10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(3):256-266. doi:10.7150/ijbs.5898 This issue Cite
Research Paper
ATF4 Promotes β-Catenin Expression and Osteoblastic Differentiation of Bone Marrow Mesenchymal Stem Cells
1. Department of Medicine, University of Pittsburgh, Pittsburgh, PA 15240, USA.
2. College of Life Sciences, Nankai University, Tianjin 300071, China.
3. Department of Biochemistry, Rush University Medical Center, Chicago, IL 60612, USA.
4. Department of Surgery, University of Pittsburgh, Pittsburgh, PA 15240, USA.
Received 2013-1-17; Accepted 2013-2-21; Published 2013-2-27
Abstract
Bone marrow mesenchymal stem cells (MSCs) can differentiate into multiple cell types including osteoblasts. How this differentiation process is controlled, however, is not completely understood. Here we show that activating transcription factor 4 (ATF4) plays a critical role in promoting bone marrow MSC differentiation towards the osteoblast lineage. Ablation of the Atf4 gene blocked the formation of osteoprogenitors and inhibited osteoblast differentiation without affecting the expansion and formation of MSCs in bone marrow cultures. Loss of ATF4 dramatically reduced the level of β-catenin protein in MSCs in vitro and in osteoblasts/osteoprogenitors located on trabecular and calvarial surfaces. Loss of ATF4 did not decrease the expression of major canonical Wnt/β-catenin signaling components such as Wnt3a, Wnt7b, Wnt10b, Lrp5, and Lrp6 in MSCs. Furthermore, shRNA knockdown of ATF4 expression decreased the level of β-catenin protein in MC-4 preosteoblasts. In contrast, overexpression of ATF4 increased β-catenin protein levels in MC-4 cells. Finally, ATF4 and β-catenin formed a protein-protein complex in COS-7 cells coexpressing both factors or in MC-4 preosteoblastic cells. This study establishes a new role of ATF4 in controlling the β-catenin protein levels and MSC differentiation towards the osteoblast lineage.
Keywords: ATF4, β-catenin, MSC, osteoblast, bone.
Introduction
Osteoblasts, the bone-forming cells, originate from multipotential mesenchymal stem cells (MSCs), which are primarily present in bone marrow (12). MSCs proliferate and differentiate into osteoprogenitors and subsequently osteoblasts, which synthesize and deposit the mineralizing extracellular matrix of bone. MSC differentiation towards the osteoblast lineage is controlled by specific key transcription factors (16). One is Runx2, which is a runt domain-containing transcription factor identified as a transcriptional activator of osteoblast differentiation and master gene for bone development (3, 9, 14, 15, 17, 19, 20). Runx2 is expressed in mesenchymal condensations during early development at E 11.5 and plays a critical role in control of MSC differentiation towards the osteoblast lineage (9). Runx2 knockout mice die at birth and lack mature osteoblasts (15, 17). Another key regulator of osteoblast differentiation is Osterix (Osx) (18, 32, 34). The facts that Runx2 is normally expressed in Osx null mice and that Osx is not expressed in Runx2 null mice suggest that Osx acts as a downstream of Runx2 (18). Osx controls differentiation of preosteoblasts to mature osteoblasts, which express high levels of osteocalcin (18, 32, 34). Similar to Runx2 null mice, Osx knockout mice die at birth and lack both skeletal ossification and mature osteoblasts (18).
The canonical WNT/β-catenin signaling is the most important pathway for control of bone mass in humans and vertebrates (5, 6). Mesenchymal stem cells lacking β-catenin do not differentiate into osteoblasts, indicating that expression of β-catenin is critical for osteoblast differentiation (7). The level of β-catenin protein is largely controlled by the Wnt proteins, a family of at least 19 members in humans and vertebrates (22). In the absence of Wnt ligands, a complex that contains glycogen synthase kinase 3β (GSK-3β), adenomatous polyposis coli (APC) and axin binds to β-catenin, which results in β-catenin phosphorylation, ubiquitination and degradation by proteasome. In contrast, binding of Wnt proteins to the Frizzled receptors and co-receptors such as Lipoprotein Receptor-related Protein (LRP)-5 and LRP-6 inhibits the formation of the GSK2-APC-Axin complex. As a result, β-catenin cannot be phosphorylated and degraded by proteasome, which allows β-catenin accumulation and translocation into the nucleus, where it interacts with TCF/LEF and activates transcription of downstream target genes such as those encoding c-Myc and cyclin D1 (2, 22). β-Catenin regulation is complex as other proteins can also influence β-catenin stability and activity, thereby allowing the integration of inputs from different signaling pathways into the β-catenin signaling (1, 2, 22, 35).
Activating transcription factor 4 (ATF4) is a basic leucine-zipper transcription factor of the ATF/CREB protein family. ATF4 is critical for bone homeostasis and the ATF4 knockout mice have dramatically reduced bone mass and bone formation rate (26). ATF4 is critical for osteoblast proliferation and survival (28, 33). ATF4 promotes osteoblast-specific osteocalcin gene expression and osteoblast differentiation (25, 26). ATF4 mediates parathyroid hormone (PTH)-induced osteoblast differentiation and bone formation (13, 28, 29). Interestingly, ATF4 also promotes osteoclast differentiation directly or through indirect up-regulation of RANKL expression in osteoblasts (4, 10, 11). These studies demonstrate a critical role of ATF4 in promotion of osteoblast and osteoclast differentiation. However, potential role of ATF4 in regulation of MSC differentiation is not known.
