10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(4):331-342. doi:10.7150/ijbs.6022 This issue Cite
Research Paper
Identification, mRNA Expression, and Functional Analysis of Chitin Synthase 1 Gene and Its Two Alternative Splicing Variants in Oriental Fruit Fly, Bactrocera dorsalis
Key Laboratory of Entomology and Pest Control Engineering, College of Plant Protection, Southwest University, Chongqing 400716, P. R. China.
Received 2013-2-2; Accepted 2013-3-31; Published 2013-4-5
Abstract
Two alternative splicing variants of chitin synthase 1 gene (BdCHS1) were cloned and characterized from the oriental fruit fly, Bactrocera dorsalis (Hendel). The cDNA of both variants (BdCHS1a and BdCHS1b) consisted of 5,552 nucleotides (nt), with an open reading frame (ORF) of 4,776 nt, encoding a protein of 1,592 amino acid residues, plus 685- and 88-nt of 5′- and 3′-noncoding regions, respectively. The alternative splicing site was located between positions 3,784-3,960 and formed a pair of mutually exclusive exons (a/b) that were same in size (177 nt), but showed only 65% identity at the nucleotide level. During B. dorsalis growth and development, BdCHS1 and BdCHS1a were both mainly expressed during the larval-pupal and pupal-adult transitions, while BdCHS1b was mainly expressed during pupal-adult metamorphosis and in the middle of the pupal stage. BdCHS1a was predominately expressed in the integument whereas BdCHS1b was mainly expressed in the trachea. The 20-hydroxyecdysone (20E) induced the expression of BdCHS1 and its variants. Injection of dsRNA of BdCHS1, BdCHS1a, and BdCHS1b into third-instar larvae significantly reduced the expression levels of the corresponding variants, generated phenotypic defects, and killed most of the treated larvae. Furthermore, silencing of BdCHS1 and BdCHS1a had a similar result in that the larva was trapped in old cuticle and died without tanning completely, while silencing of BdCHS1b has no effect on insect morphology. These results demonstrated that BdCHS1 plays an important role in the larval-pupal transition and the expression of BdCHS1 in B. dorsalis is regulated by 20E.
Keywords: Chitin synthase 1, Alternative splicing, Bactrocera dorsalis, RNA interference, 20-hydroxyecdysone.
Introduction
Chitin, a linear polymer of β-(1, 4)-linked N-acetylglucosamines, is one of the most crucial component in the cuticle, cuticular lining of the foregut, hindgut, trachea, and peritrophic membrane (PM) in insects [1-2], and widely distributed in fungi, nematodes and other arthropods [3]. During each molting cycle in insect development, part of the old cuticle is digested, while new chitin is synthesized and deposited [4]. Chitin synthase (CHS) is a crucial enzyme involved in the final step of the chitin biosynthetic pathway. CHSs are large proteins embedded in the plasma membrane and belong to family 2 of the glycosyltransferases, which catalyze the transfer of sugar moieties from activated sugar donors to specific acceptors resulting in a glycosidic bond [5].
Based on amino acid sequence similarity, distribution, and function, insect chitin synthase occurs in two types: CHS1 and CHS2 (also known as CHS-A and CHS-B) [6-9]. These two genes are closely related, but belong to different phylogenetic clades [2]. CHS1 and CHS2 have different functions during insect growth and development. CHS1 is responsible for production of chitin required in cuticle and cuticular lining of the foregut, hindgut and trachea, whereas CHS2 specializes in chitin synthesis in the PM [1, 10]. However, only CHS1 has been found in hemipterans such as pea aphids, triatomine bugs, and brown planthoppers, while the CHS2 gene was absent, consistent with the lack of PM in these insects [11, 12]. The hemipterans may have lost CHS2 during their evolution. In addition, insect CHS1 consists of two alternative splicing variants, CHS1a and CHS1b, which differ in a 177-nt region that encodes 59 amino acids forming the predicted second to last transmembrane helix [7, 13-15]. No alternative splicing variants have been reported for CHS2 in insects [3, 16].
Due to the fact that chitin is absent in plants and vertebrates [5], CHS represents a potential target for selective insecticidal agents. Many insect growth regulators, such as diflubenzuron and chlorfluazuron, have been reported to disrupt chitin synthesis in insects [17, 18]. In addition, 20-hydroxyecdysone (20E) and the ecdysone agonist RH5992 may be able to interfere with insect cuticle formation by inhibiting CHS1 [19]. In a recent study, serious cuticular malformations were observed and CHS1 expression was up-regulated as a result of 20E treatment of a lepidopteran [20]. There have been no reports on the effects of 20E on CHS1 in tephritid fruit flies to date.
The oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae), is one of the most economically important pests in tropical and subtropical areas of the world, where it causes serious damage to fruit production [21, 22]. Females typically oviposit in fruit, and the developing larvae tunnel through the fleshy mesocarp on which they feed, causing fruit damage and drop [23]. Current control techniques mainly rely on spraying chemical insecticides. Because of the high economic and environmental costs of chemical control, together with the evolution of insecticide-resistant populations, there is an urgent need to develop new pest management strategies. In recent years, RNA interference (RNAi)-based technology has shown great potential in controlling insect pests by silencing vital genes [24, 25]. RNAi has also been successfully used to investigate the developmental function of CHS1 in holometabolous and hemimetabolous insects, such as Tribolium castaneum (Coleoptera) [26], Drosophila melanogaster (Diptera) [27, 28], Spodoptera exigua (Lepidoptera) [29], Ostrinia furnacalis (Lepidoptera) [30], Locusta migratoria manilensis (Orthoptera) [14], and Nilaparvata lugens (Hemiptera) [11]. These results showed that CHS1 is required for survival, egg hatching, and molting. Therefore, clarifying the functions of CHS1 in B. dorsalis will aid in identifying a suitable gene for developing RNAi-based technology for pest control.
In this study, we reported (1) a full-length cDNA encoding chitin synthase 1 (BdCHS1) from B. dorsalis; (2) two alternative splicing variants of BdCHS1 (BdCHS1a and BdCHS1b); (3) the expression patterns of BdCHS1 and two alternative variants at different developmental stages and in different tissues; (4) the effects of 20E on the expression of BdCHS1 and its variants; and (5) a functional analysis of BdCHS1 and its variants by RNAi.
