10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(4):382-390. doi:10.7150/ijbs.5510 This issue Cite
Research Paper
Dpysl4 Is Involved in Tooth Germ Morphogenesis through Growth Regulation, Polarization and Differentiation of Dental Epithelial Cells
1. Department of Biological Science and Technology, Graduate school of Industrial Science and Technology, Tokyo University of Science, Chiba 278-8510, Japan;
2. Research Institute for Science and Technology, Tokyo University of Science, Chiba 278-8510, Japan;
3. Organ Technologies Inc., Tokyo 101-0048, Japan.
Received 2012-11-9; Accepted 2013-4-9; Published 2013-4-26
Abstract
Dihydropyrimidinase-related protein 4 (Dpysl4) is a known regulator of hippocampal neuron development. Here, we report that Dpysl4 is involved in growth regulation, polarization and differentiation of dental epithelial cells during tooth germ morphogenesis. A reduction in Dpysl4 gene expression in the tooth germ produced a loss of ameloblasts, resulting in the decrease of synthesis and secretion of enamel. The inhibition of Dpysl4 gene expression led to promotion of cell proliferation of inner enamel epithelial cells and inhibition of the differentiation of these cells into pre-ameloblasts, which was confirmed by analyzing cell polarization, columnar cell structure formation and the expression of ameloblast marker genes. By contrast, overexpression of Dpysl4 in dental epithelial cells induces inhibition of growth and increases the expression of the inner enamel epithelial cell marker gene, Msx2. These findings suggest that Dpysl4 plays essential roles in tooth germ morphogenesis through the regulation of dental epithelial cell proliferation, cell polarization and differentiation.
Keywords: Development, cell polarization, tooth germ, differentiation, Dpysl4.
Introduction
The development of ectodermal organs such as the tooth germ and hair follicles is a complex process that involves reciprocal epithelial and mesenchymal interactions [1-3]. Tooth germ morphogenesis begins with a thickening of the epithelium to form the dental epithelium during the lamina and placode stages [2]. During the bud stage, the dental epithelium invaginates to form the dental mesenchyme and then progresses to the cap stage [2]. In the bell stage of the tooth germ, epithelial and mesenchymal cells undergo stepwise histodifferentiation into enamel-secreting ameloblasts and dentin-secreting odontoblast cell lineages, respectively [2]. Dental epithelial cells consist of four distinct cell populations, including stellate reticulum (SR), outer enamel epithelium (OEE), Notch1-expressing stratum intermedium (SI) and fibroblast growth factor 9 (Fgf9)- and homeobox, msh-like 2 (Msx2)-positive inner enamel epithelium (IEE) [4-7]. IEE cells differentiate into columnar pre-ameloblasts with their nuclei shifted away from the basement membrane and that express enamel proteins, including Ameloblastin (Ambn) [4, 8]. These morphological, functional and genetic changes are thought to be associated with the differentiation of ameloblasts.
Cell polarity plays roles in a wide variety of biological phenomena including cell movement, differentiation and the transportation of secretory vesicles [9-11]. Mature epithelial cells in ectodermal secretory organs such as salivary and lacrimal glands have common characteristics, including a polarized columnar cell shape, close cell-cell adhesion, specialized intercellular junctions and an apico-basal polarity [9, 11]. It has been reported that the formation of epithelial polarity is regulated by several signaling mechanisms including Phosphatidylinositol 3,4,5-trisphosphate (PIP3) kinase, Glycogen synthase kinase 3 beta (GSK3β), Partitioning-defective protein (PAR) complex and the Ras homolog gene family (Rho) of proteins through the rearrangement of the actin cytoskeleton and microtubules [9]. Functional ameloblasts also have specific characteristics, including a columnar cell shape and the localization of molecules such as Par3, zonula occludens (ZO) and filamentous actin (F-actin) at their apical edge [12]. These cells synthesize and secrete enamel-specific proteins from their apical ends through the transportation of secretory vesicles along microtubules [13, 14]. These findings suggest that the polarization of dental epithelial cells is related to ameloblast differentiation and enamel matrix secretion. However, the molecular mechanisms underlying ameloblast polarization remain to be determined.