Materials and Methods
Reagents
Tissue culture media and fetal bovine serum were obtained from HyClone (Logan, UT). Other reagents were obtained from the following sources: Antibody against ATF4 and horseradish peroxidase-conjugated goat anti-rabbit IgG from Santa Cruz (Santa Cruz, CA), antibody against β-catenin from Abcam (Cambridge, MA), mouse monoclonal antibody against β-actin, alizarin red (AR-S), L-ascorbic acid, and β-glycerophosphate from Sigma (St Louis, MO). All other chemicals were of analytical grade.
Atf4-deficient mice
Breeding pairs of Atf4 heterozygous mice (Swiss black) were described previously (29) and used to generate ATF4 wild-type (WT) and knockout (KO) mice for this study. Six- to eight-week-old mice were sacrificed for bone marrow cells. All research protocols were approved by the Institutional Animal Care and Use Committee of the Rush University Medical Center.
Colony forming unit-fibroblast (CFU-F) assay and colony forming unit-osteoblast (CFU-OB) assay
The CFU-F assay was performed using the Mesencult Proliferation Kit (Mouse) (Stemcell Technologies) for the expansion and enumeration of the mesenchymal stem cells (MSCs) as previously described (31). Briefly, 1x106 bone marrow nucleated cells per 35-mm dish were seeded and cultured at 37°C in 5% CO2 for 10 days, followed by Giemsa staining. The numbers of CFU-Fs were counted under a microscope. CFU-OB assay was performed as previously described (23). Briefly, 1x106 bone marrow nucleated cells per 60-mm dish were seeded and cultured for the indicated days in differentiation medium (α-MEM containing 10% FBS, 1% penicillin/streptomycin, 50 µg/ml L-ascorbic acid and 2.0 mM β-glycerophosphate). Media were changed every another day. Alizarin red staining was used to identify the colonies containing mineralized bone matrix, which were designated as CFU-OB colonies.
Alkaline Phosphatase (ALP) staining and ALP activity assay
ALP staining of the CFU-OB cultures was performed using an ALP staining kit from Sigma (St. Louis, MO) according to the manufacturer's instructions. Cells were fixed by 10% formalin for 1h at room temperature before the staining. ALP assay was performed as previously described (23). Briefly, BMSCs were differentiated for 7 days and harvested in 1x Passive Buffer (Promega, Madison, WI). Lysates were clarified by centrifugation (20 min, 13,000 x g, 4°C). Five µl of cell extracts were added to each well (96-well plate) containing 150 µl p-nitrophenyl phosphate at 37 oC for 10-60 min depending on the ALP activity in the extracts. ALP activity was determined by absorbance measurement at 405 nm on a 96-well plate reader. ALP activity was normalized to total protein.
RNA isolation, reverse transcription (RT), and quantitative real-time PCR (qPCR)
RNA isolation, RT, and quantitative real-time PCR were performed to measure the relative mRNA levels using SYBR Green kit (Bio-Rad Laboratories Inc.) as previously described (29). Samples were normalized to Gapdh expression. The DNA sequences of mouse primers used in this study are summarized in Table 1.
Western blot analysis
Western blot analysis was performed as previously described (30). Tibias were frozen in liquid nitrogen and ground into powder using a mortar and pestle. Whole cells or bone tissue powder were extracted by RIPA buffer on ice as described (33) and total proteins were separated by electrophoresis on 8% SDS polyacrylamide gels. The proteins were transferred electrophoretically to nitrocellulose membranes and staining with Ponceau Red to ensure that comparable amounts of proteins were loaded and the transfer was efficient. The membranes were blocked with 5% nonfat milk in TBST for 1h at room temperature and immunobloted with β-catenin (Abcam), ATF4 (Santa Cruz) or c-Myc (Abcam) antibodies. β-Actin immunoblot was used as internal control.
Immunohistochemistry (IHC)
Tibiae were fixed in 10% formalin at 4 oC for 24h, decalcified in 10% EDTA (pH 7.4) for 10-14 days, and embedded in paraffin. Five-μm sections of tibiae were immunohistochemically stained with an antibody against β-catenin or control IgG using the EnVision+System-HRP (DAB) kit (Dako North America, Inc) as described previously (28, 31).
Statistical analysis
Data was analyzed with a GraphPad Prism software (4.0). A one-way ANOVA analysis was used followed by the Tukey test. Students' t test was used to test for differences between two groups of data as needed. Results were expressed as means ± standard deviation (S.D.). Differences with a P <0.05 was considered as statistically significant.
