10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(1):34-44. doi:10.7150/ijbs.5071 This issue Cite
Research Paper
Specific and Functional Diversity of Endophytic Bacteria from Pine Wood Nematode Bursaphelenchus Xylophilus with Different Virulence
1. Institute of Forest Protection, College of Forest Resources and Environment, Nanjing Forestry University, Nanjing, China,
2. Jiangsu Key Laboratory for Prevention and Management of Invasive Species, Nanjing Forestry University, Nanjing, China.
Received 2012-8-20; Accepted 2012-11-23; Published 2012-12-19
Abstract
Pine wilt disease (PWD) caused by the pine wood nematode (PWN), Bursaphelenchus xylophilus, is one of the most devastating diseases of Pinus spp. The PWN was therefore listed as one of the most dangerous forest pests in China meriting quarantine. Virulence of the PWN is closely linked with the spread of PWD. However, main factors responsible for the virulence of PWNs are still unclear. Recently epiphytic bacteria carried by PWNs have drawn much attention. But little is known about the relationship between endophytic bacteria and virulence of B. xylophilus. In this research, virulence of ten strains of B. xylophilus from different geographical areas in six provinces of China and four pine species were tested with 2-year-old seedlings of Pinus thunbergii. Endophytic bacteria were isolated from PWNs with different virulence to investigate the relationship between the bacteria and PWN virulence. Meanwhile, the carbon metabolism of endophytic bacteria from highly and low virulent B. xylophilus was analyzed using Biolog plates (ECO). The results indicated that ten strains of PWNs showed a wide range of virulence. Simultaneously, endophytic bacteria were isolated from 90% of the B. xylophilus strains. The dominant endophytic bacteria in the nematodes were identified as species of Stenotrophomonas, Achromobacter, Ewingella, Leifsonia, Rhizobium, and Pseudomonas using molecular and biochemical methods. Moreover, S. maltophilia, and A. xylosoxidans subsp. xylosoxidans were the predominant strains. Most of the strains (80%) from P. massoniana contained either S. maltophilia, A. xylosoxidans, or both species. There was a difference between the abilities of the endophytic bacteria to utilize carbon sources. Endophytic bacteria from highly virulent B. xylophilus had a relatively high utilization rate of carbohydrate and carboxylic acids, while bacteria from low virulent B. xylophilus made better use of amino acids. In conclusion, endophytic bacteria widely exist in B. xylophilus from different pines and areas; and B. xylophilus strains with different virulence possessed various endophytic bacteria and diverse carbon metabolism which suggested that the endophytic bacteria species and carbon metabolism might be related with the B. xylophilus virulence.
Keywords: pine wilt disease, Bursaphelenchus xylophilus, endophytic bacteria, virulence, Pinus massoniana, P. thunbergii.
Introduction
Pine wilt disease (PWD) caused by the pine wood nematode (PWN), Bursaphelenchus xylophilus, is the most destructive disease of pine species, causing economic losses in China estimated to be at least 2.5 billion Chinese RMB (about $400 million, U.S.) [1]. PWD was considered as “pine cancer”, because the infected pines will die rapidly and no effective measurement can be taken for treatment. B. xylophilus is believed to be native in North America, usually only damages exotic pine trees there, then spread to Asian and European countries such as Japan, China, South Korea, Portugal; until recently two loci of PWN were reported from Spain [2,3]. Because the cycle of PWD is highly complex, so far little is known about the mechanism of the disease. The virulence of PWN is closely related to the spread of PWD. Recent researches supported the hypothesis that the bacteria associated with B. xylophilus play a role in PWD [4-9]. Oku et al. [9] isolated epiphytic bacteria from PWNs that could produce toxins causing PWD. In a bacterial pathogenicity study, phytotoxins produced by PWN associated bacteria caused symptoms on pine seedlings similar to PWD [7]. In addition, some bacteria were reported to promote the reproduction of PWNs, whereas others restrained PWN-egg hatching [10]. Bacterial growth in nutrient agar plate was significantly increased by the presence of PWNs [10]. Various species of bacteria have been isolated from the surface of pine wood nematodes, B. xylophilus from different countries and regions carry distinct genera of bacteria. In Japan, the bacteria are mainly Pseudomonas sp. and Bacillus spp. [4, 6]. In addition to Bacillus spp. and Pseudomonas spp. [10-12], researchers in China isolated Pantoea, Enterobacter, Peptostreptococcus, Burkholderia [13,14]. Burkholderia sp., Brevibacterium , Enterobacter , Ewingella and Serratia were found in Korea [15]. In Portugal, the mainly associated bacteria of PWNs were Enterobacteriaceae (genera Yersinia, Serratia, Pantoea, Erwinia and Ewingella) , Burkholderia and Pseudomonas [16, 17]. All the bacteria above were isolated from the surface of B. xylophilus, it is difficult to analyze the relationship between the varying epiphytic bacteria species of PWNs with their contribution to the nutrition metabolism and virulence of PWN.