Materials and Methods
Insect culture
Bactrocera dorsalis was originally collected in August 2009 from Fujian Province, China. Larvae were reared in the laboratory at 27 ± 1°C, 70 ± 5% relative humidity, and 14:10 h light:dark on an artificial diet as described previously [31]. The developmental stages were synchronized at each molt by collecting new larvae or pupae.
cDNA cloning of BdCHS1
Total RNA was isolated from the integument of the third-instar larvae of B. dorsalis with a TRIzol kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. First-strand cDNA was synthesized using PrimeScript® 1st Strand cDNA synthesis Kit (TaKaRa, Dalian, China) with oligo (dT)18 primers and used as a template for PCR.
Based on the results of high-throughput transcriptome sequencing of B. dorsalis [32], three cDNA fragments encoding chitin synthase 1 were obtained (GenBank ID: JN036718, JN036719, and JN036720). The PCR strategies and the sequences of the gene-specific primers designed to obtain overlapping PCR products are shown in Fig. 1 and Supplementary Material: Table S1, respectively. The full-length BdCHS1 cDNA sequence was determined by sequencing four overlapping PCR fragments. The ends were amplified by 5′- and 3′-RACE using the SMARTerTM RACE cDNA Amplification Kit following the instructions of the manufacturer (Clontech, Palo Alto, CA, USA). PCR was carried out with rTaq™ polymerase (TaKaRa) in a 25 μL reaction mixture containing 1 μL cDNA templates, 1 μL each primer (10 μM), 2 μL dNTP (2.5 mM each), and 2.5 μL 10 × PCR buffer (Mg2+ plus). Thermal cycling was performed on C1000TM Thermal Cycler (BIO-RAD, Hercules, CA, USA) under the following conditions: initial denaturation at 95°C for 3 min; followed by 34 cycles of denaturation at 95°C for 30 s, annealing at 55-65°C (based on the primer annealing temperatures) for 30 s, and extension at 72°C for 2-3 min (based on the size of expected fragment); with a final extension at 72°C for 10 min. Amplified PCR fragments from each reaction were separated in an agarose gel and purified using the Gel Extraction Mini Kit (Watson Biotechnologies, Shanghai, China). Purified DNA was ligated into pGEM®-T Easy vector (Promega, Madison, WI, USA) and sequenced completely from both directions using an ABI Model 3100 automated sequencer (Life Technologies, Shanghai, China).
Identification of alternative splicing exons of BdCHS1
The expected fragments, including alternative exon sequences, were predicted by aligning them with CHS1 genes of other insect species. To identify alternative splicing exons of BdCHS1, two gene-specific primers, BdF, 5′-GGAAATGGAAGCGGAAAAG-3′, and BdR, 5′-TGCCGAAACGATGGAACAA-3′, were designed based on the full-length cDNA sequence of BdCHS1. cDNAs from the integument and trachea were used as templates for BdCHS1a and BdCHS1b, respectively. Thermal cycling conditions were: initial denaturation at 95°C for 3 min; followed by 34 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; with a final extension at 72°C for 10 min. Each PCR product amplified from either integument or trachea cDNA was purified and sequenced as described above.
Sequence and phylogenetic analyses
Sequence similarity and analysis for conserved domains were performed using NCBI BLAST programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi). ExPASy Proteomics Server (http://cn.expasy.org/tools/pi_tool.html) was used to compute the isoelectric points and molecular weights of deduced protein sequences. The signal peptide was predicated by SignalP 3.0 (http://www.cbs.dtu.dk/service/SignalP/), and transmembrane helices were analyzed using TMHMM v.2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) [33]. Putative coiled-coils sites were predicted with PAIRCOIL [34]. NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/) was used to analyze the N-glycosylation sites [35]. Alignments of nucleotide and amino acid sequences were made using ClustalW software [36]. Phylogenetic tree was constructed by the programs of MEGA5.04 [37] using the neighbor-joining method. Bootstrap values were calculated with 1000 replications.
Developmental stage and tissue-specific expression patterns of BdCHS1 and its two alternative splicing variants
B. dorsalis were sampled at 1 d intervals from newly-molted third-instar larvae to adults to investigate developmental-stage expression profiles. Integument, midgut, Malpighian tubules, fat body, and trachea were dissected from day-4 third-instar larvae to determine tissue-specific expression patterns. Total RNA was isolated from whole bodies of five insects at each stage using TRIzol reagent and treated with DNase (TaKaRa) for DNA digestion. Total RNA was isolated from isolated tissues (pooled from 10 insects) using RNeasy® Plus Micro Kit (with gDNA Elimator spin columns, Qiagen, Valencia, CA, USA). First strand cDNA was synthesized in a 10 μL reaction mixture using random hexamer primers and oligo-(dT) by PrimeScript® RT reagent Kit (TaKaRa). Primer sequences used for qPCR analysis are shown in Supplementary Material: Table S2. The qPCR was carried out in a Mx3000P thermal cycler (Stratagene, La Jolla, CA) with 20 μL reaction volumes containing 1 μL of template cDNA, 10 μL iQTM SYBR® Green Supermix (BIO-RAD), 1 μL each of forward and reverse primers (0.2 mM), and 7 μL ddH2O. Amplification conditions were: initial denaturation at 95°C for 2 min; followed by 40 cycles of denaturation at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. After each reaction, a melting curve analysis from 60-95°C was conducted to ensure consistency and specificity of the amplified product. All of these experiments involved three biological replications. α-Tubulin (GU269902) was used as an internal reference gene [38]. Relative gene expression data was analyzed using the 2-ΔΔCT method [39]. The significance of difference in the target gene expression was determined by student's t-test.
Schematic diagram of the strategy to amplify the BdCHS1 cDNA. The upper bar represents the cDNA. Lower bars indicate each of the PCR fragments listed in Supplementary Material: Table S1. Fragments 1 and 2 were amplified using specific primers. Fragments 3 and 4 were obtained by 3′-RACE and 5′-RACE, respectively. JN036718, JN036719, and JN036720 were obtained from the transcriptome data of Bactrocera dorsalis.