Results
Atf4 ablation severely impairs the ability of bone marrow MSCs to differentiate into osteoprogenitors without affecting the expansion and formation of MSCs in vitro
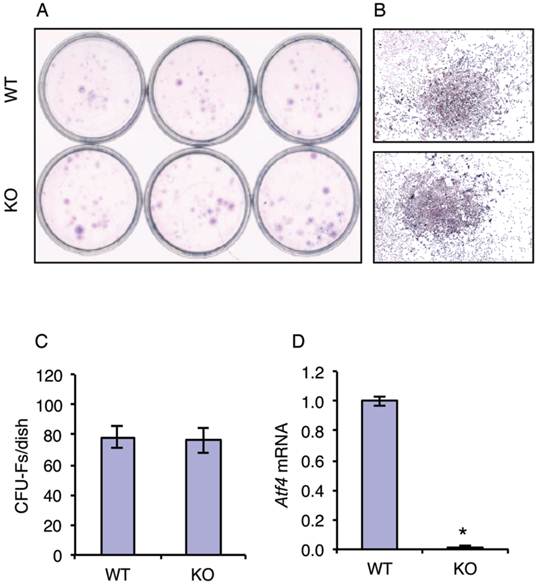
Although the role of ATF4 in regulation of terminal osteoblast differentiation and bone formation are well established, its potential role in early osteoblast differentiation (i.e., from MSCs to osteoprogenitors) has not been addressed. To test whether ATF4 plays a role in regulation of MSC differentiation towards the osteoblast lineage, we investigated the effect of ATF4 ablation on the formation of the colony-forming unit-fibroblast (CFU-F) in bone marrow MSC cultures from WT and KO mice. Results showed that inactivation of the Atf4 gene did not alter the number of CFU-Fs from bone marrow (Figure 1, A-C). Results from qPCR analysis confirmed that Atf4 mRNA expression was undetectable in the MSC cultures from KO mice (Figure 1D). These results suggest that ATF4 is not critical for the expansion and formation of MSCs.
mouse qPCR primers.
| Gene name | 5' primer | 3' primer |
|---|---|---|
| Atf4 | GAGCTTCCTGAACAGCGAAGTG | TGGCCACCTCCAGATAGTCATC |
| β-Catenin | GCGGAGACGGAGCACGGT | 5CCAAGTAAGACTGCTGCTGCCAGT |
| Gapdh | CAGTGCCAGCCTCGTCCCGTAGA | CTGCAAATGGCAGCCCTGGTGAC |
| Lrp5 | GGGTCCACAAGGTCAAGGC | GCACCCTCCATTTCCATCC |
| Lrp6 | GCCCACTACTCCCTGAATGCTG | TGTGGATAGGAAGGATGATGTCAGG |
| Wnt3a | GCACCACCGTCAGCAACAGC | CACAGGAGCGTGTCACTGC |
| Wnt5a | TCCCCCAAGGCTTAACCC | CGGTCCCCAAAGCCACTC |
| Wnt7b | CGTGTTTCTCTGCTTTGGCGTC | GGGTCGGCTCTGGCAGAT |
| Wnt10b | CGCTGGAACTGCTCGGCAC | ATGACCCCAGCAGCCAGCA |
ATF4 ablation does not affect the expansion and formation of bone marrow MSCs. (A-C) CFU-F assay. 1x106 bone marrow nucleated cells from one-month-old WT and KO mice were seeded in 35-mm culture dishes and cultured using the Mesencult Proliferation Kit (Mouse) for 10d, followed by Giemsa staining. Representative CFU-Fs from WT and KO MSC cultures are shown (B), magnification: 40X. The numbers of CFU-Fs were counted under a microscope (C). (D) 1x106 bone marrow nucleated cells from one-month-old WT and KO mice were seeded in 35-mm culture dishes and cultured using the Mesencult Proliferation Kit (Mouse) for 10d, followed by qPCR analysis. Atf4 mRNA was normalized to Gapdh mRNA. *P <0.05 (versus WT).