The microbial community inside animals has always attracted researcher's attention, particularly their interaction with host animals. Many recent studies have indicated that the microbial communities inhabit different organs of animals [18-21]. These microbial communities play important roles in the development of their hosts. During evolution, bacteria have developed strategies to adapt to the different environments in animals. Endosymbiotic bacteria of insect pathogenic nematodes, Steinernema and Heterorhabditis, can produce toxins to kill insects and provide nutrition for the nematodes [22]. Bacteria in cysts of Heterodera glycines can promote the survival of the nematodes in soil [23]. A member of a new Wolbachia supergroup of bacteria was found as an endosymbiont in nematode Radopholus similis, suggesting an essential function of the bacteria in the nematode [24]. So far, only a few studies have been focused on endogenous bacteria of B. xylophilus. Tian et al [25] constructed the library of bacteria associated with B. xylophilus, but it is not clear if endophytic bacteria were included in the library. Recently, we observed the existence of endophytic bacteria using transmission electron microscopy and successfully isolated one bacterial species, Stenotrophomonas maltophilia, from B. xylophilus [26]. Here we aimed at determining whether PWNs from various geographic locations and different host pines contain endophytic bacteria, and whether they contribute to virulence of the PWNs. Since PWNs mainly colonize pine xylem, which is composed of cellulose and semicellulose, we are interested in the bacterial ability to utilization of various carbohydrate. Biolog Eco Plate is a fast, simple and convenient way to analyze the diversity of microflora. So we used Biolog Eco Plate to analyze carbon metabolism of PWN endophytic bacteria. In general, we attempted to find the relationship between endophytic bacteria and PWN. It would be important for further identification of the possible affected factors on the virulence of B. xylophilus.
Materials and methods
Isolation of B. xylophilus
Ten strains of B. xylophilus were isolated from naturally wilted pine trees from Anhui (P. massoniana, P. taiwanensis), Zhejiang (P. thunbergii, P. massoniana), Jiangsu (P. thunbergii), Guangdong (P. massoniana), Yunnan (P. kesiya), and Hubei provinces (P. massoniana) in China (Table 1) by the Baermann funnel extraction [27]. The nematodes were propagated on Botrytis cinerea cultured on PDA medium at 25°C.
Isolation of endophytic bacteria from B. xylophilus
The PWN strains were cultured on Botrytis cinerea grown on PDA at 25°C and extracted using the Baermann funnel method. The harvested nematodes were rinsed three times with sterilized distilled water and then soaked in 1% mercuric chloride for 30 min and in a mixture of 1% spectromycin and 1% gendamycin for 30 min. Subsequently, the nematodes were centrifuged at 1,500 g for 3 min and then washed three times with sterile deionized water. The nematode suspension was adjusted to a volume of 1 mL. Contamination was tested by dropping 100 μL of treated nematode suspension onto nutrient agar plates, which were incubated for 72 hours. Another 100 μL of surface-sterilized nematodes were ground into homogenate in sterile mortars under sterile conditions and then cultured on nutrient agar (NA) plates. The process of grinding was repeated three times. Sterile water plated on NA was used as a control (CK1) and sterile water with sterile quartz sand plated on NA was used as a second control (CK2). These plates were incubated at 30°C for 2~3 days and examined daily for appearance of bacterial colonies. Single colonies were selected based on morphology and growth rate.
Virulence test of B. xylophilus
Each of the 10 PWN strains was inoculated onto two-year-old seedlings of P. thunbergii with 10 replicates for each treatment in a greenhouse under day temperature of 28°C during the experimental period. Using sterile scalpels, the stem of each seedling was wounded 6-8 cm above the base of the root. Sterile cotton balls (0.3 cm × 0.3 cm) were placed in the wounds and sealed with parafilm to form a funnel-like shape. Nematode suspension of 500 μL (containing 2000 nematodes) was injected into the cotton balls. This inoculation method used in this research had been widely used in pine wilt disease researches [2]. In the control, sterile water of 500 μL was used for inoculation. After 60 days, the mortality of P. thunbergii was recorded.
Biochemical and physiological identification of endophytic bacteria of B. xylophilus
For each chosen bacterial isolate, the morphological characteristics of the bacterial colony on a NA plate were recorded, including the shape, color, transparency, prominence, edge, and viscosity. Biochemical tests included Gram reaction, oxidase, catalase, methyl red test, Voges-Proskauer test, gelatin liquefaction, starch hydrolysis, nitrate reduction, indole production, citrate utilization, ammonia and hydrogen sulfide production and litmus milk [28, 29]. In addition, each isolate was analyzed for its carbon substrate oxidation pattern according to user instruction (BIOLOG, Hayward, Calif.).
The software for the BIOLOG GN and GP databases was version 3.5. A similarity index of ≥0.500 was considered to indicate a good species match. Where three similarity indices of ≥0.500 were recorded after 6h and 24 h of incubation, the highest similarity index was used [30].
16S rDNA analysis of the endophytic bacteria
Genomic DNAs of the endophytic bacteria of B. xylophilus were extracted using the method described by Kwon et al. [15] and then quantified with a NanoDrop ND-1000 spectrophotometer. The primers 27F (5´-AGAGTTTGATCMTGGCTCAG-3´) and 1493R (5′-ACGGCTACCTTGTTACGACT-3′) were used to generate 16S rRNA gene fragments of ca. 1,500 bp. A total of 0.1 μL of template DNA was used for a 50 μL PCR amplification reaction. The PCR amplification was performed as follows: one cycle of 5 minutes at 94°C, followed by 30 cycles of 30 seconds at 94°C, 30 seconds at 56°C, and 1 minute at 72°C, followed by one cycle of 5 minutes at 72°C [31]. Possible amplification of contaminants was determined with a negative PCR control in which the template DNA was replaced with sterile H2O. Purified PCR fragments were sequenced at Jinsite Ltd (Nanjing, China). The acquired sequences were aligned against the NCBI GenBank database by using BLAST to identify sequences with high similarity. Phylogenetic analyses were conducted by MEGA 4.1 using the neighbor-joining method. Bootstrap analysis of 1000 replicates was performed to validate the reproducibility of the branching pattern.
