10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(3):303-312. doi:10.7150/ijbs.6214 This issue Cite
Research Paper
Development of a miR-26 Companion Diagnostic Test for Adjuvant Interferon-alpha Therapy in Hepatocellular Carcinoma
1. Liver Carcinogenesis Section, Laboratory of Human Carcinogenesis, Center for Cancer Research, National Cancer Institute, Bethesda, MD, USA;
2. Liver Cancer Institute and Zhongshan Hospital, Fudan University, Shanghai, China;
3. Department of Hepato-Biliary-Pancreatic Surgery (U.T., K.S.) and Department of Diagnostic Pathology (W.K.), Osaka City University Graduate School of Medicine, Osaka, Japan;
4. ShanghaiBio Corporation, Shanghai, China & North Brunswick, New Jersey, USA.
Received 2013-3-8; Accepted 2013-3-15; Published 2013-3-16
Abstract
Background & Aims: Adjuvant therapies for hepatocellular carcinoma (HCC) such as interferon-alpha are effective only in a subset of patients. Previously we found that HCC patients with low level of miR-26 have survival benefits from interferon-alpha. The purpose of this study is to develop a standardized miR-26 diagnostic test (referred as MIR26-DX) to assist identification of candidate HCC patients for adjuvant interferon-alpha therapy.
Methods: We developed a multiplex reverse-transcription quantitative polymerase-chain-reaction assay to determine the levels of two HCC-related miR-26 transcripts along with six small RNA reference transcripts. We evaluated archived paraffin-embedded tissues from three cohorts of HCC patients (n=248) who underwent radical resection at three different clinical centers. Fifty-two percent of them underwent adjuvant interferon-alpha therapy. We used Cox-Mantel log-rank test to evaluate patient survival.
Results: We found that the multiplexing assay was stable and reproducible regardless of differences in sample preparations and operators. We developed a matrix template and a scoring algorithm based on a training cohort (n=129) to assign HCC patients, and then applied the template in two test cohorts (n=119). The proportions of HCC patients assigned as low miR-26 by this algorithm were 68, 4, and 63 percent in the training cohort and two test cohorts, respectively. Consistently, HCC with low miR-26 had a favorable response to interferon-alpha with improved median overall survival (≥3year).
Conclusions: MIR26-DX is a simple and reliable companion diagnostic test to select HCC patients for adjuvant interferon-alpha therapy.
Keywords: miR-26, hepatocellular carcinoma, interferon-alpha, adjuvant therapy, companion diagnostics.
Introduction
Hepatocellular carcinoma (HCC) is the most prevalent form of liver cancer and the second most frequent cause of cancer death worldwide in men (1). It mainly occurs in Asia and Africa, but liver cancer incidence and mortality have risen in most industrialized countries, including the United States, in the past decade (2). Resection or liver transplantation is the best option for a potential cure. However, only about 10-20% of patients with HCC are currently eligible for surgical intervention and a majority of them can develop recurrence after 5 years. Therapies such as transcatheter arterial chemoembolization and sorafenib can prolong survival in some patients (3-7), while the overall response is not satisfactory. In recent years, several clinical trials have suggested that adjuvant interferon-alpha (IFNα) therapy could improve survival of HCC patients after curative resection (5,6). However, some other studies show a limited effect of IFNα (8). We reasoned that a difference in HCC tumor biology may contribute to the overall suboptimal response and suggested that a diagnostic tool that reflects tumor biology can help selecting a subset of patients for a particular therapy and improving treatment outcome.
Molecular-based companion diagnostics linked to gene activities can improve clinical outcomes of cancer patients. For example, the HER2 amplification test can guide the use of trastuzumab treatment in breast cancer patients (9,10). Our previous study on HCC molecular pathogenesis revealed that the clinical outcome of HCC patients treated with adjuvant IFNα therapy is significantly improved when a tumor has a reduced level of miR-26 but not in a tumor with high miR-26 (11). Our results were validated in HCC tumor samples derived from two randomized control trials (RCT) to evaluate the efficacy of IFNα as an adjuvant therapeutic modality (5,6). These results indicate that miR-26 is a predictive biomarker useful to select candidate HCC patients for this treatment modality. Therefore, the establishment of a simple and standardized test to evaluate miR-26 as a diagnostic tool that can be used in a clinical diagnostic lab is warranted.