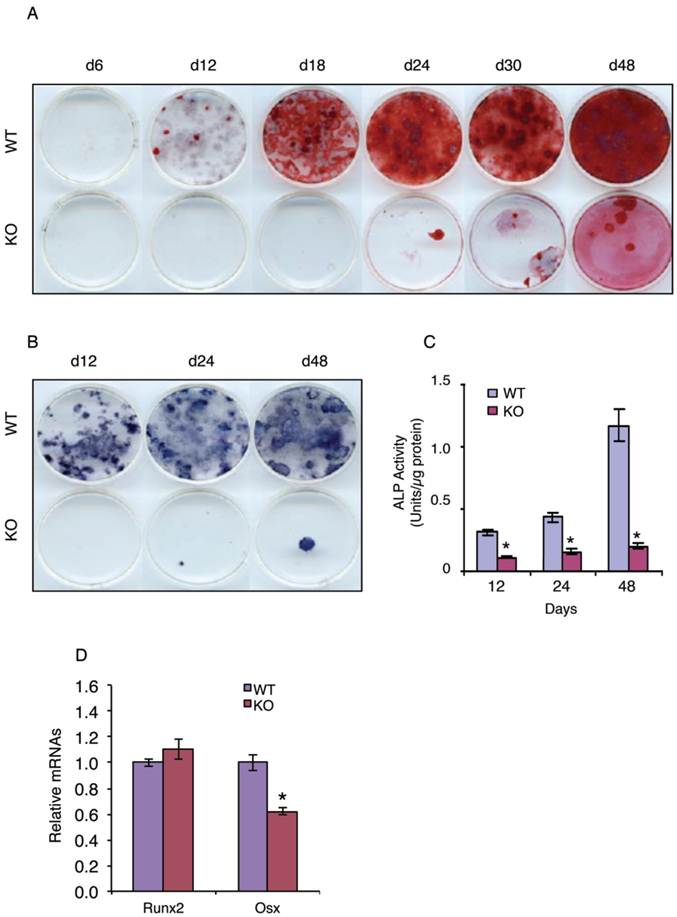
We next tested whether ATF4 plays a role in regulation of MSC differentiation towards the osteoblastic lineage by determining the effect of ATF4 ablation on the development and formation of osteoprogenitors in primary MSC cultures from WT and KO bone marrows. The results revealed that the ability of KO MSCs to form the colony-forming unit-osteoblasts (CFU-OBs) was dramatically decreased and largely delayed compared to that of WT MSCs (Figure 2A). Because each CFU-OB colony is derived from a single osteoblast progenitor, the number of CFU-OB colonies reflects the number of mesenchymal progenitors present in the original bone marrow isolate that are capable of differentiating into osteoblast. Thus, these results suggest that deletion of the Atf4 gene impairs the formation of osteoprogenitors in bone marrow. To further support this conclusion, the number of ALP-positive colonies and ALP activity were dramatically reduced in bone marrow MSCs cultures from KO mice compared to those from WT mice (Figure 2, B and C). Results from qPCR analysis revealed that Osx but not Runx2 mRNA expression was significantly decreased in the KO versus WT MSC cultures (Figure 2D).
Atf4 ablation reduces β-catenin expression in bone marrow MSCs
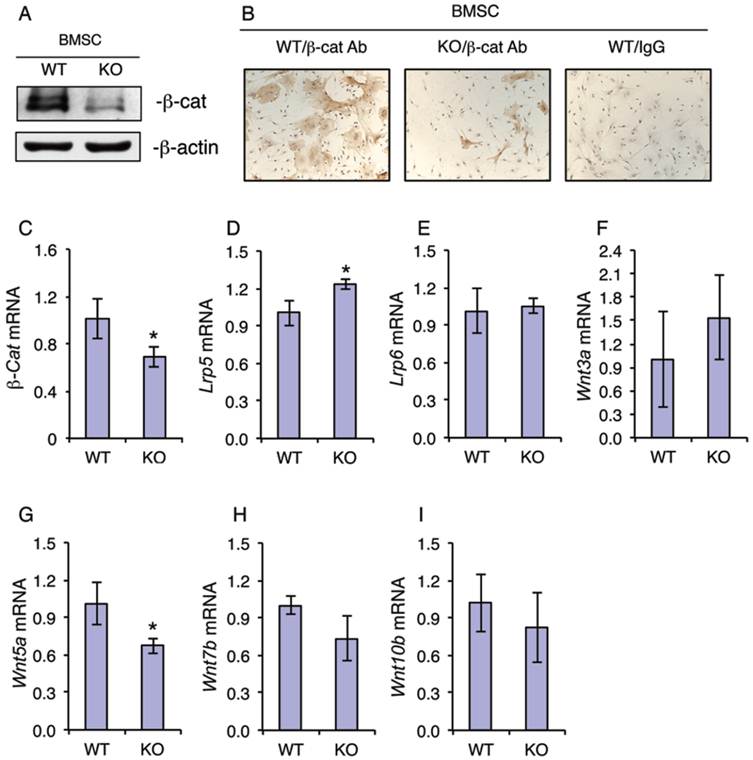
Because β-catenin is known to play a critical role in regulation of the osteoblastic differentiation of MSCs (7), we next investigated whether ATF4 ablation alters the expression of β-catenin in MSCs. Western blot analysis revealed that the level of β-catenin protein was dramatically decreased in KO MSCs compared to that from WT MSCs (Figure 3A). Immunohistochemical (IHC) staining showed that there are many β-catenin-positive cells in WT MSC cultures, which was markedly decreased in KO MSC cultures (Figure 3B). It is interesting to note that the cytoplasm of β-catenin-positive MSCs from WT mice was highly spread. However, the level of β-catenin mRNA was only slightly decreased in KO versus WT MSC cultures (Figure 3C). Since the level of β-catenin protein is largely controlled by Wnts, we next determined if loss of ATF4 affects the expression of major Wnts and their co-receptors. Results showed that the mRNA levels of major Wnt/β-catenin signaling components including Wnt3a, Wnt7b, Wnt10b, Lrp5 and Lrp6 were not decreased by the loss of ATF4 in the MSC cultures (Figure 3, D-F, H and I). Note: the level of Wnt5a mRNA was slightly reduced in KO relative to WT MSC cultures (Figure 3G).