Origin of Bursaphelenchus xylophilus strains, sampling areas, and mortality of inoculated Pinus thurnbergii seedlings.
| Number | Nematode Strain | Pine host | Sampling area | Susceptible cases of P. thunbergii seedlings | |
|---|---|---|---|---|---|
| The first brown needles (d) | Mortality (%) / Symptom Grade* | ||||
| 1 | AMA3 | Pinus massoniana | Maanshan, Anhui Province | 20 | 50 / III |
| 2 | ZL1 | P. massoniana | Linhai, Zhejiang Province | 26 | 40 / III |
| 3 | HE2 | P. massoniana | Enshi, Hubei Province | 33 | 40 / III |
| 4 | GF3 | P. massoniana | Fengkai, Guangdong Province | 23 | 40 / III |
| 5 | ZZ1 | P. thunbergii | Zhoushan, Zhejiang Province | 22 | 30 / II |
| 6 | AN19 | P. massoniana | Guangde, Anhui Province | 22 | 30 / II |
| 7 | JWB2 | P. thunbergii | Wuxi, Jiangsu Province | 26 | 20 / II |
| 8 | AA3 | P. taiwanensis | Anqing, Anwei Province | 45 | 20 / II |
| 9 | JY20 | P. thunbergii | Yangzhou, Jiangsu Province | 29 | 10 / I |
| 10 | YW4 | P. kesiya | Dehong, Yunnan Province | 23 | 10 / I |
| CK | -- | -- | -- | -- | 0 / 0 |
*Mortality was determined 60 days after inoculation.
Biolog-Eco measurement of endophytic microbial community of nematode
Functional diversity of endophytic microbial community of nematode was measured with Biolog Eco-microplates. The highly virulent strain ZL1 and low virulent strain AA3 were surface-sterilized and then suspended in 900μL (50,000 nematodes) of sterile water, which was subsequently ground with aseptic motor and silica, and volumed to 10 ml. A microplate, each well added with 150 μL of the sample, was incubated at 25°C for 192 h. Optical absorbance was measured at 590 nm and 750nm with an Emax precision microplate reader every 24h.
(1) Average well color development (AWCD), (2) Shannon index (H), (3) Simpson index (D), (4) Evenness index (E) of measurement at 96h were analyzed as previously described [30, 31]. AWCD indicates the metabolic activity of microbial community. AWCD (590-750 nm) = Σ (ni) / 31, H = -Σ ( Pi × lnPi), D=1-ΣPi2, Pi = C590 - 750 /Σ (C590 - 750 ), E=H/lnS, where ni is the metabolic activity on each substrate, Pi is the ratio of ni to the sum of activities on all substrates, and S is species richness of community [33].
Statistical analysis
The formula below was used for calculating the isolation frequency of each bacterial species. Isolation frequency (%) = (the number of occurrences of the bacterial species) ×100 / (the number of strains of B. xylophilus). In Biolog Eco plate experiment, the data were subjected to analysis of variance using the SPSS package, version 13.0. t-test was used for analysis of functional diversity of endophytic microbial community. Differences with p < 0.05 were considered significant.
Results
Virulence of different strains of B. xylophilus
Two-year-old seedlings of P. thunbergii were inoculated with the ten PWN strains. The initial symptom of P. thunbergii seedlings caused by PWNs was needle browning, which developed 20~45 days after inoculation with B. xylophilus and continued to progress. The degree of brown needles caused by B. xylophilus depended on the nematode strain. Sixty days after inoculation, a range of mortality of pines was observed while no pine died in the control group (Table 1). The virulence of the ten nematode strains could be categorized into three classes: strong virulence causing 40% ~ 50% mortality (Class I), moderate virulence causing 20% ~ 30 mortality (Class II), and weak virulence causing 10% mortality (Class III) (Table 1). Most of the PWNs collected from P. massoniana fell into Class I, while the PWNs from P. taiwanensis and P. kesiya fell into Class III. The PWNs from P. thunbergii showed a wide virulence range spanning all three classes. No significant association was found between the PWNs virulence and their geographical locations.
Endophytic bacteria of B. xylophilus
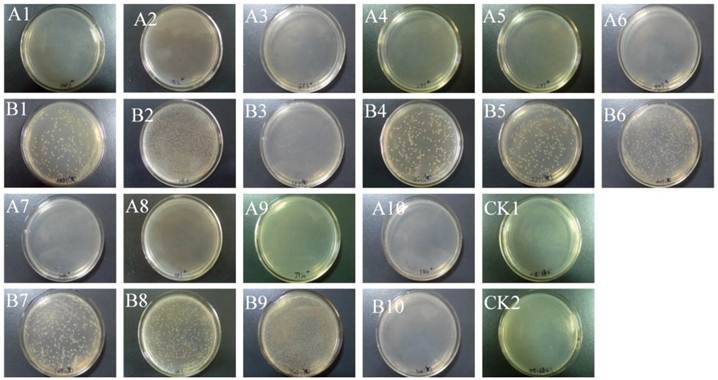
Altogether, 15 endophytic bacterial strains were isolated from the ten strains of B. xylophilus (Fig. 1 B1- B10). No microorganism were observed from surface-sterilized nematodes without grinding the nematodes (Fig. 1 A1-A10).
The highest number of bacteria was obtained from strain JY20, originally collected from infected P. thunbergii, whereas only a few bacteria were found in strain AA3 from P. taiwanensis and in strain GF3 from P. massoniana. No bacterium was isolated from strain YW4, collected from P. kesiya in Yunnan (Fig. 1 B10). In general, PWNs from P. thunbergii and P. massoniana manifested a higher isolation rate of endophytic bacteria than those from P. taiwanensis and P. kesiya. The number and abundance of endophytic bacterial species isolated from each B. xylophilus strain are shown in Table 2.