We developed a multiplex-reverse transcription (RT) quantitative polymerase chain reaction (qPCR) assay, which we referred as MIR26-DX, by examining the levels of HCC-associated miR-26a and miR-26b transcripts along with six small RNA reference transcripts in formalin-fixed paraffin-embedded (FFPE) tumor specimens derived from three independent patient populations (n=248). We first investigated the accuracy of the multiplex-RT qPCR assay by determining correlation between multiplex- and singleplex-RT qPCR. Second, we built a scoring algorithm based on a training cohort as a template for the selection of patients with a favorable response to adjuvant IFNα therapy in terms of survival and then tested its accuracy for predicting treatment response in two additional test cohorts. We found that this scoring algorithm was successful in selecting HCC patients with improved survival following adjuvant IFNα therapy in three independent cohorts from three clinical centers. We suggest that MIR26-DX is a useful companion diagnostic test in HCC patients for adjuvant IFNα therapy.
Patients and Methods
Clinical samples. We used archival FFPE specimens from three separate cohorts (Table 1) consisting of 248 patients with HCC who had undergone radical resection between 1993 and 2007 at the Liver Cancer Institute (LCI) of Fudan University, the University of Hong Kong (HKU) or Osaka City University (OCU), with the specimen collection protocols approved by the corresponding Institutional Review Boards. Cohort 1 contained cases (n=129) from LCI and cohort 2 contained cases (n=79) from HKU. These two cohorts were a part of prospective RCTs of adjuvant therapy with IFNα (5,6). Cohort 3 contained cases (n=40) from OCU, of which 22 received adjuvant IFNα therapy (8). Cohort 1 was used as a template to establish a scoring algorithm, i.e., MIR26-DX, for selection of HCC patients with a favorable response to adjuvant IFNα therapy in terms of overall survival. Cohort 2 and 3 were used to validate the selection performance of MIR26-DX.
The patient accrual criteria of Cohort 1 and 2 were similar and they were from two RCT studies. Patients from these cohorts showed no sign of portal vein tumor thrombosis, no extrahepatic metastasis and good hepatic reserve. These samples were used in our pervious study (11). Therefore, the cases with available RNAs from both cohorts were all included for this study. Furthermore, we applied these criteria to collect Cohort 3. We used overall survival for the clinical endpoint for all three cohorts as described (11).
Patient heterogeneity among three cohorts is evident as clinical variables were different, with the exception of sex (87.9 % men), multinodular status (83.1% single nodule) and status with respect to adjuvant IFNα therapy (52.4% receiving IFNα) (Table 1). Majority of HCC patients in Cohort 1 and 2 were chronic carriers of hepatitis B virus (HBV) (99.3%), while all patients in Cohort 3 were chronic hepatitis C virus (HCV) carriers. More patients in Cohort 1 had early-stage tumors compared to Cohort 2 and 3, as indicated by TNM staging. The degree of hepatic injury in patients with virus-related HCC, as indicated by ALT levels, was significantly higher in Cohort 3 than in Cohort 2 and 1. Moreover, tumor size was smaller, serum level of alpha-fetoprotein was lower, and patients were older in Cohort 3 compared to Cohort 1 and 2. In addition, the clinical outcome was better in Cohort 2 than in Cohort 1 and 3 (Table 1, Additional File 1: Supplementary Figure 1).
Tumor and non-tumor regions of FFPE tissues were separated by macrodissection, and used for total RNA isolation following the methodologies essentially as described (11).
Singleplex-RT qPCR and Multiplex-RT qPCR. Singleplex-RT PCR was performed as previously described according to the standard procedure (11,12). Briefly, a singleplex RT reaction was performed in 15 µl containing 1× RT Buffer, 2nM dNTPs, 50 units of MultiScribe Reverse Transcriptase, 4 units of RNase Inhibitor, 3 µl of single RT primer and 25 ng of total RNAs using microRNA reverse transcription kit (Applied Biosystems, Foster City, CA). The real-time PCR with 1 µl of RT product per reaction was then performed to measure gene expression using Taqman MicroRNA Assays (Applied Biosystems).
Clinical Characteristics of Patients in Three Independent Cohorts.
