both full length sodium channel transcripts encoding entire ORFs of sodium channels proteins (Figs. 4 and 5). CxNa-Lv7 and CxNa-Lv8 shared very high sequence similarity (99%), except for an alternative splicing of exon 21i present in CxNa-Lv8 (Figs. 4 and 5). The above results indicate that multiple ~full length transcripts presented in the mosquitoes, with at least two transcripts in each mosquito strain, had entire ORFs. The other transcripts found in both S-Lab and HAmCqG0 incorporated in-frame premature stop codons and, as such, any resulting proteins would be truncated from those regions onward and thus less likely to be functional transcripts.
The sequences of the CxNa-S transcripts with the ~4.0 kb sized sodium channels in each of the mosquito strains were found to be similar to those of the full length CxNa-L sequences; i.e., one or more transcripts were present in each of the mosquito strains. The main difference in the CxNa-S transcripts compared with those of the full length sequences were the internal exons missing through the alternative splicing, along with some minor short deletions or insertions. In the S-Lab strain, only one transcript was observed, CxNa-Sv1, containing a single in-frame stop codon at the IIS6 region in the sequence (Fig. 2). However, the CxNa-Sv1 transcript also lacked exons 2, 5 to18, 21i, and 22 as a result of the alternative splicing, and thus had a short sodium channel sequence. Two transcripts, CxNa-Sv2 and CxNa-Sv3, were identified in the HAmCqG0 strain (Fig. 3). Both of these sequences exhibited alternative splicing of exons, in-frame stop codons, and short deletions and insertions. The CxNa-Sv2 sequences were found to have alternative splicing of exons 2, 12-26, whereas the CxNa-Sv3 sequences lacked exons 2-15, and parts of exons 21 and 22 (Fig. 3) due to the alternative splicing, once again resulting in a short sodium channel sequence. The HAmCqG8 strain contained 2 transcripts, CxNa-Sv4 and CxNa-Sv5, with entire ORFs, encoding 1356 and 1329 amino acid residues (Accession numbers: JX424549 and JX424550), respectively, and sharing 98% sequence similarity (Fig. 4). Compared with the PRFs of the CxNa-L transcripts, these two CxNa-S transcripts lacked exons 5-18 as a result of the exon alternative splicing (Figs. 4 and 5). Thus, among all the CxNa-S transcripts identified in the tested mosquitoes, only CxNa-Sv4 and CxNa-Sv5 in the highly resistant HAmCqG8 mosquitoes contained the entire ORFs of the sodium channels.
Expression analysis of sodium channel transcripts in Culex mosquitoes
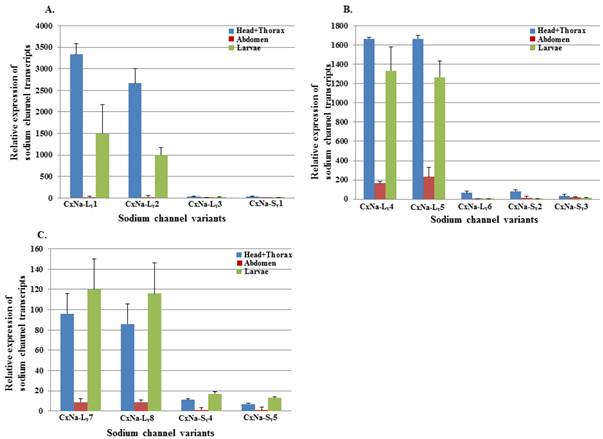
The extent of the variation in alternative transcript expression was also addressed by determining the levels of expression of individual sodium channel transcripts in the 4th instar larvae and different tissues from the adult mosquitoes in each strain using qRT-PCR. Characterizing the developmental and regional expression of the sodium channel transcripts in mosquitoes is critical to our understanding of their relative biological importance. We therefore determined the relative expression levels of sodium channel RNAs