Nematodes isolated from infected pines and the abundance of endophytic bacteria.
| Nematode strain | number of nematodes | CFU* number per nematode |
|---|---|---|
| YW4 | 53 | 0 |
| GF3 | 110 | 0.05±0.01 |
| AA3 | 132 | 1.9±1.42 |
| WB2 | 109 | 2.42±1.41 |
| ZZ1 | 58 | 3.33±0.48 |
| AMA3 | 104 | 4.08±2.75 |
| ZL1 | 78 | 4.45±1.99 |
| HE2 | 163 | 5.19±1.71 |
| AN19 | 58 | 5.87±0.80 |
| JY20 | 80 | 19.3±2.93 |
Isolation of endophytic bacteria from surface-sterilized B. xylophilus. Surface-sterilized nematodes that were homogenized (B1-B10) or intact (A1-A10) were spread onto nutrient agar and incubated for 2 or 3 days. CK1 and CK2 were sterile water without or with grinding, respectively.
Identification of endophytic bacteria of B. xylophilus
All the bacteria isolated were rods with varying size. Except NSBx.29 (ZZ1), all the isolates were Gram-negative. All the isolates were oxidase-negative, catalase-positive, starch hydrolysis-negative. Other biochemical characteristics of the fifteen endophytic bacterial strains are shown in Table 3. According to the 16S rDNA sequence analysis, the bacterial strains belonged to six genera: Stenotrophomonas, Achromobacter, Ewingella, Leifsonia, Rhizobium, and Pseudomonas (Fig. 2). The Biolog Microbial Identification System results, PROB, SIM and DIS15 of 15 isolated B. xylophilus strains were shown in Table 3.
As shown in Table 3, endophytic bacteria NSBx.2, NSBx.9, NSBx.14, NSBx.23 and NSBx.25 were S. maltophilia; NSBx.6, NSBx.10, NSBx.11, NSBx.19 and NSBx.22 were A. xylosoxidans ss. xylosoxidans; NSBx.16 and NSBx.18 were E. americana; NSBx.27 was R. radiobacter, which is the synonym of A. tumefaciens obtained from 16 rDNA sequence analysis; NSBx.29 was Leifsonia aquatica and NSBx.31was P. fluorescens. According to the SIM and DIS, the bacteria from the 15 B. xylophilus strains were classified as six genera, which were in line with the results of 16 rDNA sequence analysis.
The frequency of each bacterial genus among the nematodes was 50%, 50%, 20%, 10%, 10%, and 10%, respectively. S. maltophilia and A. xylosoxidans subsp. xylosoxidans were the most abundant bacterial species. We found 80% of the strains isolated from P. massoniana contained S. maltophilia or A. xylosoxidans subsp. xylosoxidans. In the strains isolated from P. thunbergii, 66% of them contained S. maltophilia, while 33% of them contained A. xylosoxidans subsp. xylosoxidans.
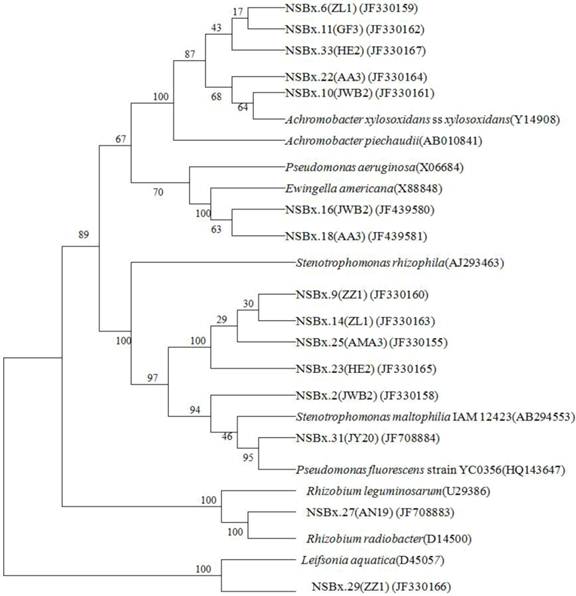
Relationship between the endophytic bacteria and virulence of PWNs
The correlation between the mortality of the inoculated P. thunbergii (Table 1) and the endophytic bacteria isolated from the ten PWN strains was analyzed. PWN strains with strong virulence contained at least one of the following two species: S. maltophilia and A. xylosoxidans subsp. xylosoxidans. Neither of the two bacterial species was found in PWN strains with weak virulence (causing 10% mortality). The PWN strains with moderate virulence (20% to 30% mortality) contained other bacteria species in addition to S. maltophilia and A. xylosoxidans subsp. xylosoxidans. Variation in virulence among different PWN strains was related to the composition of endophytic bacteria (Fig. 3). Nematode strains ZZ1 and AMA3 both contained S. maltophilia, but strain ZZ1 had an additional bacterial species, Leifsonia aquatica, which may account for the lower virulence of strain ZZ1 compared to strain AMA3. Differences in the proportions of endophytic bacterial species may also account for the variation in virulence between strains HE2 and JWB2, as well as the virulence difference between strains GF3 and AA3. Compared with HE2, strain JWB2 had an additional bacterium, Ewingella americana, and had 50% lower virulence than HE2. Similarly, despite the presence of A. xylosoxidans subsp. xylosoxidans in both GF3 and AA3, strain AA3 also had E. americana and had a 50% lower virulence than strain GF3. These observations suggested that endophytic bacteria may influence the virulence of PWN strains.