Effect of 20E treatment on the expression of BdCHS1 and its two alternative splicing variants
A 10 μg/μL stock solution of 20E (Sigma-Aldrich, St. Louis, MO, USA) dissolved in 95% ethanol was diluted to 1 μg/μL with distilled water as a working solution. The 20E treatment group of 2-d-old third-instar larvae was injected with 20E solution at doses of 500 ng/larva. The control group was injected with an equivalent volume of 0.1% ethanol. Five injected larvae were randomly selected from each group at 1, 4, 8, and 12 h after injection, and total RNA was isolated to analyze relative expression levels by using qPCR, as described above. Each group contained 30 individual larvae, and the experiment was repeated three times.
Specificity of RNAi for BdCHS1 and its two alternative splicing variants
RNAi was applied to further explore the biological functions of BdCHS1 and its two alternative splicing variants, BdCHS1a and BdCHS1b. The most unique nucleotide regions of BdCHS1, BdCHS1a, and BdCHS1b were selected for specific double-stranded RNA (dsRNA) synthesis (the synthesized regions are shown in Fig. 2 and 3), and dsGFP was used as a negative control. The primers used to synthesize dsRNA are listed in Supplementary Material: Table S2. All of the reagents and enzymes used for the dsRNA synthesis were from the MEGAscript® RNAi kit (Ambion, Carlsbad, CA, USA). The size of the dsRNA products was confirmed by electrophoresis on a 1% agarose gel. The final concentration of dsRNA was 1.2 μg/μL.
To detect the knock-down effect of dsRNA for BdCHS1, BdCHS1a, and BdCHS1b, approximately 1 μg of dsRNA was slowly injected into the antepenultimate abdominal segments of 2-d-old third-instar larvae with a Nanoject II Auto-Nanoliter Injector (Drummond Scientific, Broomall, PA, USA). Control larvae were injected with equivalent volumes of dsGFP. Injected larvae were reared on an artificial diet under conditions described above until pupation, and morphological changes were observed. Photos were taken using a Leica M165C microscope (Leica Microsystems, Wetzlar, Germany). Forty-five larvae from each group were treated with three replications. Total RNA was isolated from five randomly-selected insects from each (treated and control) group after injection, and the levels of transcripts were measured using qPCR as described above.
Results
cDNA cloning and sequence analysis of BdCHS1
The full-length cDNA sequence of BdCHS1 was obtained from DNA fragments amplified by PCR from cDNA from the integument and from 5′ and 3′-RACE (Supplementary Material: Table S1). The full-length cDNA consisted of 5,552 nucleotides (nt) with an open reading frame (ORF) of 4,776 nt encoding a protein of 1,592 amino acid residues. The BdCHS1 cDNA included a 5′-untranslated region (UTR) located 685 nt upstream of the start codon (ATG) and a 3′ UTR of 88 nt ending in a poly (A) tail. The complete nucleotide and deduced amino acid sequences of BdCHS1 are shown in Fig. 2. Based on the deduced amino acid sequence, BdCHS1 had a theoretical molecular mass of 180.7 kDa, an isoelectric point of 6.50, and no signal peptide. BdCHS1 was predicted to contain three domains: an N-terminal domain (domain A) with 10 transmembrane helices; a highly conserved central domain (domain B) with two motifs, EDR and QRRRW, regarded as signatures of chitin synthases; and a C-terminal domain (domain C) with an additional seven transmembrane helices. PAIRCOIL identified one coiled-coil, a principal subunit oligomerization domain in proteins and a feature not seen in CHS2 proteins. Four potential N-glycosylation sites (N-X-S/T) at positions 21, 911, 955, and 1311 were predicted using NetNGLyc 1.0 software from the ExPASy Proteomics website.
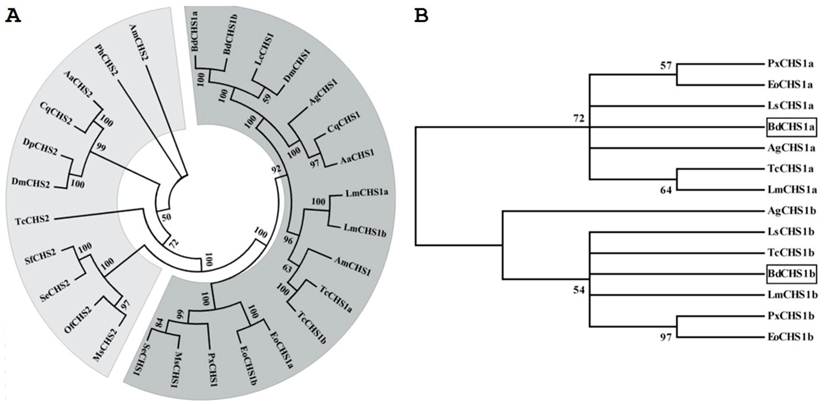
Multiple protein alignments showed that BdCHS1 had homology to known and predicated CHS1 in other insects. For example, BdCHS1 shared 88.0%, 87.8%, 78.2%, 68.7%, and 67.2% identity with the CHS1 of Lucilia cuprina (AAG09712; Diptera), D. melanogaster (NP_524233), Anopheles quadrimaculatus (ABD74441; Diptera), S. exigua (AAZ03545), and L. migratoria manilensis (ACY38588), respectively. Phylogenetic analysis showed that BdCHS1 was grouped into the CHS1 family, but the two splicing variants, BdCHS1a and BdCHS1b, fell into distinct phylogenetic groups (CHS1a and CHS1b) (Fig. 3).
Comparative analysis of alternative splicing exons of BdCHS1
Analysis of the BdCHS1 sequence and its corresponding cDNA sequence revealed two alternative splicing variants that were highly conserved among CHS1 genes of other insect species. These two alternative splicing exons, BdCHS1a (JN207848) and BdCHS1b (JX170758), were equal in length (177 nt), encoded a 59-amino acid residue, and occurred at identical consensus cDNA alignment positions (3784-3960 nt; Fig. 4). BdCHS1a and BdCHS1b shared 65% identity.