for all the transcripts identified in all three mosquito strains, S-Lab, HAmCqG0 and HAmCqG8. Total RNAs were extracted from whole bodies of 4th instar larvae, as well as the head+thorax, and abdomen tissues of 2-3 day old adults. The expression levels were determined using qRT-PCR and the expression ratios for the head + thorax and larval samples were then calculated relative to the quantity of the transcript expression in the corresponding abdomen samples for each strain (Fig. 6). The results show that the sodium channel expression in all three strains shared a number of common features. The expression levels were relatively high in the head + thorax tissues compared to the abdomen tissues; the full length sodium channel transcripts of CxNa-L with an ORF of ~6.5 kb had abundant expression compared with those of CxNa-S ~4.0 kb transcripts, the transcripts with in-frame-stop codons, and CxNa-Lv5 with in-frame-stop codons (Fig. 6). Comparing the transcripts with the full length ORFs in each of the three strains, even though the transcripts had undergone alternative splicing events the expression levels were similar, suggesting that the variants may have equivalent functional importance in the tissues and the mosquitoes. Indeed, the transcripts with in-frame stop codons were detected in both the S-Lab and HAmCqG0 mosquitoes, but at extremely low levels. The difference in the sodium channel expression between the CxNa-L and CxNa-S transcripts was particularly pronounced for S-Lab and HAmCqG0, where the CxNa-S transcripts were expressed at levels more than 1000-fold lower than the CxNa-L sodium channel transcripts (Fig. 6a, b). In contrast, only about a 10-fold difference in expression between the CxNa-L and CxNa-S transcripts was identified in HAmCqG8 (Fig. 6c). This feature, plus the markedly higher expression in HAmCqG8, might reflect their function in HAmCqG8.
Alignment of deduced amino acid transcript sequences of the para-type sodium channel transcripts (Cx-Na) in S-Lab Culex mosquitoes. Transmembrane segments are indicated on the line over the sequence. Exons are indicated above the sequence with solid triangle symbols to indicate the boundaries between exons. The differences in the aa sequences are indicated by shading. A stop codon is marked by an asterisk (*). - indicates deletions. ∆ indicates insertions with the sequences of ∆1: P; ∆2: VSEITRTTAPTATAAGTAKARKVSA; ∆3: GAIIVPVYYANL; ∆4:*I; ∆5: VSVYYFPT; ∆6: GPFR; ∆7: E; ∆8:*; ∆9: **SSR**VR; ∆10: *HCQY; ∆11:*; ∆12: G; ∆13: R; ∆14: R; ∆15: RRR; ∆16: T; ∆17: R; ∆18: A; ∆19: G; ∆20:**
Alignment of deduced amino acid transcript sequences of the para-type sodium channel reanscripts (Cx-Na) in HAmCqG0 Culex mosquitoes. Transmembrane segments are indicated on the line over the sequence. Exons are indicated above the sequence with solid triangle symbols to indicate the bounderies between exons. The differences in the aa sequences are indicated by shading. A stop codon is marked by an asterisk (*). - indicates deletions. ∆ indicates insertions with the sequences of ∆1: VSEITRTTAPTATAAGTAKARKVSA; ∆2: AA; ∆3: R; ∆4: *F; ∆5: L; ∆6: G.