Identifications of endophytic bacteria of B. xylophilus based on Biochemical characteristics and carbon substrate oxidation pattern (BIOLOG). An identification is considered a good match if the similarity index is ≥0.500 for BIOLOG analysis.
| Bacterial strain | Host nematode | Physical and Biochemical tests | Biolog Identification | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram-stain reaction | Bacterial size (μm) | M.R. test | V.P. test | Indole test | Gelatin hydrolysis | Citrate utilization | H2S production | NH2 production | Nitrate reduction | PROB (%) | SIM | DIS | Speices | ||
| NSBx.2 | JWB2 | - | 0.28~0.4 × 0.5~1.2 | - | - | - | + | - | + | + | + | 100 | 0.776 | 3.37 | S. maltophilia |
| NSBx.6 | ZL1 | - | 0.27~0.35 × 0.4~1.1 | - | - | + | _ | + | - | + | + | 97 | 0.620 | 5.53 | Achromobacter xylosoxidans ss.xylosoxidans |
| NSBx.9 | ZZ1 | - | 0.28~0.4 × 0.5~1.2 | - | - | + | + | - | + | + | + | 100 | 0.776 | 3.37 | S. maltophilia |
| NSBx.10 | JWB2 | - | 0.27~0.35 × 0.4~1.1 | - | - | + | - | - | - | + | + | 100 | 0.764 | 3.53 | A. xylosoxidans ss.xylosoxidans |
| NSBx.11 | GF3 | - | 0.27~0.35 × 0.4~1.1 | - | - | + | - | - | - | + | + | 88 | 0.666 | 3.64 | A. xylosoxidans ss.xylosoxidans |
| NSBx.14 | ZL1 | - | 0.28~0.4 × 0.5~1.2 | - | - | + | + | - | + | + | + | 100 | 0.530 | 7.44 | S. maltophilia |
| NSBx.16 | JWB2 | - | 0.34~0.6 × 0.25~0.37 | + | + | - | + | + | + | + | + | 100 | 0.868 | 1.97 | Ewingella americana |
| NSBx.18 | AA3 | - | 0.34~0.6 × 0.25~0.37 | + | + | - | - | + | + | + | + | 100 | 0.781 | 3.29 | E. americana |
| NSBx.19 | HE2 | - | 0.27~0.35 × 0.4~1.1 | - | - | + | + | + | - | + | + | 93 | 0.730 | 3.23 | A. xylosoxidans ss.xylosoxidans |
| NSBx.22 | AA3 | - | 0.27~0.35 × 0.4~1.1 | - | - | + | - | + | - | + | + | 100 | 0.828 | 2.56 | A. xylosoxidans ss.xylosoxidans |
| NSBx.23 | HE2 | - | 0.28~0.4 × 0.5~1.2 | - | - | + | + | - | + | + | + | 100 | 0.569 | 6.76 | S. maltophilia |
| NSBx.25 | AMA3 | - | 0.28~0.4 × 0.5~1.2 | - | - | + | + | - | + | + | + | 97 | 0.629 | 5.00 | Stenotrophomonas maltophilia |
| NSBx.27 | AN19 | - | 0.8~0.36×0.33~0.23 | - | - | + | - | - | + | + | + | 100 | 0.587 | 6.41 | Rhizobium radiobacter |
| NSBx.29 | ZZ1 | + | 0.8~0.36 × 0.35~0.26 | - | - | - | + | - | - | - | - | 100 | 0.738 | 3.95 | Leifsonia aquatica |
| NSBx.31 | JY20 | - | 1.1~0.37 × 0.33~0.26 | - | - | + | + | - | + | + | + | 100 | 0.570 | 4.32 | Pseudomonas fluorescens |
Neighbor-joining phylogenetic trees of endophytic bacteria isolated from B. xylophilus based on 16S rDNA. Support values lower than 50% are not indicated. The number at each branch point is the percentage supported by bootstrap. Genbank accession numbers are shown in parentheses.
Virulence and endophytic bacterial species of different strains of B. xylophilus.
Dynamic features of endophytic microbes in different virulent B. xylophilus to utilize carbons
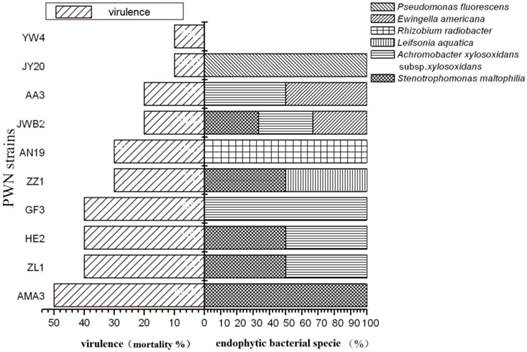
AWCDs of endophytic bacteria in highly virulent B. xylophilus ZL1 and low virulent B. xylophilus AA3 varied with time (Fig.4). In Figure 4, the ability of endophytic bacteria in B. xylophilus increased gradually along time. There was a difference between the total carbon amount which was utilized by AA3 and ZL1. The endophytic bacteria from AA3 showed a higher carbon utilization than ZL1. This indicated that the endophytic bacteria associated with carbon utilization in PWNs with different virulence were distinct in categories and abundance.
AWCD of endophytic microbial community from B. xylophilus with different virulence.