Nucleotide and deduced amino acid sequences of BdCHS1a cDNA from Bactrocera dorsalis (JN207848). The start codon is indicated in bold and the stop codon in bold with an asterisk. The two chitin synthase signature motifs (EDR and QRRRW) are boxed, and the putative N-glycosylation sites are underlined. Transmembrane regions predicted are shaded. The putative coiled-coil site is indicated in bold with a dotted line. Primers of BdCHS1 for dsRNA synthesis are in white with black background.

Phylogenetic trees of the known insect chitin synthases. (A) Tree of the known chitin synthases. (B) Tree of the alternative exons of insect CHS1s. The tree was constructed with MEGA 5.04 using the neighbor-joining method. Bootstrap support values are indicated on branches. The following insect chitin synthases sequences were used: Aedes aegypti (Aa), Anopheles gambiae (Ag), Apis mellifera (Am), Culex quinquefasciatus (Cq), Drosophila melanogaster (Dm), Daphnia pulex (Dp), Ectropis oblique Prout (Eo), Lucilia cuprina (Lc), Locusta migratoria manilensis (Lm), Laodelphax striatella (Ls), Manduca sexta (MS), Ostrinia furnacalis (Of), Pediculus humanus corporis (Ph), Plutella xylostella (Px), Spodoptera exigua (Se), Spodoptera frugiperda (Sf), Tribolium castaneum (Tc). GenBank ID are as follows: AaCHS1 (XP_001651163), AaCHS2 (EAT46081), AgCHS1a (XP_321336.5), AgCHS1b (XP_321336.4), AmCHS1 (XP_395677), AmCHS2 (XP_001121152), CqCHS1 (XP_001866798), CqCHS2 (XP_001864594), DmCHS1 (NM_079509), DmCHS2 (NM_079485), DpCHS2 (EFX80669), EoCHS1a (ACA50098), EoCHS1b (ACD10533), LcCHS1 (AF221067), LmCHS1a (GU067730), LmCHS1b (GU067731), LsCHS1a (JQ040012), LsCHS1b (JQ040011), MsCHS1 (AY062175), MsCHS2 (AY82156), OfCHS2 (ABB97082), PhCHS2 (XP_002423604), PxCHS1a (AB271784), PxCHS1b (AB281490), SeCHS1 (DQ062153), SeCHS2 (EU622827), SfCHS2 (AY525599), TcCHS1a (AY291475), TcCHS1b (AY291476), and TcCHS2 (AY291477).
Comparative analysis of alternative splicing exons of BdCHS1 in Bactrocera dorsalis. (A) Schematic diagram of the structure of BdCHS1 cDNA. The black box indicates nucleotide positions 3784-3960 that splice alternatively. ClustalW alignment of nucleotide (B) and deduced amino acid (C) sequences of the alternative exons a and b of BdCHS1. Symbols below the alignments indicate identical (*), highly conserved (:), and conserved (.) residues. Primers for dsRNA synthesis are highlighted in dark (for BdCHS1a) and light (for BdCHS1b) gray. Primers for qPCR analysis are underlined.

Developmental-stage and tissue-specific expression patterns of BdCHS1 and its alternative splicing variants
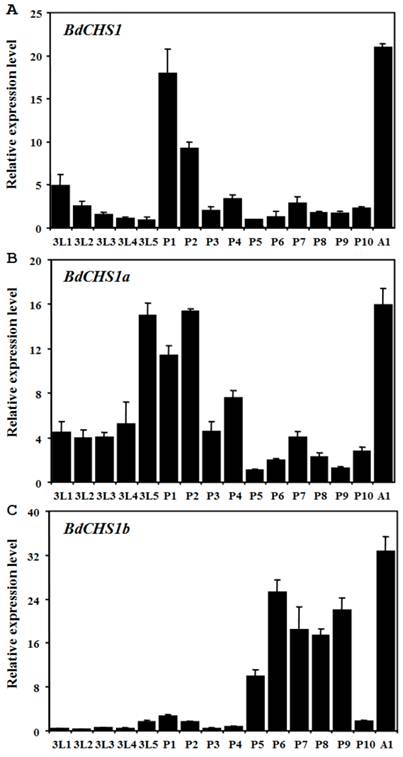
Expression levels of BdCHS1 and two alternative variants in different developmental stages and tissues were examined by qPCR. The results showed that BdCHS1 and its alternative variants mRNA expression levels were constant from day-1 third-instar larvae to day-1 adults. However, BdCHS1 was mainly expressed during the larval-pupal and pupal-adult transitions, with the highest expression level in day-1 adults (Fig. 5A). During the feeding stage of third-instar larvae and in day 2-4 pupae, BdCHS1 was expressed, but at lower levels. BdCHS1a had a similar expression pattern, but a high expression level was observed in day-5 third-instar larvae (Fig. 5B). In contrast, BdCHS1b was expressed differently, mainly during pupal-adult metamorphosis and in the middle of pupal stage (Fig. 5C).
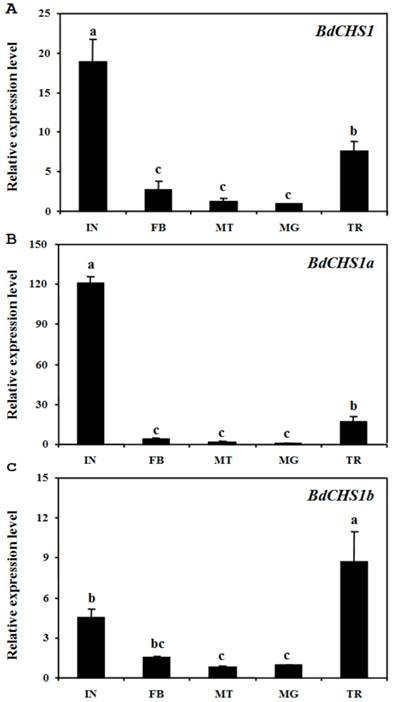
Analyses of different tissues of third-instar larvae showed that BdCHS1 was highly expressed in the integument, followed by the trachea, then the midgut, fat body, and Malpighian tubules. BdCHS1a was also mainly expressed in the integument, whereas BdCHS1b was predominantly in the trachea (Fig. 6).