| Clinical variable | Cohort 1 (n=129) | Cohort 2 (n=79) | Cohort 3 (n=40) | p value |
|---|---|---|---|---|
| Gender | ||||
| Female | 12 | 14 | 4 | |
| Male | 117 | 65 | 36 | 0.177a |
| Age-year | ||||
| Median (range) | 50 (20-77) | 52 (24-75) | 66 (54-81) | <0.001b |
| Alanine transaminase (ALT) | ||||
| Normal (≤50U/L) | 107 | 44 | 10 | |
| Abnormal (>50U/L) | 18 | 35 | 30 | <0.001a |
| Missing data | 4 | 0 | 0 | |
| HBV | ||||
| Negative | 5 | 9 | 40 | |
| Positive | 124 | 70 | 0 | <0.001a |
| HCV | ||||
| Negative | 129 | 74 | 0 | |
| Positive | 0 | 3 | 40 | <0.001a |
| Missing data | 0 | 2 | 0 | |
| Tumor size-cm | ||||
| <=3 | 49 | 22 | 25 | |
| >3 | 80 | 57 | 15 | 0.001a |
| Multinodular | ||||
| No | 110 | 63 | 33 | |
| Yes | 19 | 16 | 7 | 0.585a |
| TNM Stage | ||||
| I | 79 | 7 | 7 | |
| II | 28 | 34 | 24 | |
| III-IV | 14 | 38 | 9 | <0.001a |
| Missing data | 8 | 0 | 0 | |
| Alpha fetoprotein (AFP) | ||||
| Negative (≤20ng/ml) | 49 | 33 | 28 | |
| Positive (>20ng/ml) | 80 | 46 | 12 | 0.002a |
| IFNa Therapy | ||||
| Yes | 69 | 39 | 22 | |
| No | 60 | 40 | 18 | 0.794a |
| Survival-month | ||||
| Median (range) | 60 (2-119) | >116 (5-116) | 85 (10-165) | 0.010c |
aChi-square test; bOne way ANOVA; cLog-rank test.
An optimized eight-gene multiplex RT reaction was performed in 30 µl containing 1× RT Buffer, 2 nM dNTPs, 250 units of MultiScribe Reverse Transcriptase, 9.3 units of RNase Inhibitor, 15.2 µl of eight RT primers (1.9 µl for each) and 120 ng of total RNAs using microRNA reverse transcription kit. The real-time PCR with 0.57 µl of RT products per reaction was further performed to measure microRNA expression using Taqman MicroRNA Assays.
The PCR reaction was done in 384-well plates with the use of Prism 7900HT instruments (Applied Biosystems) (each microRNA per reaction). The expression for each microRNA was measured in triplicate.
Gene expression data collection. After real-time PCR reaction, the raw Ct value was collected with the threshold setting “0.1” as the gene expression results. If the standard deviation of Ct value from the triplicate was higher than 0.2, this reading was considered as an invalid reading and it was required to re-run the PCR. If Ct value of reference gene is higher than 36, this reading was considered as a missing value.
miR-26 expression scoring algorithm. The miR-26 reading was the average of miR-26a and miR-26b expression. It was then normalized with levels of reference transcripts. The miR-26 expression scoring algorithm was derived from six reference-normalized miR-26 readings in Cohort 1. The six medians of normalized miR-26 levels by each reference gene in Cohort 1 were set as standard miR-26 cutoff points or thresholds. Reference-normalized miR-26 readings from test samples were then compared with the corresponding miR-26 thresholds. For each test case, there are six reference-normalized readings. If there are ≥2 readings below the corresponding median levels (i.e., ≤4 out of 6 readings above the median levels), the case was assigned to the low miR-26 reading group. If there are ≥5 readings above median levels, the case was assigned to the high miR-26 category (Additional File 1: Supplementary Figure 2A). This could allow maximizing the case number in low miR-26 group which is the IFNα treatment arm for future.
Statistical analysis. Kaplan-Meier survival analysis was used to compare patient survival based on the assignment by MIR26-DX, using GraphPad Prism 5.0 (GraphPad Software, San Diego, CA) with statistical p values generated by the Cox-Mantel log-rank test. The results were considered statistically significant if they have a p value of < 0.05. The hazard ratio of death was calculated between the IFNα treatment group and control group using GraphPad Prism 5.0. The prognostic receiver operating characteristic (ROC) curves were computed using the IFNα treatment status, as well as patient survival times and death status. Area under curve (AUC) was calculated in GraphPad Prism 5.0.
Results
Selection of microRNA references. A robust biomarker assay requires that it can tolerate day-to-day variability and sample variability under typical clinical scenarios such as the use of FFPE tissues for molecular diagnosis yet still maintain analytic sensitivity, specificity and precision (13). One successful experimental design for measuring gene expression in archival FFPE samples is to incorporate multiple reference genes for data normalization in cross comparison of specimens thereby reduce assay variability (14,15). We previously showed that the RNU6B normalized miR-26 expression levels as determined by singleplex-RT qPCR can be used to identify HCC patients who had improved overall survival following adjuvant IFNα therapy (11). Considering singleplex-RT qPCR assays with multiple transcripts could introduce variability due to preparatory procedures on multiple singleplex-RT reactions, a multiplex-RT qPCR assay to run a single multiplex-RT reaction with all transcripts at once was introduced.