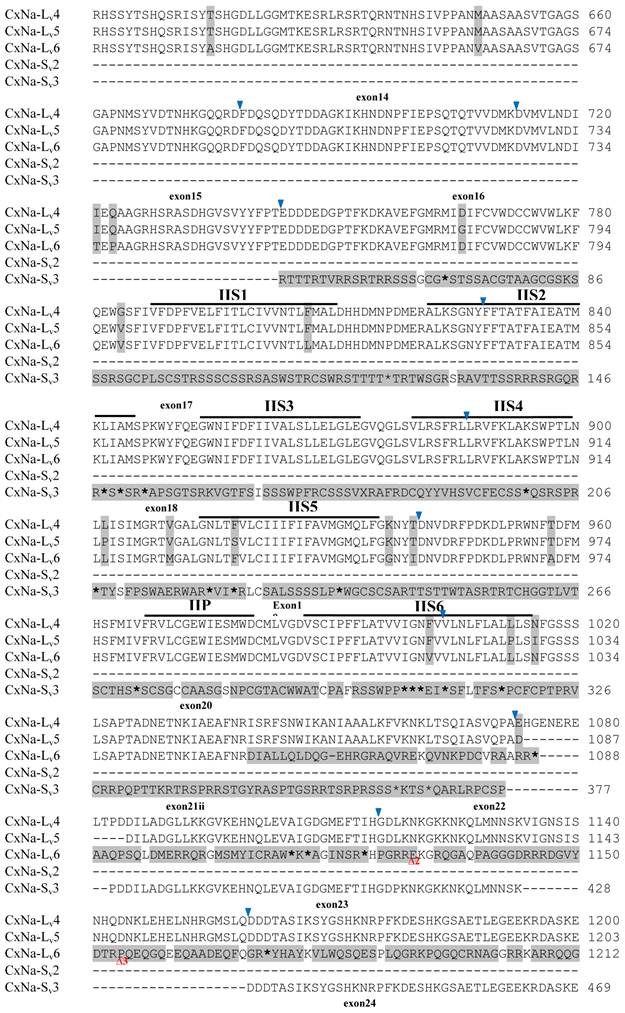

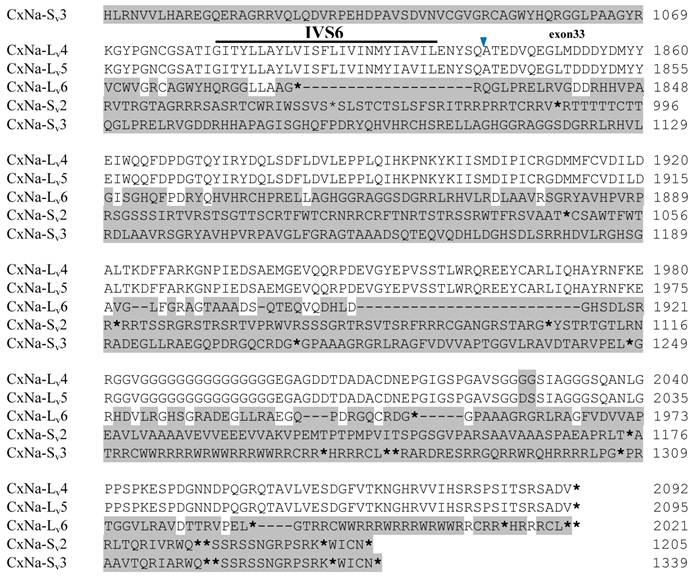
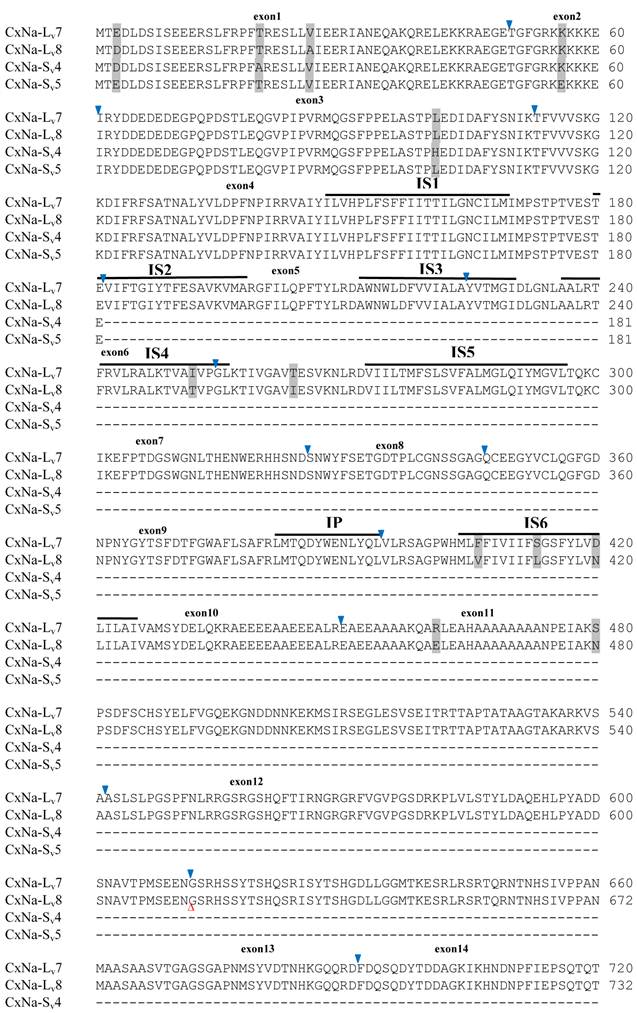
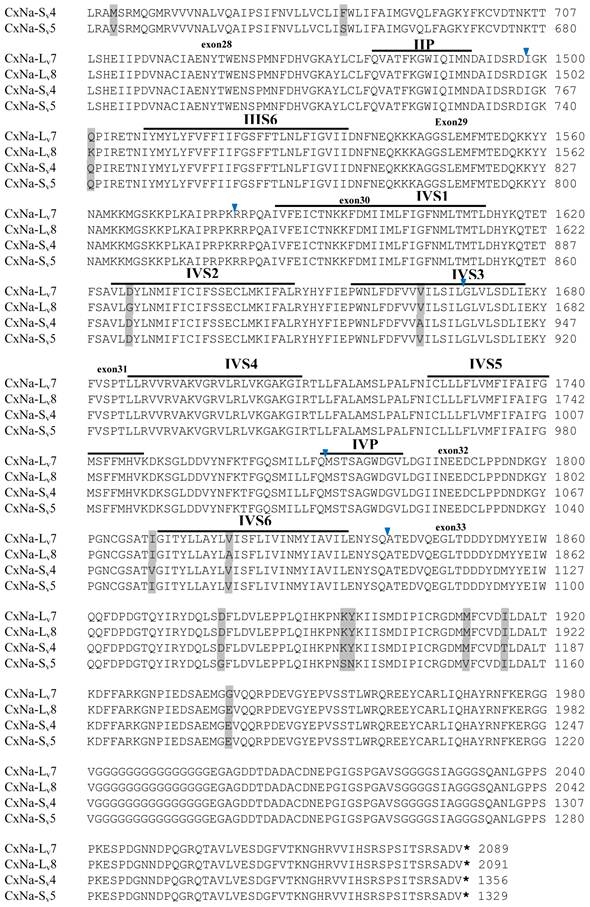
Alignment of deduced amino acid transcript sequences of the para-type sodium channel transcripts (Cx-Na) in HAmCqG8 Culex mosquitoes. Transmembrane segments are indicated on the line over the sequence. Exons are indicated above the sequence with solid triangle symbols to indicate the bounderies between exons. The differences in the aa sequences are indicated by shading. A stop codon is marked by an asterisk (*). - indicates deletions. ∆ indicates insertions with the sequences of ∆1: GAIIVPVYYANL ∆2: GEQHSHLSWIWSE; ∆3: GEQHNHLSWIWSE; ∆4: VIGNSISNHQDNKLEHELNHRGMSLQ.

Alternative splicing of Cx-Nav from mosquitoes Culex quinquefasciatus. Boxes represent exons. The junctions of exons are indicated with straight lines or bridge lines. The schematic of the predicted 6 segments (S1 to S6) in each of the 4 domains (I, II, III, and IV) in the structure of Cx-Nav protein are shown. *The transcript had an entire ORF.