Effects of 20E treatment on the expression of BdCHS1 and its alternative splicing variants
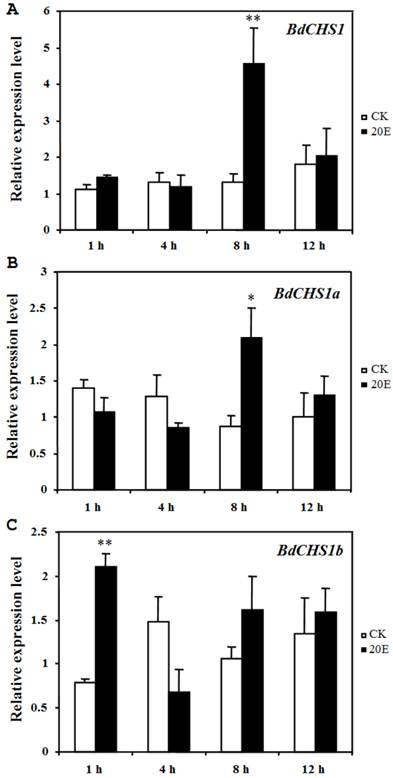
Application of 20E to day-2 third-instar larvae caused a precocious and incomplete molt within 25 h, serious malformations, and death. However, in the control groups, the larvae grew normally and developed into pupae after 3 d. After 20E injection, the relative expression levels of BdCHS1 and its alternative variants were determined by qPCR. The results showed that both BdCHS1 and BdCHS1a were up-regulated dramatically at 8 h post-injection compared with control insects, whereas the expression of BdCHS1b was up-regulated as early as at 1 h post-injection (Fig. 7), indicating that BdCHS1b quickly responded to 20E.
Specificity of RNAi for BdCHS1 and its alternative splicing variants
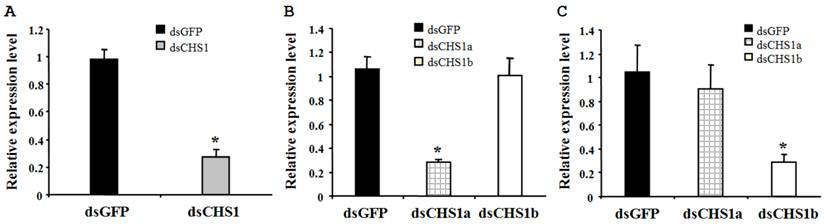
To verify the specificity of RNAi for BdCHS1 and its alternative variants, corresponding sequence-specific dsRNAs were synthesized and injected into B. dorsalis. Subsequently, qPCR was carried out using templates prepared from insects injected with BdCHS1, BdCHS1a, or BdCHS1b dsRNA. The results showed that RNAi effectively inhibited the expression of each corresponding transcript compared to control groups injected with dsGFP (Fig. 8). Furthermore, RNAi of BdCHS1a or BdCHS1b did not decrease the expression level of the other variant (Fig. 8).
Relative expression levels of BdCHS1 and its two alternative splicing variants in different developmental stages of Bactrocera dorsalis. Expression levels at 16 different time points in third-instar larvae, pupae, and adults were detected by qPCR. (A) BdCHS1; (B) BdCHS1a; (C) BdCHS1b. α-Tubulin was used as an internal reference gene. Data are means ± SE of three biological replications. The relative expression was calculated based on the value of the lowest expression which was ascribed an arbitrary value of 1. The age in days of the insects is indicated, e.g., 3L1, first day of third-instar larvae; P1, first day of pupae; A1, first day of adults.
Relative expression levels of BdCHS1 and its two alternative splicing exons in different tissues of Bactrocera dorsalis. Expression levels in integument (IN), trachea (TR), midgut (MG), Malpighian tubules (MT), and fat body (FB) were detected by qPCR. (A) BdCHS1; (B) BdCHS1a; (C) BdCHS1b. α-Tubulin was used as an internal reference gene. Data are means ± SE of three biological replications. The relative expression was calculated based on the value of the lowest expression which was ascribed an arbitrary value of 1. Different letters above the bars indicate significant differences among tissues (P < 0.05, t test).
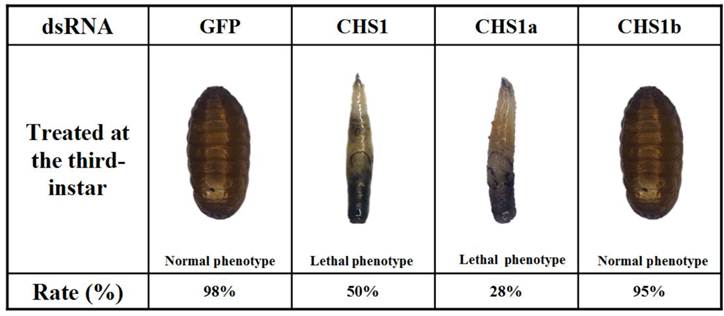
dsRNAs were injected into day-2 third-instar larvae to investigate their effects on larval-pupal transition. When injected with dsRNA of BdCHS1, 50 ± 2.3% of individuals failed to develop into larvae and later died. They were trapped in old cuticle, died without tanning completely, had abnormal pigmentation (Fig. 9). Following injection with BdCHS1a dsRNA, 28 ± 3.5% of larvae died and exhibited the same phenotypic malformations as those injected with dsBdCHS1. However, treatment with BdCHS1b dsRNA caused no visible abnormal morphological changes, and the larvae molted to pupae. No change in phenotype was observed in the dsGFP control.
Influence of 20E on expression of BdCHS1 and its two alternative splicing exons. The insects were injected with 20-hydroxyecdysone (20E) at day-2 of third-instar larvae, and five individuals each were analyzed 1, 4, 8, and 12 h after treatment using qPCR. α-Tubulin was used as an internal reference gene. Data are means ± SE of three biological replications. (A) BdCHS1; (B) BdCHS1a; (C) BdCHS1b. CK: control insects; 20E: insects injected with 20E. Significantly differences between treatment and control are indicated with * (P < 0.05) or ** (P < 0.01, t test).