In this vein, we searched for reference non-coding RNA transcripts that can be incorporated into a multiplex assay together with two HCC-related miRNAs, i.e., miR-26a and miR-26b. We identified six reference transcripts, i.e., U6 SnRNA, RNU44, RNU24, RNU6B, RNU38B and RNU48, recommended as internal controls for non-coding RNA analysis by Applied Biosystems. Moreover, potential mature miRNAs were also explored as internal controls since they have similar length and reaction efficacy as the target miRNA ideal for multiplexing. The ideal reference mature miRNAs should express in high abundance without any differential expression among HCC and paired non-tumor specimens. Accordingly, we analyzed microRNA microarray dataset (GSE6857) that consists of 241 HCC samples and paired non-tumor tissues (16). Class comparison analysis using paired t-statistics used successfully in the past (17,18) was used to identify microRNAs without differential expression between tumors and surrounding non-cancerous tissues. Furthermore, the variance for ratio of tumor vs. non-tumor was calculated for each microRNA. From theses analyses, we selected miR-331-5p as a reference miRNA since it has the highest abundance and lowest variance with minimum expression difference between tumor and non-tumor specimens (Additional File 1: Supplementary Table 1, Additional File 1: Supplementary Figure 3). Comparison of multiplexing and singleplexing data derived from two HCC cell lines revealed that among seven reference transcripts, only RNU48 showed a disconcordant expression pattern between singleplexing and multiplexing (data not shown), suggesting that this probe was not suitable for multiplexing. Therefore, we decided to use six reference transcripts, i.e., U6 SnRNA, RNU44, RNU24, RNU6B, RNU38B and miR-331-5p, together with miR-26a and miR-26b in a multiplexing assay.
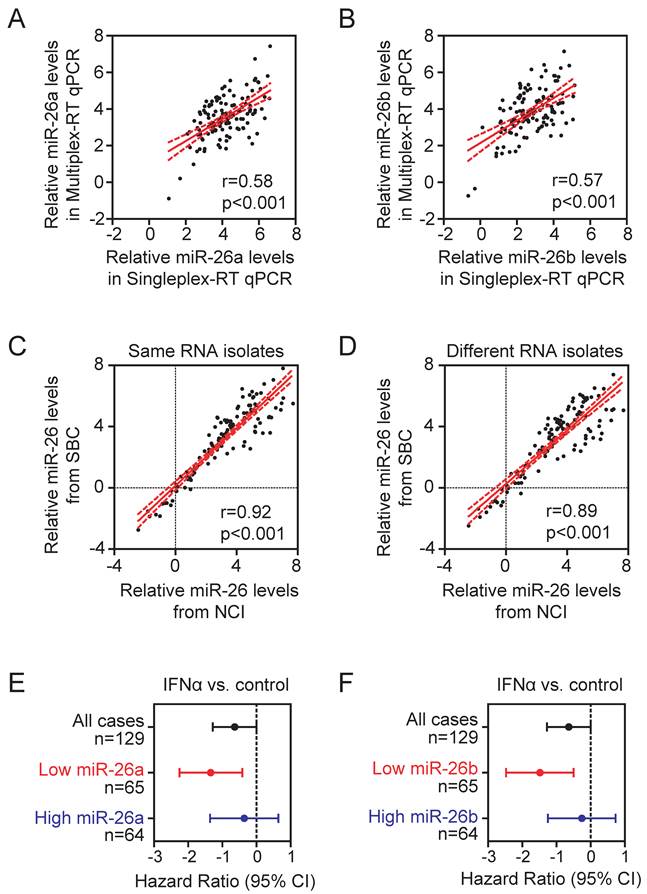
An eight-gene multiplex-RT qPCR assay. To determine the performance of the eight-gene multiplexing assay, we first carried out eight-gene multiplex-RT PCR reactions in FFPE samples from cohort 1, in which the U6B-normalized miR-26a and miR-26b data are available from previous singleplex-RT PCR assay. We found that the U6B-normalized miR-26 readings from multiplex-RT PCR assays were significantly correlated with that of singleplex-RT PCR assays (p<0.001) (Figure 1A-B). To further examine assay reproducibility, we performed multiplexing reactions of the same RNA isolates in 10 FFPE samples derived from 10 randomly selected HCC patients by two laboratories, i.e., National Cancer Institute (NCI) and ShanghaiBio Corporation (SBC), using two different batches of assay reagents. In addition, we also isolated tumor cells from two different parts of tumor FFPE samples and performed RNA isolation and multiplexing PCR assays independently by two laboratories. We found that data generated from the two laboratories using the same RNA isolates (Figure 1C) or different RNA isolates from different tumor areas (Figure 1D) were highly correlated, indicating that the assay is stable and reproducible.