Expression of Cx-Nav in larvae and head+ thorax and abdomen tissues of 2-3 day-old female adult Culex mosquitoes. The relative level of gene expression shown along the Y axis represents the ratio of the gene expression in each tissue from the adults or larvae compared to that measured in the abdomen tissue of the same strain (ratio=1 indicates equal amounts). The experiments were performed three times. The results are shown as the mean ± S.E. No significant difference (P≤0.05) in the levels of sodium channel transcript expression was found in samples labeled with the same alphabetic letter (i.e., a, b, or c).
Discussion
Voltage-gated sodium channels are essential for the action potential generation of the neuron membrane and play a critical role in membrane excitability [6-7]. Over the last few years, a great deal of evidence has accumulated that supports the expression of diverse distinct sodium channel variants in insects through extensive alternative splicing of a single gene [16-23, 34]. The growing interest in alternative splicing of the sodium channels is propelled by its prominent contribution as a key mechanism generating the structural and functional diversity of sodium channels [15, 19]. Following the first reported cloning, sequencing and characterization of multiple variant transcripts from Drosophila melanogaster [32], the alternative splicing of sodium channels has now been characterized in many medically or agriculturally important insect and arachnid pest species, including Drosophila melanogaster [17, 20], the house fly Musca domestica [16], German cockroach Blattella germanica [19], the mosquito Anopheles gambiae [34], diamondback moth Plutella xylostella [22], silkworm Bombyx mori [35], and varroa mite Varroa destructor [36]. The current study represents the first investigation of the transcripts of sodium channels in Cx. quinquefasciatus and has revealed multiple variants of sodium channels generated from extensive alternative splicing and small deletions/ insertions, which is consistent with the results of the previous studies of the sodium channels of other insect species.
These multiple variants of para-type sodium channel transcripts presented in the mosquito Cx. quinquefasciatus can be classified in terms of two categories, CxNa-L and CxNa-S, based on their distinguishing sizes of ~6.5 kb and ~4.0 kb, which were present in all three mosquito strains tested - the susceptible S-Lab strain, the low resistant HAmCqG0 strain, and the highly resistant HAmCqG8 strain. The main difference in the sequences obtained for these two subcategories is the presence of multiple internal exons obtained through alternative splicing. In all, nine alternatively splice variants were identified in Culex mosquitoes. In the CxNa-L sodium channel category, four splice variants were identified, of which three were full length variants with three optional exons (2, 5, and 21i) and one incorporated in-frame-stop codons. Exon 2 is located in the N-terminus, which is an optional exon corresponding to optional exon 2 of the sodium channel in the silkworm and optional exon j of the para in Drosophila and which is also conserved in other insect sodium channel genes [35]. Exon 5 is located between IS2 and IS3. Interestingly, skipping of exon 5 also occurs in the silkworm [35], German cockroaches [54] and the mosquito Anopheles gambiae [34], suggesting that exon 2 and exon 5 may be a conserved optional exon in insects. Exon 21 is located in the intracellular linker connecting domains II and III of the Culex mosquito sodium channels. The 5' portion of exon 21, named 21i, is optional in Culex mosquitoes. Exon 21i corresponds to optional exon f in the para gene of Drosophilar and exon 22i in the silkworm [35]. These variants with optional exons 2, 5, and/or 21i are all entire ORFs of sodium channels, which may suggest the functional importance of these transcripts in mosquitoes. It has been reported that, when expressed in Xenopus oocytes, the alternative splicing variants could exhibit different gating properties and generate sodium channel proteins with differing sensitivities to pyrethroids [18-19]. Whether these variants identified in the Culex mosquitoes also have different protein properties and different responses to pyrethroids remains to be seen.