Relative expression levels of BdCHS1 in third-instar larvae after RNAi. Expression levels of (A) BdCHS1 in larvae injected with GFP or BdCHS1 dsRNA; (B) BdCHS1a in larvae injected with GFP, BdCHS1a, or BdCHS1b dsRNA; and (C) BdCHS1b in larvae injected with GFP, BdCHS1a, or BdCHS1b dsRNA. α-Tubulin was used as an internal reference gene. Data are means ± SE from three independent experiments. Significant differences between treatment and control are indicated with * (at P < 0.05, t test).
Representative phenotypes of Bactrocera dorsalis after injection of day-2 third-instar larvae with BdCHS1, BdCHS1a and BdCHS1b dsRNA. Injection of BdCHS1 and BdCHS1a dsRNAs resulted in the same lethal phenotype: the larva was trapped in old cuticle and died without tanning completely, whereas insects injected with BdCHS1b dsRNA seemed normal.
Discussion
Chitin synthases play major roles in chitin formation during insect growth and development. Many CHSs have been cloned and characterized in various insect species [11]. To date, most insects are known to possess two different chitin synthases, CHS1 and CHS2. CHS1 is mainly expressed in the epidermis and tracheal cells, while CHS2 is expressed in the midgut epithelial cells [7, 40-41]. To explore the potential of chitin-synthesis inhibition as a pest management tool, CHSs have received much attention in recent years. In this study, we reported the first full-length cDNA encoding chitin synthase 1 from tephritid fruit flies.
Alternative splicing expands protein diversity and vastly augments the coding potential via complex regulation of gene expression [42]. Investigations of many insects have shown that CHS1 contains two alternative exons and produces two splicing variants, unlike CHS2. Specifically, an alternative site in the middle of the ORF has been reported in T. castaneum [3], Manduca sexta (Lepidoptera) [16], Plutella xylostella (Lepidoptera) [43], and L. migratoria manilensis [14]. In addition to the known alternative splicing exons, another recently discovered site involving the second exon was found in lepidopterans such as O. furnacalis and Bombyx mori [20]. Interestingly, no alternate forms had been identified the genome of pea aphid; only exon b was found. Whether the function of exon a has been taken over by exon b or whether exon a is not necessary for chitin synthesis in pea aphid is unknown [12]. In this paper, our finding of the two alternative splicing exons of CHS1 in B. dorsalis supports the presence of only one splicing site in the middle of the ORF.
The different expression patterns of the two variants of CHS1 indicated that they might play distinct roles during insect growth and development. In O. furnacalis, OfCHSA-2a was mainly expressed in newly-laid eggs and during larval-larval molting and larval-pupal transformation, while OfCHSA-2b was expressed only during larval-larval molting [30]. In recent studies of N. lugens, transcription of NlCHS1a reached its highest level just after molting, whereas NlCHS1b expression peaked 1-2 days before molting [11]. Furthermore, the developmental expression pattern of LmCHS1A was reported to be similar to that of LmCHS1, but different from that of LmCHS1B [14]. The expression levels of LmCHS1A were much higher than those of LmCHS1B during most developmental stages. Our results suggested that BdCHS1a and BdCHS1b likely play different roles in chitin synthesis during molting, when the insects needed significantly more exoskeletal chitin to form new cuticle.
Expression of BdCHS1 and its two alternative forms was also observed in different tissues. Our data indicated that BdCHS1 expression was high in the integument and tracheae and lower in the midgut, fat body, and Malpighian tubules. BdCHS1a was highly expressed in the integument, whereas BdCHS1b was predominantly expressed in the trachea, consistent with observations in L. migratoria manilensis, M. sexta, and T. castaneum [14, 16, 26]. Taken together, these data demonstrated that CHS1a and CHS1b are associated with chitin synthesis in the integument and trachea, respectively. However, in Anopheles gambiae larvae, AgCHS1A and AgCHS1B showed the same expression patterns and were predominately expressed in the carcass [15]. Further investigations are required to clarify the precise roles of the two alternative variants in different insects.
In insects, 20E mediates a cascade of gene regulatory events that control molting and metamorphosis [44]. Previous studies found that CHS1 expression could be regulated by 20E in several insect species. Here, substantially up-regulation of BdCHS1 and BdCHS1a was detected at 8 h and BdCHS1b was significantly up-regulated at 1 h after 20E injection into larvae. This suggested that 20E acted as a positive regulator of BdCHS1, BdCHS1a, and BdCHS1b expression during the larval-pupae molting. In agreement with our present study, significant up-regulation of CHS1 expression by 20E has also been observed in other insects, including D. melanogaster, S. exigua and O. furnacalis [20, 45-46]. Moreover, up-regulation of CHS1 in D. melanogaster by 20E may be due to an indirect interaction between the ecdysone receptor complex and ecdysone-responsive elements within the CHS1 promoters [45]. However, 20E could also suppress the expression of CHS1 in Choristoneura fumiferana (Lepidoptera), suggesting that 20E may facilitate the digestion and separation of old cuticle from the epidermis during molting [19]. Further studies are required to confirm the precise relationship between chitin synthase genes and hormone regulation of insect molting.