Diversity and evenness of microbial metabolism in B. xylophilus with different virulence
Diversity and evenness of microbes in two different PWN strains were shown in Table 4. The Shannon-Wiener index, Simpson index and Evenness index of microbes in ZL1 were significantly lower than those in AA3. The lower Simpson and Shannon-Wienner index in ZL1 suggests fewer bacterial strains were present in ZL1 than in AA3. The lower eveness index suggests that there may be dominance of some strains in ZL1 (they are not evenly represented). So maybe the highly virulent nematodes have fewer but better adapted bacteria.
Utilization of different carbon by microbes in B. xylophilus with different virulence
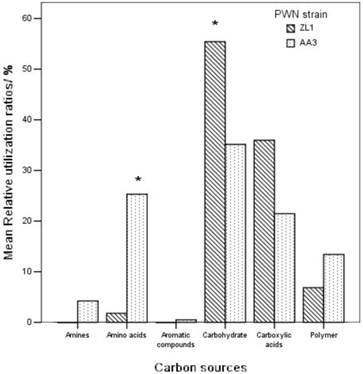
The 31 kinds of carbon on Biolog ECO plates were classified as 6 categories according to the chemical groups, they were aromatic (3 kinds), polymers (4 kinds), carbohydrate (7 kinds), carboxylic acids (9 kinds), amino acids (6 kinds) and amine (2 kinds) [33]. Figure 5 showed the utilization of carbon on Biolog ECO plates by the microbes in PWNs. Highly virulent B. xylophilus ZL1 had a high utilization rate of carbohydrate (55.4%) and carboxylic acids (35.9%). The utilization rate of carbohydrate by AA3 was 35.1%, which was much lower than ZL1. The utilization of other carbon was less than 10%. The low virulent B. xylophilus AA3 mainly took use of carbohydrate, carboxylic acids and amino acids, and the rate were 35.1%, 21.5% and 25.2%, respectively. Both ZL1 and AA3 had low utilization rates of aromatic, polymers and amine. This indicated that microbes from different PWNs had a different ability to use various carbons: ZL1 possessed a better utilization of carbohydrate and carboxylic acids, while AA3 was better at using amino acids.
Functional diversity of endophytic microbial community from B. xylophilus with different virulence.
| B. xylophilus | Diversity index | ||
|---|---|---|---|
| Shannon-Wiener | Simpson | Evenness | |
| Highly virulent ZL1 | 1.646±0.103b | 0.763±0.030b | 0.705±0.040b |
| Low virulent AA3 | 2.769±0.151a | 0.930±0.008a | 0.978±0.044ab |
The values followed by different letters are significantly different according to t-test.
Relative utilization ratios of six groups of carbon sources in Biolog Eco plate by the endophytic microbes from B. xylophilus.
Discussion
In previous work we demonstrated the presence of endophytic bacteria in the plant-parasitic nematode B. xylophilus [26]. In current study, we have investigated virulence variation among the ten strains of B. xylophilus and isolated the endophytic bacterial species from 10 PWN strains. The nematodes from P. massoniana showed a higher virulence than nematodes from other pines. The bacterial species of Stenotrophomonas, Achromobacter, Ewingella, Leifsonia, Rhizobium, and Pseudomonas were isolated. Most of the nematode strains with strong virulence contained either S. maltophilia or A. xylosoxidans subsp. xylosoxidans. S. maltophilia and A. xylosoxidans subsp. xylosoxidans were the dominant species.
S. maltophilia was most frequently isolated and was somewhat correlated with the virulence of B. xylophilus. S. maltophilia, commonly found in close association with plants [36], has an extraordinarily high hydrolytic potential and produce diverse proteases, chitinases, glucanases and lipases [37]. Chitinase plays a physiological role in the growth of many organisms. Glucanase belongs to the cellulase family and can be produced by PWNs to destroy cell wall and cell membrane of parenchymatous plant cells [38]. S. maltophilia was also found to be a dominant bacterial species in gut flora of Ips pini and can promote host intestinal absorption [39]. Another dominant species, A. xylosoxidans subsp. xylosoxidans was found in about 50% of the PWN strains. Ying-Ning et al. [40] reported that A. xylosoxidans F3B found in reed (Phragmites australis), Ipomoea aquatica, and vetiver (Vetiveria zizanioides (L.) Nash) help plants cope with pollutant stress and improves phytoremediation. Further investigation of the roles of S. maltophilia and A. xylosoxidans subsp. xylosoxidans in nematode-pine interactions is still necessary. Other species such as E. americana, L. aquatica, R. radiobacter, and P. fluorescens were also isolated from PWN strains. L. aquatica was originally isolated from Poa annua root galls [41]. However, little is known about the relationship between these bacteria and nematodes. Han et al. [42] had suggested that P. fluorescens carried by PWNs may be associated with nematode virulence due to its capability to causing browning of calluses and wilting of pine seedlings. In our study P. fluorescens was isolated in a lower rate; it's difficult to evaluate its effect on the virulence of PWN.
PWN virulence in the presence of either S. maltophilia or A. xylosoxidans varied in our study when more bacteria co-occurred in the nematode. Except the isolated bacteria mentioned above, there are still many bacteria presented in PWNs which are hard to be cultured or isolated. Thus, further investigations are still needed.
BIOLOG is widely used to study the metabolic diversity of heterotrophic microbes. In BIOLOG, variation of metabolic diversity is not only related to the group members and density of the inoculated microbes, but also to the physiological activity [32]. This research firstly analyzed the culturable bacteria in B. xylophilus with different virulence in terms of carbon metabolism using Biolog-Eco. The results showed that there was a big difference of AWCD and diversity between high and low virulent PWNs. As time went by, the AWCD and metabolic diversity index of AA3 were always higher than ZL1, indicating that the diversity of cultural bacteria in AA3 was higher than ZL1, which were in line with the identification of the isolated bacteria. This suggested that the parasitism of AA3 might be weaker than ZL1. The high metabolic diversity of AA3 might be due to its utilization of more carbon categories. Besides carbohydrate and carboxylic acids, AA3 also made a better use of amino acids than ZL1.