Atf4 ablation reduces β-catenin protein expression in osteoblasts/osteoprogenitors located on trabecular and calvarial bone surfaces
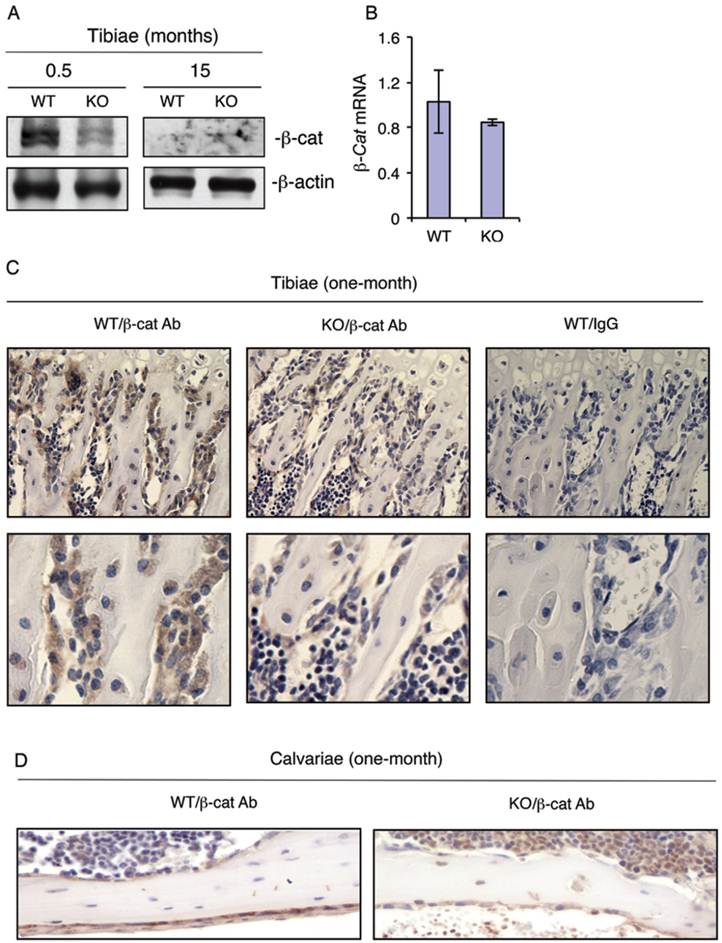
To determine whether loss of ATF4 decreases the level of β-catenin expression in bone, we performed Western blot analysis using protein extracts from tibiae of WT and KO mice. Results showed that loss of ATF4 dramatically decreased the levels of β-catenin protein in 2-week-old KO versus WT tibiae (Figure 4A). β-Catenin protein level was low but not reduced by the loss of ATF4 in tibiae of 15-month-old mice (Figure 4A). The level of β-catenin mRNA was not decreased in 2-week-old KO relative to WT tibiae (Figure 4B). To determine whether loss of ATF4 decreases the level of β-catenin protein in osteoblasts/osteoprogenitors in vivo, we performed IHC staining of tibial and calvarial sections from one-month-old WT and KO mice. Results revealed that β-catenin protein was strongly detected in osteoblasts/osteoprogenitors on trabecular bone surfaces of WT tibiae, which was markedly decreased in osteoblasts/osteoprogenitors of KO tibiae (Figure 4C). Loss of ATF4 also decreased the level of β-catenin protein in calvarial osteoblasts/osteoprogenitors in vivo (Figure 4D). The IHC staining was highly specific since no signal was detected in the IgG control group (Figure 4C).
shRNA knockdown of Atf4 expression reduces and overexpression of ATF4 increases β-catenin protein levels and β-catenin-dependent transcriptional activity and ATF4 and β-catenin form a protein-protein complex
To further determine the role of ATF4 in regulation of β-catenin protein levels in osteoblasts, we knocked down the expression of ATF4 in MC-4 cells, a well-known preosteoblastic cell line (21, 24). Results showed that knocking down ATF4 expression significantly reduced the level of β-catenin protein in MC4 cells (Figure 5A). In contrast, overexpression of ATF4 dose-dependently increased the level of β-catenin protein in MC-4 cells without significantly affecting the level of β-catenin mRNA (Figure 5, B and C). Overexpression of ATF4 dramatically increased the β-catenin-dependent TOPflash/FOPflash ratio in vitro (Figure 5D). Finally, Co-immuonoprecipitation assays showed that ATF4 and β-catenin formed a protein-protein complex in COS-7 cells coexpressing both factors (Figure 5E, top) or MC-4 cells (Figure 5E, bottom) which express high levels of endogenous ATF4 and β-catenin proteins.