RNAi has been successfully used to ascertain the function of CHS1 during the development of many insects. To identify functional difference among BdCHS1 and two variants, three specific dsRNAs targeting BdCHS1, BdCHS1a, and BdCHS1b were injected into third-instar larvae. During the larval-pupal transformation, both BdCHS1 and BdCHS1a dsRNA-mediated silencing affected larval growth and development, leading to a lethal phenotype. However, no visible morphological abnormality was observed when BdCHS1b dsRNA was injected compared with the GFP dsRNA control. This result suggested that BdCHS1 and BdCHS1a were essential for larval-pupal growth and development, an observation that is partly supported by phenotypes observed in N. lugens [11]. In T. castaneum, TcCHS1a dsRNA disrupted larval-larval, larval-pupal, and pupal-adult moltings, while TcCHS1b was required only for pupal-adult molting [7, 26]. In L. migratoria manilensis, LmCHS1A dsRNA resulted in three different lethal phenotypes, including translucent new cuticle, trouble shedding old cuticle, and stunted development, whereas injection of LmCHS1B dsRNA caused crumpled cuticle [14]. In O. furnacalis, silencing of OfCHSA-2a caused incomplete molting, while silencing of OfCHSA-2b influenced only the formation of head cuticle in third-instar larvae [30]. Thus, dsRNAs of CHS1 can disrupt its function, leading to severe disorders in insects. However, ingestion of bacteria-expressed dsSeCHSA by S. exigua during larval-pupal and pupal-adult phases caused no significant phenotypic changes [29], either because the amount of ingested dsRNA was not sufficient or because RNAi in this species is not effective. Future studies will be needed to show how knockdown of BdCHS1 and its variants affects other developmental stages of B. dorsalis.
Outbreaks of B. dorsalis have occurred with increasing frequency in south China, causing significant losses in citrus yield. Currently, control of oriental fruit fly relies on spraying chemical insecticides and induces the evolution of resistance [47, 48]. In addition to traditional pesticides, chitin synthase genes could be targeted through an RNAi-based approach. Injection of dsRNA for doublesex gene in female flies led to the under-development of ovary and reduced the number of matured eggs, suggesting that administering dsRNAs corresponding to important genes may be a means for pest control [49]. However, continuously feeding dsRNA led to over-expression of target genes in the oriental fruit fly [50], so not all dsRNAs effectively suppress the expression of target genes. Therefore, there is an urgent need to clarify the functions of vital candidate genes. In summary, injecting dsRNA of BdCHS1 and its variants into B. dorsalis larvae not only resulted in a strong decline in target gene expression, but also affected larval growth and development, leading to an increasing mortality rate over time. These results can advance our understanding of the biological functions of CHS1 in B. dorsalis, and showed that CHS1 dsRNA may be useful as a means of insect pest control.
Abbreviations
CHS: Chitin synthase; PM: Peritrophic membrane; qPCR: Quantitative real-time PCR; UTR: untranslated region; dsRNA: double-strand RNA; RNAi: RNA interference.
Supplementary Material
Tables S1-S2.
Acknowledgements
This work was supported in part by the National Basic Research Program of China (2009CB125903), Natural Science Foundation of Chongqing (CSTC, 2013jjB80005, JJA80020), the Program for Changjiang Scholars and Innovative Research Team in University (IRT0976), and the Earmarked Fund for Modern Agro-industry (Citrus) Technology Research System of China.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kramer KJ, Muthukrishnan S. Chitin metabolism in Insects. In: (ed.) Gilbert L.I, Iatrou K, Gill S. Comprehensive Molecular Insect Science; Vol. 4, Biochemistry and Molecular Biology, Chapter 3. Oxford, UK: Elsevier Press. 2005:111-144
2. Lehane MJ. 1997. Peritrophic matrix structure and function. Annu Rev Entomol. 1997;42:525-550
3. Arakane Y, Hogenkamp DG, Zhu YC. et al. Characterization of two chitin synthase genes of the red flour beetle, Tribolium castaneum, and alternate exon usage in one of the genes during development. Insect Biochem Mol Biol. 2004;34:291-304
4. Nation JL. Insect Physiology and Biochemistry; second ed. Boca Raton, FL, USA: CRC press. 2008
5. Merzendorfer H. Insect chitin synthases: a review. J Comp Physiol B Biochem Syst Environ Physiol. 2006;176:1-15
6. Tellam RL, Vuocolo T, Johnson SE. et al. Insect chitin synthase: cDNA sequence, gene organization and expression. Eur J Biochem. 2000;267:6025-6043
7. Arakane Y, Muthukrishnan S, Kramer KJ. et al. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Mol Biol. 2005;14:453-463
8. Bolognesi R, Arakane Y, Muthukrishnan S. et al. Sequences of cDNAs and expression of genes encoding chitin synthase and chitinase in the midgut of Spodoptera frugiperda. Insect Biochem Mol Biol. 2005;35:1249-1259
9. Moussian B. Recent advances in understanding mechanism of insect: structure, function and regulation of chtin synthases and chitinases. Insect Biochem Mol Biol. 2010;40:364-375
10. Zimoch L, Merzendorfer H. Immunolocalization of chitin synthase in the tobacco hornworm. Cell Tissue Res. 2002;308:287-297
11. Wang Y, Fan HW, Huang HJ. et al. Chitin synthase 1 gene and its two alternative splicing variants from two sap-sucking insects, Nilaparvata lugens and Laodelphax striatellus (Hemiptera: Delphacidae). Insect Biochem Mol Biol. 2012;42:637-646
12. Bansal R, Mian MA, Mittapalli O. et al. Characterization of a chitin synthase encoding gene and effect of diflubenzuron in soybean aphid, Aphis glycines. Int J Biol Sci. 2012;8:1323-1334
13. Rezende GL, Martins AJ, Gentile C. et al. Embryonic desiccation resistance in Aedes aegypti: presumptive role of the chitinized serosal cuticle. BMC Dev Biol. 2008;8:82
14. Zhang JZ, Liu XJ, Zhang JQ. et al. Silencing of two alternative splicing-derived mRNA variants of chitin synthase 1 gene by RNAi is lethal to the oriental migratory locust, Locusta migratoria manilensis (Meyen). Insect Biochem Mol Biol. 2010;40:824-833