Dihydropyrimidinase-related protein (Dpysl), which consists of five family members, is involved in the development of hippocampal neurons [15]. Dpysl2 has been reported to regulate neuronal polarity and axon elongation in hippocampal neurons [16, 17]. Dpysl2 also regulates neural differentiation, as it induces the expression of SRY-box containing gene 1 and Nestin, which is an early neuronal marker in ES cells [18]. Dpysl1, 3 and 5 are thought to be involved in neuronal polarity, as several knockout mice studies have been observed to have impairments in dendritic patterning and spine development in the hippocampus [15]. Dpysl4 has important roles in dendrite arborization, guidepost navigation and neuronal plasticity and has been shown to be involved in neural polarity and differentiation based on the results observed in Dpysl4-deficient mice [19]. Furthermore, it has been suggested that Dpysl2, which is expressed in the lung tissue of the airway epithelium and is activated by phosphorylation by Neuropilin 1, could play a role in alveolarization during lung development [20]. It has also been suggested that the migration and actin cytoskeleton of carcinoma cells are regulated by Dpysl2-Rho-associated coiled-coil containing protein kinase 2 (ROCK II) interactions [21]. Therefore, it is thought that Dpysl4 family members could potentially regulate the polarity and differentiation of epithelial cells during organogenesis [19].
In this study, we show that Dpysl4 is involved in the differentiation of ameloblasts during tooth germ morphogenesis through the regulation of cell growth, cell polarization and differentiation of dental epithelial cells. The expression of Dpysl4 was observed in enamel knots and the IEE, pre-ameloblast cell lineage of dental epithelial cells of both incisors and molars during tooth germ development. The reduction of Dpysl4 gene expression resulted in the overgrowth of IEE cells and the inhibition of their differentiation into pre-ameloblasts, which was detected via their polarization and the expression of ameloblast marker genes. These results suggest that Dpysl4 plays essential roles in cellular polarization and differentiation of dental epithelial cell lineage during tooth germ morphogenesis.
Materials and methods
Animals
C57BL/6 mice were purchased from SLC Inc (Shizuoka, Japan). All mouse care and handling was in compliance with the NIH guidelines for animal research. All experimental protocols were approved by the Tokyo University of Science Animal Care and Use Committee.
In situ hybridization
In situ hybridizations were performed using 10- or 12-μm frozen sections as described previously [22]. Digoxygenin-labeled probes for specific transcripts were prepared by PCR with primers designed using published sequences (Dpysl4; NM_011993, Fgf9; NM_013518, Msx2; NM_013601, Ambn; NM_009664, Notch1; NM_008714, Enam; NM_017468, Sox2; NM_011443). Specific primers for mouse Dpysl4 (forward, 5'- ccctccccataagctctctc -3'; reverse, 5'- ctgcaccttagcaacatgga -3'), Fgf9 (forward, 5'- gtcttcatcagtaacgaccttgg -3'; reverse, 5'- taagatggctacatggatttgct -3'), Msx2 (forward, 5'- tttatgtataaatatataat -3'; reverse, 5'- cttcacttgcaagaatctgt -3'), Ambn (forward, 5'- agccactgctacctggaaac -3'; reverse, 5'- cagggttttccaccaatcac -3'), Notch1 (forward, 5'- ctaggtgctcttgcgtcactt -3'; reverse, 5'- tcactcaggtcagggagaactac -3'), Enam (forward, 5'- tcatacagagatctgcacaccag -3'; reverse, 5'- aacaaaagcagctatttcagacg -3') and Sox2 (forward, 5'- acatgtgagggctggactg -3'; reverse, 5'- ggacatttgattgccatgt -3') were used. The transcripts were visualized by immunoreactivity with anti-digoxigenin alkaline phosphatase-conjugated Fab-fragments (Roche, Basel, Switzerland) according to the manufacturer's instructions.