Discussion
ATF4 is critical for postnatal bone mass accrual by promotion of osteoblast functions and bone formation. Studies from this and other groups demonstrate that ATF4 promotes osteoblast differentiation through at least three distinct molecular mechanisms. First, ATF4, through interactions with Runx2, a master regulator of osteoblast formation and differentiation, directly activates the expression of the osteocalcin gene, an osteoblast differentiation marker gene, and promotes bone formation (8, 25, 27). Because of the critical role of Runx2 in mesenchymal condensations during early development, it is possible that the ATF4-Runx2 interactions also play a role in control of MSC differentiation towards the osteoblast lineage. Future studies will test this exciting possibility. Second, our recent studies demonstrate that ATF4 is an upstream transcriptional activator of the Osx gene and increases Osx expression by interactions with an ATF4-binding site of the proximal Osx gene promoter (28). Interestingly, the PTH-dependent increase in osteoblast differentiation in bone is correlated with ATF4-dependent up-regulation of Osx (28). PTH induction of Osx gene expression in osteoblasts requires the ATF4-binding site and is associated with increased binding of ATF4 to a chromatin fragment of the Osx promoter (28). Finally, results from the present study demonstrate that ATF4 additionally promotes osteoblast differentiation by favoring MSC differentiation towards the osteoblast lineage by, at least in part, up-regulation of the level of β-catenin protein. Thus, these results add a new layer of control to osteoblast differentiation from MSCs by ATF4 and β-catenin.
ATF4 ablation blocks bone marrow MSC differentiation towards the osteoblast lineage. (A-C) CFU-OB assay. 1x106 bone marrow nucleated cells from WT and KO mice were seeded in 60-mm culture dishes in osteoblast differentiation media (α-MEM containing 10% FBS, 1% PS, 50 µg/ml L-ascorbic acid and 2.0 mM β-glycerophosphate). Media were changed every 2d. At indicated times, cultures were used for Alizarin Red staining (A), which was used to identify the colonies containing mineralized bone matrix, which were designated as CFU-osteoblast (CFU-OB) colonies. (B and C) ALP staining and ALP activity assay. 1x106 bone marrow nucleated cells from WT and KO mice were differentiated as in (A), followed by ALP staining (B) or ALP activity assay as described in Materials and Methods. Bars represent means ± S.D. from three independent experiments. *P <0.05 (versus WT). (D) 1x106 bone marrow nucleated cells from one-month-old WT and KO mice were seeded in 35-mm culture dishes and differentiated for 10d, followed by qPCR analysis. Runx2 and Osx mRNAs were normalized to Gapdh mRNA. *P <0.05 (versus WT).
ATF4 ablation decreases the level of β-catenin protein in primary bone marrow MSCs. (A-I) MSCs from WT and KO mice were cultured in proliferation media (α-MEM containing 10% FBS and 1% PS), followed by Western blot analysis (A), IHC staining (B) or qPCR analysis (C-I). β-Actin was used as a loading control for Western blot analysis. mRNA expression was normalized to Gapdh mRNA for qPCR analysis. Bars represent means ± S.D. from three independent experiments. *P <0.05 (versus WT).
ATF4 ablation decreases the levels of β-catenin protein in osteoblasts/osteoprogenitors located on trabecular and calvarial bone surfaces. (A and B) Total RNAs and protein extracts were prepared from tibiae of WT and KO mice of the indicated ages and used for Western blot (A) and qPCR (B) analyses for β-catenin. (C and D) IHC staining. Tibial (C) and calvarial (D) sections from one-month-old WT and KO mice were used for IHC staining using antibody against β-catenin or control IgG. β-Catenin-positive osteoblasts/osteoprogenitors on trabecular and calvarial surfaces were stained brown. β-Catenin-negative cells were stained blue. Original magnification, x100 (top), x200 (bottom).
shRNA knockdown of ATF4 expression decreases and overexpression of ATF4 increases β-catenin protein levels and β-catenin-dependent transcriptional activity and ATF4 and β-catenin form a protein-protein complex. (A) ATF4 shRNA knockdown. MC4 cells were transfected with ATF4 (20, 40, and 80 nM) or control siRNA (80 nM) as described previously (29). The ATF4 siRNA and control transfectants were analyzed by Western blot analysis with antibodies recognizing ATF4, β-catenin or β-actin. (B and C) MC-4 cells were co-transfected with expression vectors for ATF4 and Myc-tagged β-catenin, followed by Western blot analysis with antibodies recognizing ATF4, c-Myc or β-actin or qPCR for β-catenin mRNA. β-actin was used as a loading control for Western blots. mRNA expression was normalized to Gapdh mRNA for qPCR analysis. (D) COS-7 cells were transfected with TOPflash or FOPflash reporter plasmids, pRL-SV40 (for normalization), and expression vector for β-catenin in the presence and absence of expression vector for ATF4, followed by dual luciferase assays. Bars represent means ± S.D. from three independent experiments. The amount of the plasmid DNAs was balanced as necessary with a control plasmid expressing β-galactosidase (β-gal) such that the total amount was constant in each group. *P <0.05 (versus FOPflash). (E) IP assays: Whole cell extracts from COS-7 cells coexpressing ATF4 and Myc-tagged β-catenin (top) or MC-4 cells (bottom) were immunoprecipitated with an anti-ATF4 antibody or control IgG, followed by Western blot analysis for c-Myc (top) or β-catenin (bottom) as described previously (25, 30).