We also tested whether data derived from multiplex-RT qPCR in cohort 1 could predict HCC cases for their responses to IFNα. Consistent with our previous data (11), when cases were divided into low and high miR-26a groups based on the median level of U6B-normalized miR-26a in tumor, patients with low miR-26a had improved survival from adjuvant IFNα therapy while patients with high miR-26a did not (Figure 1E). Similar results were obtained with U6B-normalized miR-26b data (Figure 1F). Taken together, these results indicate that the multiplex-RT qPCR assay is comparable with singleplex-RT qPCR.
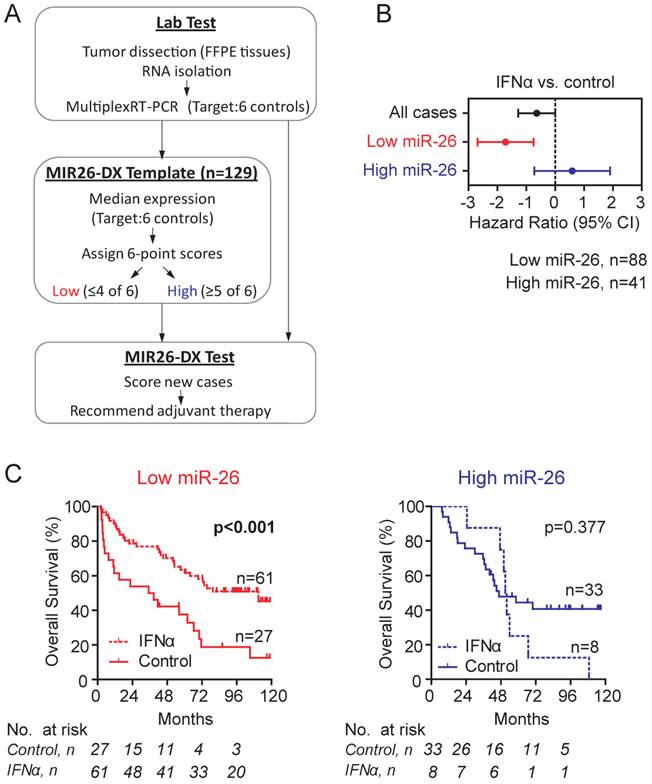
Establishment and validation of a scoring algorithm of MIR26-DX. We developed a simple scoring algorithm for assigning cases into high miR-26 or low miR-26 group. It was based on the average expression of miR-26a and 26b normalized to six reference genes in tumor from cohort 1. By six individual reference-normalized miR-26 expression data points, the scoring algorithm assigned HCC cases into the low miR-26 group when ≥2 out of 6 readings are below the corresponding median expression (in other words, ≤4 of 6 readings above their medians), and the high miR-26 group when ≥5 readings are above the corresponding median expression (Figure 2A, Additional File 1: Supplementary Figure 2A). Based on this scoring algorithm, we found that 68.2% of cases in cohort 1 were assigned as low miR-26 cases, and that low miR-26 cases, but not high miR-26 cases, had survival benefits from adjuvant IFNα therapy (p<0.001) (Figure 2B-C). As a result, this algorithm follows a 2:1 case allocation ratio (i.e., 67% cases in the treatment group and 33% cases in the control group), a typical strategy used for the assignment of patients in RCT studies for maximizing cases in the treatment arm.