Plasmid construction
Dpysl4-specific knockdowns in molar germs were performed by the expression of hairpin siRNA using a pBAsi-mU6 vector (Takara Bio, Shiga, Japan). To trace the transfected cells, a CMV-EGFP-polyA fragment was subcloned downstream of the shRNA expression cassette (pBAsi-mU6-CMV-EGFP). To generate the Dpysl4-shRNA (shDpysl4) plasmid, annealed dsDNA (sense, 5'-GATCCGAATTTGTTTAATCAGACCGTCTTCAAGAGAGACGGTCTGATTAAACAAATTCTTTTTT-3' and antisense, 5'-CTAGAAAAAAGAATTTGTTTAATCAGACCGTCTCTCTTGAAGACGGTCTGATTAAACAAATTCG-3') was cloned into the BamHI/XbaI site of pBAsi-mU6-CMV-EGFP. To generate the shDpysl4 cosmid vector, mU6-shDpysl4-CMV-EGFP fragment was subcloned into SmiI site of the pAxwtit2 (Takara Bio). To generate the pcDNA3.1/Zeo(+)-IV (mock) vector, the IRES-Venus fragment was subcloned into XbaI/PmeI sites of pcDNA3.1/Zeo(+) vector (Invitrogen, CA, USA). To generate the Dpysl4 expression plasmid vector, amplified Dpysl4 fragment was subcloned into NotI/EcoRI sites of pcDNA3.1/Zeo(+)-IV. Finally, to generate the mock and Dpysl4 cosmid vector, IRES-Venus or Dpysl4-IRES-Venus fragments were subcloned into SmiI site of pAxCAwtit2 (Takara Bio).
Generation and preparation of adenovirus
Recombinant adenovirus was constructed by homologous recombination between the expression cosmid cassette (pAxwtit2 or pAxCAwtit2) and the parental virus genome in 293 cells according to the manufacturer's instructions. High-titer adenoviruses (1010 - 1011 ifu/ml) were prepared by centrifugation at 3000 rpm for 24 hours, and the pellets were suspended in formulation buffer (2.5% glycerol (w/v), 25 mM NaCl, and 20 mM Tris-HCl, pH 8.0).
Infection of developing tooth germ with adenovirus
Molar germs were dissected from embryonic day (ED) 13.5 mouse embryos to perform knockdown experiments. The high-titer adenoviruses were injected into the molar epithelium using micro-glass capillaries. The adenovirus-infected molar germs were then further incubated as an in vitro organ culture at 37℃ or transplanted into the subrenal capsule as described previously [23]. EGFP adenovirus was successfully infected in dental epithelium after 2 to 4 days incubation, but gradually disappeared at 6 days (Supplementary Material: Fig. S2A).
Microcomputed tomography (Micro-CT) analysis
The analysis of enamel formation in control and shDpysl4 molars was performed by x-ray using a Micro-CT device (R_mCT; Rigaku, Tokyo, Japan) with exposure at 90 kV and 150 mA. Micro-CT images were captured using i-view R (Morita, Kyoto, Japan) [23].
Histochemical and immunohistochemical analysis
Histochemical and immunohistochemical tissue analysis was performed as described previously [23]. For immunohistochemistry, the tissue sections (10 or 50 μm thick) were incubated with an anti-Ki67 primary antibody (1:100; Abcam, Cambridge, MA), anti-E-cadherin primary antibody (1:100; BD, NJ, USA), anti-β-catenin primary antibody (1:100; BD), anti-Par3 primary antibody (1:200; Invitrogen, Carlsbad, CA), anti-ZO-1 (1:100; Invitrogen), Rhodamin Phalloidin (1:100; Invitrogen) and Hoechst33342 (1:500; Invitrogen) for 2 h at room temperature. The secondary antibodies used were Alexa 555 anti-rabbit (1:250; Invitrogen) and Alexa 488 anti-mouse IgG (1:250; Invitrogen). Fluorescence microscopy images were captured using an Axiovert 200M (Carl Zeiss, Oberkochen, Germany) or a confocal microscope featuring 403-, 488-, and 543-nm laser lines (LSM510; Carl Zeiss) and processed with AxioVision software (Carl Zeiss) or Zen software. Tissue sections (10 or 12 μm thick) were stained with hematoxylin and eosin (H&E) and observed using an Axioimager A1 (Carl Zeiss) with an AxioCAM MRc5 (Carl Zeiss) microscope.
RNA preparation and real-time PCR
Total RNA was isolated from cells using the TRIzol® reagent (Invitrogen) according to the manufacturer's protocol. cDNAs were synthesized from a 200-μg aliquot of total RNA by a PrimeScript® II 1st strand cDNA Synthesis Kit (Takara Bio). mRNA expression levels were determined using SYBR® Premix Ex Taq™ II (Takara Bio), and the products were analyzed with an ABI PRISM 7000 real-time PCR system (Applied Biosystems, CA, USA). Specific primers for mouse Dpysl4 (forward, 5'- ccccagctgatgatttctgt -3'; reverse, 5'- accagcgtcaggaaacacat -3'), Msx2 (forward, 5'- aattccgaagacggagcac -3'; reverse, 5'- ggttggtcttgtgtttcctcag -3') were used for real-time PCR.