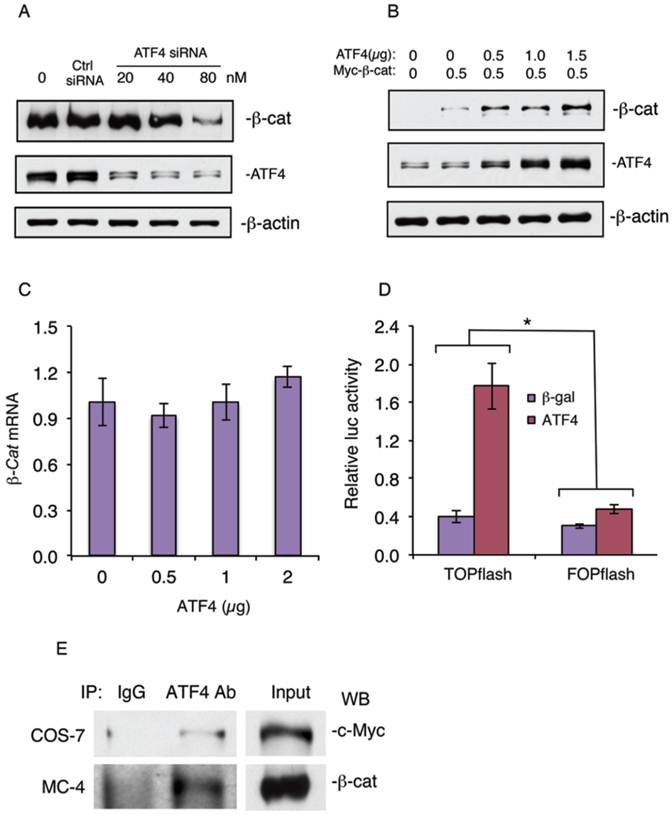
The fact that loss of ATF4 dramatically decreased the level of β-catenin protein with a minimal effect on the level of β-catenin mRNA suggests that this regulation is mainly through a post-transcriptional mechanism. In further support of this notion, we found that ATF4 and β-catenin forms a protein-protein complex in MC-4 preosteoblasts or when coexpressed in COS-7. ATF4 does not seem to increases the β-catenin protein levels by activating upstream components of the canonical WNT/β-catenin pathways because the expression levels of major Wnts (Wnt3a, Wnt7b and Wnt10b) and their co-receptors (Lrp5 and Lrp6) were not significantly decreased by the loss of ATF4. Increased Topflash/Fopflash ratio in Figure 5D could be due to the increased β-catenin protein level by ATF4 and/or potential synergistic effects of the ATF4-β-catenin interactions. Future studies will determine the molecular mechanism whereby ATF4 modulates the level of β-catenin protein in greater detail.
In summary, this study demonstrates a new role of ATF4 in regulation of the β-catenin protein levels in MSCs and osteoblasts/osteoprogenitors and the osteoblastic differentiation of bone marrow MSCs.
Abbreviations
ALP: alkaline phosphatase; ATF4: activating transcription factor 4; CFU-F: colony-forming unit-fibroblast; CFU-OB: colony-forming unit-osteoblast; GSK-3β: glycogen synthase kinase 3β; LRP: lipoprotein receptor-related protein; MSC: mesenchymal stem cell; Osx: Osterix; PTH: parathyroid hormone; Runx2: Runt-related transcription factor 2.
Acknowledgements
This work was supported by Chinese Ministry of Science and Technology Grant 2009CB918902 and NIH Grant AR059647.
Competing Interests
The authors have declared that no competing interests exist.
References
1. Aktary Z, Chapman K, Lam L, Lo A, Ji C, Graham K.. et al. Plakoglobin interacts with and increases the protein levels of metastasis suppressor Nm23-H2 and regulates the expression of Nm23-H1. Oncogene. 2010;29:2118-2129
2. Augello A, De Bari C. The regulation of differentiation in mesenchymal stem cells. Hum Gene Ther. 2010;21:1226-1238
3. Banerjee C, McCabe L.R, Choi J.Y, Hiebert S.W, Stein J.L, Stein G.S.. et al. Runt homology domain proteins in osteoblast differentiation: AML3/CBFA1 is a major component of a bone-specific complex. J Cell Biochem. 1997;66:1-8
4. Cao H, Yu S, Yao Z, Galson D.L, Jiang Y, Zhang X.. et al. Activating transcription factor 4 regulates osteoclast differentiation in mice. J Clin Invest. 2010;120:2755-2766
5. Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469-480
6. Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192-1205
7. Day T.F, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739-750
8. Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I.. et al. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971-986
9. Ducy P, Zhang R, Geoffroy V, Ridall A.L, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation [see comments]. Cell. 1997;89:747-754
10. Elefteriou F, Ahn J.D, Takeda S, Starbuck M, Yang X, Liu X.. et al. Leptin regulation of bone resorption by the sympathetic nervous system and CART. Nature. 2005;434:514-520
11. Elefteriou F, Benson M.D, Sowa H, Starbuck M, Liu X, Ron D.. et al. ATF4 mediation of NF1 functions in osteoblast reveals a nutritional basis for congenital skeletal dysplasiae. Cell Metab. 2006;4:441-451
12. Franceschi R.T, Ge C, Xiao G, Roca H, Jiang D. Transcriptional regulation of osteoblasts. Cells Tissues Organs. 2009;189:144-152
13. Jiang D, Franceschi R.T, Boules H, Xiao G. Parathyroid Hormone Induction of the Osteocalcin Gene: REQUIREMENT FOR AN OSTEOBLAST-SPECIFIC ELEMENT 1 SEQUENCE IN THE PROMOTER AND INVOLVEMENT OF MULTIPLE SIGNALING PATHWAYS. J Biol Chem. 2004;279:5329-5337
14. Komori T. Requisite roles of Runx2 and Cbfb in skeletal development. J Bone Miner Metab. 2003;21:193-197
15. Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K.. et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts [see comments]. Cell. 1997;89:755-764
16. Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2012;13:27-38