Further studies about utilization degree of carbohydrate by microbes in B. xylophilus found that, the higher virulent a PWN was, the better utilization of carbohydrates it made. PWNs ingest parenchyma cells in trees, and the components of parenchyma cells are mainly cellulose, a few semicellulose and pectin, which are all belong to carbohydrate. Endophytic microbes in highly virulent B. xylophilus have a relative high utilization rate of carbohydrate, suggesting that they could take use of carbohydrate in cell walls faster and better. This might increase its parasitic adaptability in trees and the damage levels to pines, which might be the reason why these nematodes have high virulence. Endophytic microbes in low virulent B. xylophilus have a low utilization of carbohydrate, which might be the reason for its low virulence. Meanwhile, low virulent B. xylophilus can use all kinds of tested carbon source, indicating that its specificity to the nutrients of hosts may be not very strict in trees.
The research showed that endophytic bacteria existed in B. xylophilus from different pines and areas; the species and carbon metabolism of cultural bacteria in PWNs were related to the virulence of B. xylophilus. This provides reference for studying the effects of endophytic bacteria community in B. xylophilus on the growth and development, ecological adaptability and virulence of the hosts. As Biolog only showed the activity of fast-growing or eutrophic microbes instead of slowly-growing or uncultured microbes [43, 44], further studies about the relationship between the diversity of uncultured bacteria in PWNs and the virulence of B. xylophilus are still needed.
Acknowledgements
This work was supported by the National Nature Science Fund of China (No.31270683), the Project for Natural Science Research of Jiangsu University in China (No.11KJA220002), the Major State Basic Research Development Program of China, 973 Program (2009CB119200), National key Technology R&D Program (2012BAD19B0703) and a Project Founded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). We are grateful to Dr. Hui Sun for help in preparation of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Yang BJ, Pan HY, Tang J. et al. Pine Wood Nematode Disease. Beijing, China: Forestry Publishing House. 2003:45-48
2. Zhao BG, Futai K, Sutherland JR, Takeuchi Y. Pine wilt disease. Japan: Springer. 2008
3. Vicente C, Espada M, Vieira P, Mota M. Pine Wilt Disease: a threat to European forestry. Eur J Plant Pathol. 2012;133:89-99
4. Oku H, Shiraishi T, Ouchi S. Pine wilt toxin, the metabolite of a bacterium associated with a nematode. Naturwissenschaften. 1980;67:198-199
5. Kawazu K, Kaneko N. Asepsis of the pine wood nematode isolate OKD-3 causes it to lose its pathogenicity. Jpn J Nematol. 1997;27:76-80
6. Kawazu K. Isolation of pine wilting bacteria accompanying pine wood nematode, Bursaphelenchus xylophilus, and their toxic metabolite. Scientific Reports of the Faculty of Agriculture, Okayama University. 1998;87:1-7
7. Zhao BG, Li L. Observation of Wilting Symptoms Caused by Cell Free Filtration of the Culture in Pseudomonas fluorescence. Acta Agric Univ Jiangxiensis. 2008;30:575-580
8. Zhao BG, Guo DS, Gao R. et al. A Preliminary Study on the Relationship Between the Bacterium Isolate B619 and Pine Wilt Disease. J Nanjing For Univ. 2004;24:72-74
9. Oku H, Shiraishi T, Kurozumi S. Participation of toxin in wilting of Japanese pine caused by a nematode. Naturwissenschaften. 1979;66:210
10. Zhao BG, Lin F. Mutualistic symbiosis between Bursaphelenchus xylophilus and bacteria of the genus Pseudomonas. For Pathol. 2005;35:339-345
11. Tan JJ, Wang XR, Feng ZX. A preliminary study on the relationship between the bacterium accompanying Bursaphelenchus xylophilus and pine wilt disease. Plant Quarantine. 2001;15:326-328
12. Hong YD; Cao Y; Zhao BG. et al. Studies on Identification and Toxicity of Bacteria Carried by Pine Wood Nematodes. J Nanjing For Univ. 2002;26:37-40
13. Wang HL, Han SF, Zhao BG. Distribution and pathogenicitiy of bacteria carried by pine wood nematode in epidemic regions and hosts. J Nanjing For Univ. 2004;26:48-53
14. Zhao BG; Guo DS. Isolation and pathogenicity of a bacterium strain carried by pine wood nematode. J Nanjing For Univ. 2004;26:57-60
15. Kwon H, Choi G, Choi Y. et al. Suppression of pine wilt disease by an antibacterial agent, oxolinic acid. Pest Manag Sci. 2010;66:634-639
16. Proenca DN, Francisco R, Santos CV. et al. Diversity of bacteria associated with Bursaphelenchus xylophilus and other nematodes isolated from Pinus pinaster trees with pine wilt disease. PLoS ONE. 2010;5(12):e15191
17. Vicente CSL, Nascimento F, Espada M. et al. Bacteria associated with the pinewood nematode Bursaphelenchus xylophilus collected in Portugal. Antonie van Leeuwenhoek. 2011;100:477-481
18. Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512-519