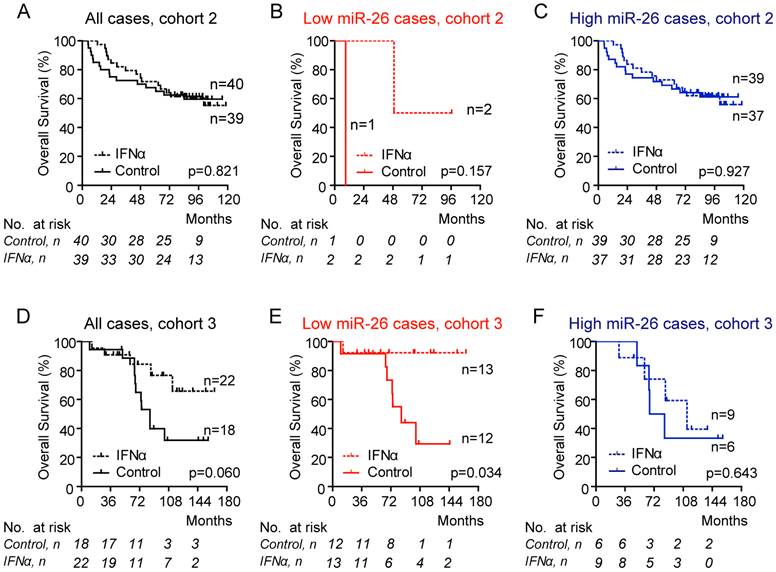
Reference-normalized miR-26 readings in each test case were compared with the corresponding threshold derived from cohort 1. Using these thresholds, we found that 3.8% of Cohort 2 patients were classified as low miR-26 cases. Consistently, HCC patients with low miR-26 had improved overall survival following adjuvant IFNα therapy (Figure 3A-C). However, the difference is statistically insignificant (p=0.157). To further validate if MIR26-DX is predictive of IFNα response in additional HCC cases especially those with HCV infection, we identified a cohort consisting of HCV-positive Japanese HCC patients who underwent curative resection (8). We identified 40 cases with available FFPE samples without portal vein thrombosis. We found that MIR26-DX identified 62.5% patients as low miR-26. Consistently, Japanese HCC cases with low miR-26, but not high miR-26, had a favorable response to adjuvant IFNα therapy in terms of overall survival (p=0.034) (Figure 3D-E).
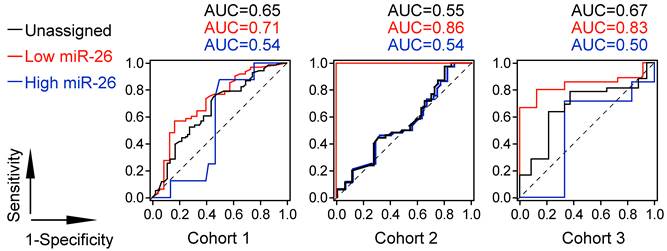
Prognostic ROC curves showed that patients with low level of miR-26 selected by MIR26-DX had increased sensitivity and specificity in response to IFNα treatment at each survival time points compared to patients with high level of miR-26 with selection or patients without selection by MIR26-DX. The AUC value representing the cumulative measurement of IFNα effect on survival for HCC patients with low level of miR-26 was increased by an average of 27.5% compared to AUC in patients with high level of miR-26 when selected by MIR26-DX, or increased by an average of 17.7% compared to AUC in patients without selection in three different cohorts (Figure 4).
We compared the clinical characteristics among low and high miR-26 HCC cases. No statistical significant difference on clinical characteristics was found between these two groups among three independent cohorts (Additional File 1: Supplementary Table 3). Therefore, MIR26-DX appeared effective in identifying HCC candidates for adjuvant IFNα therapy.
The stability and reproducibility of multiplex-RT qPCR. (A, B) Pearson correlation of U6B-normalized miR-26a (A) and miR-26b (B) readings from multiplex- and singleplex-RT qPCR in cohort 1 (n=129). (C, D) Pearson correlation of six individual reference-normalized miR-26 readings derived from SBC and NCI. The multiplex-RT qPCR assay was performed in the same RNA isolates (n=10) (C) or different RNA isolates from two different tumor regions of the same FFPE specimens (n=10) (D). (E, F) Forest plots showing hazard ratios in log2 scale for the overall survival of HCC patients from cohort 1 treated with IFNα versus controls without or with selection by U6B-normalized miR-26a (E) or miR-26b (F). The median of U6B-normalized miR-26a and miR-26b were used as cut-off points.
Development of a template MIR26-DX to identify candidate HCC patients for adjuvant IFNα therapy based Cohort 1. (A) A schematic diagram of the MIR26-DX. A 6-point scoring system was developed based on the median miR-26 expression normalized to six reference transcripts in the Cohort 1 as described in the Method section. (B) The forest plot showing hazard ratios in log2 scale for the overall survival of HCC patients from Cohort 1 treated with IFNα versus controls without or with selection by the 6-point score system. (C) Kaplan-Meier survival curves showing the overall survival of HCC patients with low miR-26 (left) or high miR-26 (right) from Cohort 1 treated with or without IFNα. The number of cases under risk was listed.
Association between miR-26 expression in tumors and overall survival of HCC patients treated with IFNα. Data were shown in Cohort 2 (panels A-C) or in Cohort 3 (panels D-F). The case assignment of high or low miR-26 was based on the 6-point score system. No assignment (A, D); cases assigned as low miR-26 (B, E); cases assigned as high miR-26 (C, F). The number of cases under risk was listed.