17. Mundlos S, Otto F, Mundlos C, Mulliken J.B, Aylsworth A.S, Albright S.. et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia [see comments]. Cell. 1997;89:773-779
18. Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng J.M, Behringer R.R.. et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17-29
19. Otto F, Thornell A.P, Crompton T, Denzel A, Gilmour K.C, Rosewell I.R.. et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development [see comments]. Cell. 1997;89:765-771
20. Schroeder T.M, Jensen E.D, Westendorf J.J. Runx2: a master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Res C Embryo Today. 2005;75:213-225
21. Wang D, Christensen K, Chawla K, Xiao G, Krebsbach P.H, Franceschi R.T. Isolation and characterization of MC3T3-E1 preosteoblast subclones with distinct in vitro and in vivo differentiation/mineralization potential. J Bone Miner Res. 1999;14:893-903
22. Willert K, Nusse R. Wnt proteins. Cold Spring Harb Perspect Biol. 2012;4:a007864
23. Xiao G, Cheng H, Cao H, Chen K, Tu Y, Yu S.. et al. Critical role of filamin-binding LIM protein 1 (FBLP-1)/migfilin in regulation of bone remodeling. J Biol Chem. 2012;287:21450-21460
24. Xiao G, Cui Y, Ducy P, Karsenty G, Franceschi R.T. Ascorbic acid-dependent activation of the osteocalcin promoter in MC3T3-E1 preosteoblasts: requirement for collagen matrix synthesis and the presence of an intact OSE2 sequence. Mol Endocrinol. 1997;11:1103-1113
25. Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H.. et al. Cooperative Interactions between Activating Transcription Factor 4 and Runx2/Cbfa1 Stimulate Osteoblast-specific Osteocalcin Gene Expression. J Biol Chem. 2005;280:30689-30696
26. Yang X, Karsenty G. ATF4, the osteoblast accumulation of which is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J Biol Chem. 2004;279:47109-47114
27. Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka H.C, Schinke T.. et al. ATF4 Is a Substrate of RSK2 and an Essential Regulator of Osteoblast Biology; Implication for Coffin-Lowry Syndrome. Cell. 2004;117:387-398
28. Yu S, Franceschi R.T, Luo M, Fan J, Jiang D, Cao H.. et al. Critical role of activating transcription factor 4 in the anabolic actions of parathyroid hormone in bone. PLoS One. 2009;4:e7583
29. Yu S, Franceschi R.T, Luo M, Zhang X, Jiang D, Lai Y.. et al. Parathyroid hormone increases activating transcription factor 4 expression and activity in osteoblasts: requirement for osteocalcin gene expression. Endocrinology. 2008;149:1960-1968
30. Yu S, Jiang Y, Galson D.L, Luo M, Lai Y, Lu Y.. et al. General transcription factor IIA-gamma increases osteoblast-specific osteocalcin gene expression via activating transcription factor 4 and runt-related transcription factor 2. J Biol Chem. 2008;283:5542-5553
31. Yu S, Sharma R, Nie D, Jiao H, Im H.J, Lai Y.. et al. ADAR1 ablation decreases bone mass by impairing osteoblast function in mice. Gene. 2013;513:101-110
32. Zhang C, Cho K, Huang Y, Lyons J.P, Zhou X, Sinha K.. et al. Inhibition of Wnt signaling by the osteoblast-specific transcription factor Osterix. Proc Natl Acad Sci U S A. 2008;105:6936-6941
33. Zhang X, Yu S, Galson D.L, Luo M, Fan J, Zhang J.. et al. Activating transcription factor 4 is critical for proliferation and survival in primary bone marrow stromal cells and calvarial osteoblasts. J Cell Biochem. 2008;105:885-895
34. Zhou X, Zhang Z, Feng J.Q, Dusevich V.M, Sinha K, Zhang H.. et al. Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci U S A. 2010;107:12919-12924
35. Zhurinsky J, Shtutman M, Ben-Ze'ev A. Plakoglobin and beta-catenin: protein interactions, regulation and biological roles. J Cell Sci. 2000;113( Pt 18):3127-3139
Author contact
![]() Corresponding author: Dr. Guozhi Xiao, Cohn Research Building, Rm 518, Rush University Medical Center, 1735 West Harrison Street, Chicago, IL 60612, Phone: 312-942-4879, Email: Guozhi_Xiaoedu.
Corresponding author: Dr. Guozhi Xiao, Cohn Research Building, Rm 518, Rush University Medical Center, 1735 West Harrison Street, Chicago, IL 60612, Phone: 312-942-4879, Email: Guozhi_Xiaoedu.

 Global reach, higher impact
Global reach, higher impact