15. Zhang X, Zhang JZ, Zhu KY. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol Biol. 2010;19:683-693
16. Hogenkamp DG, Arakane Y, Zimoch L. et al. Chitin synthase genes in Manduca sexta: characterization of a gut-specific transcript and differential tissue expression of alternately spliced mRNAs during development. Insect Biochem Mol Biol. 2005;35:529-540
17. Meola SM, Mayer RT. Inhibition of cellular proliferation of imaginal cells epidermal cells by diflubenzuron in pupae of the stable fly. Science. 1980;207:985-987
18. Mayer RT, Chen AC, DeLoach JR. Chitin synthesis inhibiting insect growth regulators do not inhibit chitin synthesis. Experientia. 1981;37:337-338
19. Ampasala DR, Zheng SC, Zhang DY. et al. An epidermis-specific chitin synthase cDNA in Choristoneura fumiferana: cloning, characterization, developmental and hormonal-regulated expression. Arch Insect Biochem Physiol. 2011;76:83-96
20. Qu MB, Yang Q. Physiological significance of alternatively spliced exon combinations of the single-copy gene class A chitin synthase in the insect Ostrinia furnacalis (Lepidoptera). Insect Mol Biol. 2012;21:395-404
21. Clarke AR, Armstrong KF, Carmichael AE. et al. Invasive phytophagous pests arising through a recent tropical evolutionary radiation: the Bactrocera dorsalis complex of fruit flies. Annu Rev Entomol. 2005;50:293-319
22. Li WF, Yang L, Tang K. et al. Microsatellite polymorphism of Bactrocera dorsalis (Hendel) populations in China. Acta Entomol Sin. 2007;50:1255
23. Fletcher B. The biology of dacine fruit flies. Annu Rev Entomol. 1987;32:115-144
24. Baum JA, Bogaert T, Clinton W. et al. Control of coleopteran insect pests through RNA interference. Nat Biotech. 2007;25:1322-1326
25. Mao YB, Cai WJ, Wang JW. et al. Silencing a cotton bollworm P450 monooxy genase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotech. 2007;25:1307-1313
26. Arakane Y, Specht CA, Kramer KJ. et al. Chitin synthases are required for survival, fecundity and egg hatch in the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol. 2008;38:959-962
27. Ostrowski S, Dierick HA, Bejsovec A. Genetic control of cuticle formation during embryonic development of Drosophila melanogaster. Genetics. 2002;161:171-182
28. Devine WP, Lubarsky B, Shaw K. et al. Requirement for chitin biosynthesis in epithelial tube morphogenesis. Proc Natl Acad Sci USA. 2005;102:17014-17019
29. Tian HG, Peng H, Yao Q. et al. Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS One. 2009;4:e6225
30. Qu MB, Yang Q. A novel alternative splicing site of class A chitin synthase from the insect Ostrinia furnacalis -Gene organization, expression pattern and physiological significance. Insect Biochem Mol Biol. 2011;41:923-931
31. Lin C, Yang WJ, Shen GM. et al. Molecular characterization of cDNA encoding ecdysone receptor isoform B1 and its expression in the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Fla Entomol. 2012;95:650-658
32. Shen GM, Dou W, Niu JZ. et al. Transcriptome analysis of the oriental fruit fly (Bactrocera dorsalis). PLoS One. 2011;6:e29127
33. Krogh A, Larsson B, Heijne G. et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567-580
34. Berger B, Wilson DB, Wolf E. et al. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259-8263
35. Hansen JE, Lund O, Rapachi K. et al. O-GLYCBASE version 2.0: a revised database of O-glycosylated proteins. Nucleic Acids Res. 1997;25:278-282
36. Li KB. ClustalW-WPI: ClustalW analysis using distributed and parallel computing. Bioinformatics. 2003;19:1585
37. Tamura K, Peterson D, Peterson N. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731-2739
38. Shen GM, Jiang HB, Wang XN. et al. Evaluation of endogenous references for gene expression profiling in different tissues of the oriental fruit fly Bactrocera dorsalis (Dipter: Tephritidae). BMC Mol Biol. 2010;11:76
39. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402-408
40. Zimoch L, Hogenkamp DG, Kramer KJ. et al. Regulation of chitin synthesis in the larval midgut of Manduca sexta. Insect Biochem Mol Biol. 2005;35:515-527
41. Liang YR, Lin C, Wang RR. et al. Cloning and expression pattern of chitin synthase (CHS) gene in epidermis of Ectropis obliqua Prout. Afr J Biotechnol. 2010;9:5297-5308
42. Black DL. Mechanism of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291-336
43. Ashfaq M, Sonoda S, Tsumuki H. Developmental and tissue-specific expression of CHS1 from Plutella xylostella and its response to chlorfluazuron. Pestic Biochem Physiol. 2007;89:20-30
44. Henrich VC, Rybczynski R, Gilbert LI. Peptide hormones, steroid hormones, and puffs: mechanisms and models in insect development. Vitam Horm. 1998;55:73-125
45. Gagou ME, Kapsetaki M, Turberg A. et al. Stage-specific expression of the chitin synthase DmCHSA and DmCHSB genes during the onset of Drosophila metamorphosis. Insect Biochem Mol Biol. 2002;32:141-146
46. Yao Q, Zhang DW, Tang B. et al. Identification of 20-hydroxyecdysone late-response genes in the chitin biosynthesis pathway. PLoS One. 2010;2:e14058
47. Vontas J, Hernandez-Crespo P, Margaritopoulos JT. et al. Insecticide resistance in tephritid flies. Pestic Biochem Physiol. 2011;100:199-205
48. Hsu JC, Feng HT, Wu WJ. et al. Truncated transcripts of nicotinic acetylcholine subunit gene Bdα6 are associated with spinosad resistance in Bactrocera dorsalis. Insect Biochem Mol Biol. 2012;42:805-816
49. Chen SL, Dai SM, Lu KH. et al. Female-specific doublesex dsRNA interrupts yolk protein gene expression and reproductive ability in oriental fruit fly, Bactrocera dorsalis (Hendel). Insect Biochem Mol Biol. 2008;38:155-165
50. Li XX, Zhang MY, Zhang HY. RNA interference of four genes in adult Bactrocera dorsalis by feeding their dsRNAs. PLoS One. 2011;6:e17788
Author contact
![]() Corresponding author: Dr. Jin-Jun Wang, College of Plant Protection, Southwest University, Chongqing 400716, China. Tel: +86-23-68250255; Fax: +86-23-68251269; Email: jjwang7008com.
Corresponding author: Dr. Jin-Jun Wang, College of Plant Protection, Southwest University, Chongqing 400716, China. Tel: +86-23-68250255; Fax: +86-23-68251269; Email: jjwang7008com.

 Global reach, higher impact
Global reach, higher impact