The Prognostic ROC curve analysis for the survival of HCC patients in response to adjuvant IFNα therapy.
Discussion
HCC is a heterogeneous disease, which represents by its clinical presentation and tumor biology. The omics studies including genomics, transcriptomics, proteomics and metablomics in HCC revealed that tumor biology is largely variable among HCC patients (11,19-24). HCC heterogeneity makes treatment and eradication of this cancer challenging and contributes to poor clinical prognosis. Therefore, biomarker-guided patient-tailored therapy offers a hope to improve clinical outcome in patients with HCC.
The use of molecular diagnostics for detecting variations of specific genes or signatures and guiding targeted therapies is becoming increasingly common in cancer management. For example, the HER2 gene amplification test is utilized to guide the use of trastuzumab in patients with breast cancer (9,10). The KRAS mutation test is used as a companion diagnostics to guide the use of cetuximab or panitumumab for treatment of colorectal cancer (25). BRAF mutation test is useful to select melanoma patients for vemurafenib therapy (26). However, biomarker-guided therapy for HCC patients is currently not available.
Our previous results demonstrated that HCC patients with a reduced miR-26 expression in their tumor samples have significantly improved overall survival when treated with IFNα (11), indicating that miR-26 is an effective biomarker useful to select HCC patients for adjuvant IFNα therapy. In the present study, we established a simple and reliable assay to determine miR-26 expression and built a scoring algorithm useful to assign HCC cases, a method we referred as MIR26-DX, which was simple and could be easily implemented in clinical diagnostic labs.
MIR26-DX was based on an 8-gene multiplex-RT qPCR assay utilizing FFPE tumor specimens. We found that multiplex-RT PCR was reliable and provided accurate data. It is noted that the RT reaction was done in one reaction for all eight genes in the multiplex-RT PCR assay, which could further minimize data variability from multiple individual RT preparatory procedures. Consistently, the microRNA expression data from the multiplex-RT qPCR assay were significantly correlated with data from the singleplex-RT qPCR assay. To minimize assay variability, we introduced six reference genes for normalization. It should be noted that the expression levels of all six reference genes were similar between tumor and non-tumor tissues for all HCC cases (Additional File 1: Supplementary Table 2). These results suggest that all six reference genes are reliable endogenous controls for normalization of microRNA expression in HCCs. We found that the assay was reproducible regardless of the differences in sample preparation and operators. Moreover, MIR26-DX was effective in identifying HCC patients who had improved survival following IFNα therapy. In addition, only 60% of RNA and 60% or less of RT reagents was used in multiplex assays as compared to singleplex assays. The time required to prepare multiplex assay was also much shorter than that of singleplex assay. Furthermore, the assay was done reliably using RNAs isolated from FFPE samples, which makes sample preparation and transportation much easier. Taken together, we have demonstrated that MIR26-DX is an efficient and time-saving test that can produce accurate data with minimal use of specimens and reagents.
The proportions of HCC patients assigned as low miR-26 by MIR26-DX were varied among three cohorts, i.e., 68%, 4% and 63% in Cohort 1, Cohort 2 and Cohort 3, respectively. The small number of cases in Cohort 2 assigned as low miR-26 might be due to its inclusion criteria. The criteria about inclusion of cases with no postoperative residual tumors or poor hepatic functions in Cohort 2 are more stringent. In Cohort 2, postoperative residual tumors were checked by a 3-phase contrast-enhanced computed tomography at one month after resection (6). However, only ultrasonography was required in Cohort 1 (5) and there was no postoperative examination required in Cohort 3 (8). Therefore, it is possible that patients in Cohort 2 generally had better overall survival and higher level of miR-26 compared to Cohort 1 and 3 (Additional File 1: Supplementary Figure 1-2) since miR-26 expression levels are associated with HCC prognosis (11).
MIR26-DX was successful in assigning HCC patients who had improved survival following adjuvant IFNα therapy in three cohorts. These patients were from HBV and HCV positive HCC cases and came from both developing and developed countries. Therefore, our observations may be broadly applicable to other HCC populations, and suggest the potential universal utilization of MIR26-DX in patients with HCC worldwide. Because of a limited number of cases used in this study, a prospective randomized study to evaluate MIR26-DX is warranted (clinicaltrial.gov; NCT01681446).
In summary, we developed a standardized miR-26 diagnostic test which was an eight-gene multiplex-RT qPCR assay together with a scoring algorithm. Our results indicate that MIR26-DX may serve as a companion diagnostic test to assist selecting candidate HCC patients who have a favorable response to adjuvant IFNα treatment in terms of overall survival. Such a test can prevent unnecessary treatments with IFNα to those who do not have improved survival from this type of therapy so that they could gain a chance for other treatment modalities.