Investigation of the putative amino acid sequences of alternative splicing variants in the CxNa-S sodium channel category, i.e. the ~4.5 kb transcripts, revealed that in contrast to the findings of CxNa-L, the alternative splicing identified in the sodium channel of Culex mosquitoes has resulted in large size or multi-exon-splicing. All the CxNa-S splicing variants in both the susceptible S-Lab and low resistance parental HAmCqG0 strains had in-frame stop codons, suggesting that these splicing variants and any resulting proteins would be truncated from those regions onward. As it has been reported that a truncated channel does not produce any sodium current when it expressed in Xenopus oocytes [19], the transcripts identified in our study that contain in-frame stop codons may not be functional transcripts. Furthermore, the ~1000 to ~3000-fold lower expression of the splice variants with stop codons compared to the CxNa-L splicing variances may further support the conclusion that these variances in mosquitoes are relatively unimportant. Nevertheless, two alternative splicing variants of CxNa-S splicing in HAmCqG8 had no in-frame stop codons but still had ORFs encoding sodium channel transcripts lacking exons 5 to18. In addition, these two variants in HAmCqG8 had relatively high expression levels, with only ~10-fold lower expression levels compared with the CxNa-L variants. Nevertheless, these variants both lacked IS4 and IIS4 as a result of the alternative splicing. Since the S4 segments act as voltage sensors that initiate voltage-dependent activation [34-35], the issue of whether these two alternative splicing variants identified in the highly resistant HAmCqG8 strain perform some function in the sodium channels of mosquitoes requires further investigation.
Acknowledgements
This study was supported by Award Number R21AI090303 from the National Institute of Allergy and Infectious Diseases, AAES Hatch/Multistate Grants ALA08-045 to N.L., and ALA015-1-10026 to N.L. We also thank anonymous reviewers for their valuable comments and suggestions for our manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bloomquist JR. Ion channels as targets for insecticides. Annu Rev Entomol. 1996;41:163-190
2. Narahashi T. Neuronal ion channels as the target sites of insecticides. Pharmacol Toxico. 1996;7:1-14
3. Soderlund DM. Sodium channels. In: (ed.) Gilbert LI, Iatrou K, Gill SS. Comprehensive molecular insect science. New York: Elsevier Pergamon. 2005:1-24
4. Soderlund DM, Knipple DC. The molecular biology of knockdown resistance to pyrethroid insecticides. Insect Biochem Mol Biol. 2003;33:563-77
5. Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13-25
6. Narahashi T. The role of ion channels in insecticide action. In: (ed.) Narahashi T, Charnbers EJ. Insecticide action: from molecule to organism. New York: Plenum Press. 1988:55-84
7. Narahashi T. Neuroreceptors and ion channels as the basis for drug action: past, present and future. J Pharmacol Exp Ther. 2000;294:1-26
8. Smith TJ, Lee SH, Ingles PJ. et al. The L1014F point mutation in the house fly Vssc1 sodium channel confers knockdown resistance to pyrethroids. Insect Biochem Mol Biol. 1997;27:807-812
9. Davies TGE, Field LM, Usherwood PNR. et al. DDT, pyrethrins, pyrethroids and insect sodium channels. IUBMBLife. 2007;59:151-162
10. Davies TGE, O'Reilly AO, Field LM. et al. Knockdown resistance to DDT and pyrethroids: from target-site mutations to molecular modeling. Pest Manag Sci. 2008;64:1126-1130
11. Williamson MS, Martinez-Torres D, Hick CA. et al. Identification of mutations in the house fly para-type sodium channel gene associated with knockdown resistance (kdr) to pyrethroid insecticides. Mol Gen Genet. 1996;252:51-60