19. Vandekerckhove T, Coomans A, Cornelis K. et al. Use of the Verrucomicrobia-specific probe EUB338-III and fluorescent in situ hybridization for detection of "Candidatus xiphinematobacter" cells in nematode hosts. Appl Environ Microbiol. 2002;68(6):3121-3125
20. Supriya S, Khanolkar-Gaitonde G, Kenneth R. et al. Isolation of Bacteria Other than Helicobacter pylori from Stomachs of Squirrel Monkeys (Saimiri spp.) with Gastritis. Digest Dis Sci. 2000;45(2):272-280
21. Noguchi T, Hwang DF, Arakawa O.. et al. Vibrio alginolyticus, a tetrodotoxin-producing bacterium, in the intestines of the fish Fugu vermicularis vermicularis. Marine Biology. 1987;94(4):625-630
22. Forst S, Dowds B, Boemare N. et al. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu Rev Microbiol. 1997;51:47-72
23. Nour SM, Lawrence JR, Zhu H, Swerhone GD, Welsh M. et al. Bacteria associated with cysts of the soybean cyst nematode (Heterodera glycines). Appl Environ Microbiol. 2003;69:607-615
24. Haegeman A, Vanholme B, Jacob J. et al. An endosymbiotic bacterium in a plant-parasitic nematode: member of a new Wolbachia supergroup. Int J Parasitol. 2009;39:1045-1054
25. Tian XL, Zhang QL, Chen GH. et al. Diversity of bacteria associated with pine wood nematode revealed by metagenome. Acta Microbiologica Sinica. 2010;50:909-916
26. Yuan WM, Wu XQ, Ye JR. et al. Observation by transmission electron microscope and identification of endophytic bacteria isolated from Bursaphelenchus xylophilus and B. mucronatus. Acta Microbiolo Sin. 2011;51:1071-1077
27. Mamiya Y. Pathology of the pine wilt disease caused by Bursaphelenchus xylophilus. Annu Rev Phytopathol. 1983;21:201-220
28. John GH. Bergey's Manual of Determinative Bacteriology. Baltimore: Williams &Wilkins. 1994
29. Dong XZ, Cai MY. Identification system manual of common bacteria. Beijing: Science Press. 2001
30. Oka N, Hartel PG. Finlay-Moore O, et al. Misidentification of soil bacteria by fatty acid methyl ester (FAME) and BIOLOG analyses. Biol Fertil Soils. 2000;32:256-258
31. Weisburg WG, Barns SM, Pelletier DA. et al. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697-703
32. Garland J L, MillsA L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole carbon source utilization. Appl Environ Microbiol. 1991;57:2351- 2359
33. Hu JL, Lin XG, Wang JH. et al. Microbial functional diversity, metabolic quotient and invertase activity of a sandy loam soil as affected by long-term application of organic amendment and mineral fertilizer. J Soils Sediments. 2011;11:271-280
34. Choi K, Dobbs F C. Comparison of two kinds of Biolog microplates( GN and ECO) in their ability to distinguish among aquatic microbial communities. J Microbiol Meth. 1999;36(3):203- 213
35. Ling Q, Bao J, Li R, Tao Y. et al. Analysis of Carbon Metabolism Diversity Characters of Air Microbes in Huangshan Scenic Spot Using Biolog-Eco. Method. 2012;20(1):56-63
36. Ryan RP, Monchy S, Cardinale M. et al. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol. 2009;7:514-525
37. Zhang Z, Yuen GY. Effects of culture fluids and preinduction of chitinase production on biocontrol of bipolaris leaf spot by Stenotrophomonas maltophilia C3. Biol Control. 2000;18:277-286
38. Ichhihara Y, Fukuda K, Suzuki K. Migration of pine wood nematode, Bursaphelenchus xylophilus, in Japanese black pine seedling, Pinus thunbergii, interaction to early symptom of pine wilt disease. Sustainability of Pine forests in relation to pine wilt and decline proceedings of international symposium. Tokyo. 1998
39. Delalibera JI, Vasanthakumar A, Burwitz B. et al. Composition of the bacterial community in the gut of the pine engraver, Ips pini (Say) (Coloptera) colonizing red pine. Symbiosis. 2007;43:97-104
40. Ho YN, Shih CH, Hsiao SC, Huang CC. A novel endophytic bacterium, Achromobacter xylosoxidans, helps plants against pollutant stress and improves phytoremediation. Biosci Bioeng. 2009;108:94
41. Evtushenko LI, Dorofeeva LV, Subbotin SA. et al. Leifsonia poae gen. nov, sp.nov, isolated from nematode galls on Poa annua, and reclassification of 'Corynebacterium aquaticum' Leifson 1962 as Leifsonia aquatica (ex Leifson 1962) gen. nov, nom. rev, comb. nov. and Clavibacter xyli Davis et al. 1984 with two subspecies as Leifsonia xyli (Davis et al. 1984) gen. nov, comb. nov. Int J Syst Evol Microbiol. 2000;50(1):371-380
42. Han ZM, Hong YD, Zhao BG. A study on pathogenicity of bacteria carried by pinewood nematode. J Phytopathol. 2003;151:683-689
43. Li C G, Li X M, Wang J G. Effect of s oybean continuous cropping on bulk and rhizosphere soil microbial community function. Acta Ecologica Sinica. 2006;26(4):144-150
44. Konopka A, Oliver L, Turco R F. The use of carbon substrate utilization patterns in environmental and ecol ogicalmicrobiology. Microbiol Ecol. 1998;35(2):103-105
Author contact
![]() Corresponding author: xqwu_njfucn.
Corresponding author: xqwu_njfucn.

 Global reach, higher impact
Global reach, higher impact