Supplementary Material
Additional File 1Supplementary Figures 1-3, Supplementary Tables 1-3.
Acknowledgements
We thank Karen Yarrick for bibliographic assistance and the NIH Fellows Editorial Board for editing the manuscript. This work was supported in part by research grants from the Intramural Research Program of the Center for Cancer Research, the National Cancer Institute (Z01 BC 010876 and Z01-BC 010313). JZ, JF, ZYT, SF, HZ, JGJ and HCS were supported by research grants from the Ministry of Science of China (#2013ZX10002007-004) and Bureau of Science and Technology of Shanghai (#12DZ1941300) for supporting clinical development of MIR26-DX.
Conflict of interest
The authors declare that they do not have anything to disclose regarding funding from industries or conflict of interest with respect to this paper.
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90
2. El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340:745-50
3. Llovet JM, Ricci S, Mazzaferro V. et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378-90
4. Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: Chemoembolization improves survival. Hepatology. 2003;37:429-42
5. Sun HC, Tang ZY, Wang L. et al. Postoperative interferon alpha treatment postponed recurrence and improved overall survival in patients after curative resection of HBV-related hepatocellular carcinoma: a randomized clinical trial. J Cancer Res Clin Oncol. 2006;132:458-65
6. Lo CM, Liu CL, Chan SC. et al. A randomized, controlled trial of postoperative adjuvant interferon therapy after resection of hepatocellular carcinoma. Ann Surg. 2007;245:831-42
7. Clavien PA. Interferon: the magic bullet to prevent hepatocellular carcinoma recurrence after resection? Ann Surg. 2007;245:843-5
8. Uenishi T, Nishiguchi S, Tanaka S, Yamamoto T, Takemura S, Kubo S. Response to interferon therapy affects risk factors for postoperative recurrence of hepatitis C virus-related hepatocellular carcinoma. J Surg Oncol. 2008;98:358-62
9. Slamon DJ, Leyland-Jones B, Shak S. et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783-92
10. Piccart-Gebhart MJ, Procter M, Leyland-Jones B. et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659-72
11. Ji J, Shi J, Budhu A. et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437-47
12. Ji J, Wang XW. Identification of Cancer Stem Cell-Related MicroRNAs in Hepatocellular Carcinoma. Methods Mol Biol. 2012;826:163-75
13. Dancey JE, Dobbin KK, Groshen S. et al. Guidelines for the development and incorporation of biomarker studies in early clinical trials of novel agents. Clin Cancer Res. 2010;16:1745-55
14. Paik S, Shak S, Tang G. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817-26
15. Cronin M, Pho M, Dutta D. et al. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol. 2004;164:35-42
16. Budhu A, Jia HL, Forgues M. et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897-907
17. Budhu A, Forgues M, Ye QH. et al. Prediction of venous metastases, recurrence and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99-111
18. Ye QH, Qin LX, Forgues M. et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416-23
19. Ji J, Wang XW. New kids on the block: Diagnostic and prognostic microRNAs in hepatocellular carcinoma. Cancer Biol Ther. 2009;8:1686-93
20. Ji J, Yamashita T, Budhu A. et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472-80
21. Yamashita T, Ji J, Budhu A. et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012-24
22. Lee JS, Heo J, Libbrecht L. et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410-6
23. Roessler S, Long EL, Budhu A. et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957-66
24. Budhu A, Roessler S, Zhao X. et al. Integrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomes. Gastroenterology. 2013 In press
25. Allegra CJ, Jessup JM, Somerfield MR. et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091-6
26. Chapman PB, Hauschild A, Robert C. et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507-16
Author contact
![]() Corresponding author: Dr. Junfang Ji at National Cancer Institute, 37 Convent Drive, MSC 4258, Bethesda, MD 20892; Tel: 301-594-9581; Fax: 301-496-0497; Bethesda, MD 20892; Email: jijunnih.gov; or Dr. Xin Wei Wang, Tel: 301-496-2099; Email: xw3ugov.
Corresponding author: Dr. Junfang Ji at National Cancer Institute, 37 Convent Drive, MSC 4258, Bethesda, MD 20892; Tel: 301-594-9581; Fax: 301-496-0497; Bethesda, MD 20892; Email: jijunnih.gov; or Dr. Xin Wei Wang, Tel: 301-496-2099; Email: xw3ugov.

 Global reach, higher impact
Global reach, higher impact