12. Miyazaki M, Ohyama K, Dunlap DY. et al. Cloning and sequencing of the para-type sodium channel gene from susceptible and kdr-resistant German cockroaches (Blattella germanica) and house fly (Musca domestica). Mol Gen Genet. 1996;252:61-68
13. Feng Y, Zhao S, Sun W. et al. The sodium channel gene in Tetranychus cinnabarinus (Boisduval): identification and expression analysis of a mutation associated with pyrethroid resistance. Pest Manag Sci. 2011;67(8):904-912
14. Li T, Zhang L, Xu Q. et al. Multiple mutations and mutation combinations in the sodium channel of permethrin resistant mosquitoes, Culex quinquefasciatus. Sci Rep. 2012;2:781
15. Dong K. Insect sodium channels and insecticide resistance. Invert Neurosci. 2007;7:17-30
16. Lee SH, Ingles PJ, Knipple DC. et al. Developmental regulation of alternative exon usage in the house fly Vssc1 sodium channel gene. Invert Neurosci. 2002;4(3):125-133
17. Olson ROD, Liu Z, Nomura Y. et al. Molecular and functional characterization of voltage-gated sodium channel variants from Drosophila melanogaster. Insect Biochem Mol Biol. 2008;38(5):604-610
18. Lin WH, Wright DE, Muraro NI. et al. Alternative splicing in the voltage-gated sodium channel DmNav regulates activation, inactivation, and persistent current. J Neurophysiol. 2009;102:1994-2006
19. Tan J, Liu Z, Nomura Y. et al. Alternative splicing of an insect sodium channel gene generates pharmacologically distinct sodium channels. J Neurosci. 2002;22(13):5300-5309
20. Thackeray JR, Ganetzky B. Conserved alternative splicing patterns and splicing signals in the Drosophila sodium channel gene para. Genetics. 1995;141:203-214
21. O'Dowd DK, Gee JR, Smith MA. Sodium current density correlates with expression of specific alternatively spliced sodium channel mRNAs in single neurons. J Neurosci. 1995;15:4005-4012
22. Sonoda S, Tsukahara Y, Ashfaq M. et al. Genomic organization of the para sodium channel alpha-subunit genes from the pyrethroid-resistant and -susceptible strains of the diamondback moth. Arch Insect Biochem Physiol. 2008;69(1):1-12
23. Sonoda S, Igaki C, Tsumuki H. Alternatively spliced sodium channel transcripts expressed in field strains of the diamondback moth. Insect Biochem Mol Biol. 2008;38:883-890
24. Meisler MH, O'Brien JE, Sharkey LM. Sodium channel gene family: epilepsy mutations, gene interactions and modifier effects. J Physiol. 2010;588(11):1841-1848
25. Goldin AL. Resurgence of sodium channel research. Annu Rev Physiol. 2001;63:871-894
26. Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2003;4:207
27. Novak AE, Jost MC, Lu Y. et al. Gene duplications and evolution of vertebrate voltage-gated sodium channels. J Mol Evol. 2006;63(2):208-221
28. Lopreato GF, Lu Y, Southwell A. et al. Evolution and divergence of sodium channel genes in vertebrates. Proc Natl Acad Sci USA. 2001;98:7588-7592
29. Blackshaw SE, Henderson LP, Malek J. et al. Single-cell analysis reveals cell-specific patterns of expression of a family of putative voltage-gated sodium channel genes in the leech. J Neurobiol. 2003;55:355-371
30. Nagahora H, Okada T, Yahagi N. et al. Diversity of voltage-gated sodium channels in the ascidian larval nervous system. Biochem Biophys Res Commun. 2000;275:558-564
31. Okamura Y, Ono F, Okagaki R. et al. Neural expression of a sodium channel gene requires cell-specific interactions. Neuron. 1994;13:937-948
32. Loughney K, Kreber R, Ganetzky B. Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell. 1989;58:1143-1154
33. Salkoff L, Butler A, Wei A. et al. Genomic organization and deduced amino acid sequence of a putative sodium channel gene in Drosophila. Sci. 1987;237(4816):744-749
34. Davies TGE, Field LM, Usherwood PN. et al. A comparative study of voltage-gated sodium channels in the Insecta: implications for pyrethroid resistance in Anopheline and other Neopteran species. Insect Mol Biol. 2007;16(3):361-375
35. Shao YM, Dong K, Tang ZH. et al. Molecular characterization of a sodium channel gene from the Silkworm Bombyx mori. Insect Biochem Mol Biol. 2009;39:145-151
36. Wang R, Huang ZY, Dong K. Molecular characterization of an arachnid sodium channel gene from the varroa mite (Varroa destructor). Insect Biochem Mol Biol. 2003;33(7):733-739
37. Sardelis MR, Turell MJ, Dohm DJ. et al. Vector competence of selected North American Culex and Coquillettidia mosquitoes for West Nile virus. Emerg Infect Dis. 2001;7:1018-1022
38. Jones SC, Morris J, Hill G. et al. St. Louis encephalitis outbreak in Louisiana in 2001. J La State Med Soc. 2002;154:303-306
39. Arensburger P, Megy K, Waterhouse RM. et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Sci. 2010;330:86-88
40. Li T, Liu N. Genetics and inheritance of permethrin resistance in the mosquito Culex quinquefasciatus. J Med Entomol. 2010;47:1127-1134
41. Liu H, Cupp EW, Micher KM. et al. Insecticide resistance and cross-resistance in Alabama and Florida strains of Culex quinquefasciatus. J Med Entoml. 2004;41:408-413
42. Liu N, Scott GJ. Phenobarbital induction of CYP6D1 is due to a trans acting factor on autosome 2 in house flies, Musca domestica. Insect Mol Biol. 1997;6:77-81
43. Xu Q, Zhang L, Li T. et al. Evolutionary adaptation of the amino acid and codon usage of the mosquito sodium channel following insecticide selection in the field mosquitoes. PLoS One. 2012;7(10):e47609. doi:10.1371/journal.pone.0047609
44. Wittwer CT, Herrmann MG, Moss AA. et al. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques. 1997;22:130-131
45. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402-408
46. Liu N, Liu H, Zhu F. et al. Differential expression of genes in pyrethroid resistant and susceptible mosquitoes, Culex quinquefasciatus (S.). Gene. 2007;394:61-68
47. Aerts JL, Gonzales MI, Topalian SL. Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. BioTechniques. 2004;36:84-86
48. Zhu F, Liu N. Differential expression of CYP6A5 and CYP6A5v2 in pyrethroid-resistant house flies, Musca domestica. Arch Insect Biochem Physiol. 2008;34:147-161
49. Zhu F, Feng J, Zhang L. et al. Characterization of two novel cytochrome P450 genes in insecticide resistant house flies. Insect Mol Biol. 2008;20:1365-1583
50. Strode C, Wondji CS, David JP. et al. Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochem Mol Biol. 2008;38:113-123
51. Xu Q, Tian L, Zhang L. et al. Sodium channel genes and their differential genotypes at the L-to-F kdr locus in the mosquito Culex quinquefasciatus. Biochem Biophys Res Com. 2011;407:645-649
52. Knipple DC, Doyle KE, Marsella-Herrick PA. et al. Tight genetic linkage between the kdr insecticide resistance trait and a voltage-sensitive sodium channel gene in the house fly. Proc Natl Acad Sci USA. 1994;91(7):2483-2487
53. Ingles PJ, Adams PM, Knipple DC. et al. Characterization of voltage-sensitive sodium channel gene coding sequences from insecticides-susceptible and knockdown-resistant house fly strains. Insect Biochem Mol Biol. 1996;26:319-326
54. Song W, Liu Z, Tan J. et al. RNA editing generates tissue specific sodium channels with distinct gating properties. J Biol Chem. 2004;279:32554-32561
Author contact
![]() Corresponding author: Dr. Nannan Liu, Department of Entomology & Plant Pathology, 301 Funchess Hall, Auburn University, Auburn, AL 36849-5413, USA. Phone: (334) 844-2661, Fax: (334) 844-5005 E-mail: liunannedu.
Corresponding author: Dr. Nannan Liu, Department of Entomology & Plant Pathology, 301 Funchess Hall, Auburn University, Auburn, AL 36849-5413, USA. Phone: (334) 844-2661, Fax: (334) 844-5005 E-mail: liunannedu.

 Global reach, higher impact
Global reach, higher impact