Cell culture and gene transfection
Molar primary epithelial cells were taken from the molars of ED14.5 mice. Epithelial tissues were minced surgically and plated on 96-well plates in DMEM (Kohjin Bio, Saitama, Japan) supplemented with 15% fetal bovine serum (FBS). After 24 hours of culture, primary epithelial cells were infected with mock or Dpysl4 adenovirus (1x107 ifu/ml) for 3 days. For the cell proliferation assay, emtg2 cells, kindly provided from Dr. Tomooka Y of the Tokyo University of Science, were plated on 96-well plates (BD) in DMEM/F12 (Kojin Bio) supplemented with 10% FBS, insulin (10 μg/ml; Sigma-Aldrich, MO, USA) and transferrin (10 μg/ml, Sigma-Aldrich) [24]. After 24 hours of culture, emtg2 cells were infected with mock or Dpysl4 adenovirus (1x107 ifu/ml) for 2 days, and the highly expressing cells were purified using a FACS Aria III (BD).
Cell proliferation assay
Cell proliferation was assessed using a Cell Counting Kit-8 (Dojindo Labs, Tokyo, Japan) according to the manufacturer's protocol. The number of cells in each well was determined by reading the optical density at 450/630 nm with a VersaMax ELISA microplate reader (Molecular Devices, CA, USA).
3D reconstruction of cell morphology
Three-dimensional (3D) reconstructions of cell morphology were made from E-cadherin expression analyses (50-μm) sections using 3D rendering software Imaris (Bitplane, Zurich, Switzerland). After adjustment of the overall threshold of the image, the software assigns coordinates to each cell.
Results
Dpysl4 is expressed in the immature ameloblast lineage cells of the developing tooth
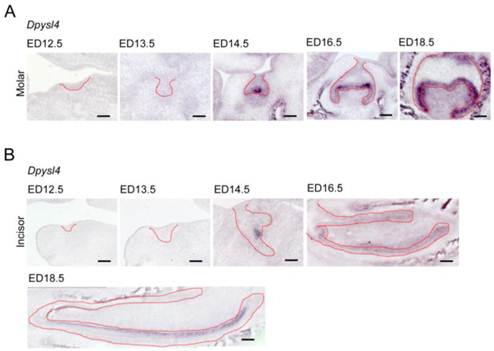
We first investigated the expression patterns of Dpysl4 in the developing molar and incisor germs. Dpysl4 mRNA-positive cells were observed in the enamel knot, which is the epithelial signaling center at ED14.5. At the bell stage, Dpysl4 transcripts were detected in the IEE and secondary enamel knots at ED16.5 and in pre-ameloblast cells of the molar germ at ED18.5 (Fig. 1A). During incisor tooth germ development, Dpysl4 mRNA was also expressed in the enamel knot at ED14.5 and the IEE cells at ED16.5 and subsequently was detected in the transit amplifying zone (T-A), IEE and pre-ameloblasts of incisor germs (Fig. 1B). Previous studies have shown that the differentiation processes of ameloblast lineage cells can be observed in the incisor tooth germ at ED 18.5. We therefore compared the expression patterns of Dpysl4 and an ameloblast lineage marker in the incisor tooth germ at ED18.5. Localization of Dpysl4-positive cells was detectable with Fgf9- and Msx2-positive IEE cells [5, 6] and Ambn-positive pre-ameloblasts [8]. However, no cells were observed in the area of Sox2-positive dental epithelial stem cells [25], Notch1-positive SI cells [7] or Ambn- and Enam-positive differentiated ameloblasts (Supplementary Material: Fig. S1A) [8]. The expression patterns of Dpysl4 and ameloblast marker genes in molar germs were almost identical to the incisor tooth germ (Supplementary Material: Fig. S1B). These results indicate that Dpysl4 is expressed in the enamel knot, IEE and pre-ameloblast during tooth germ development.
Expression patterns of Dpysl4 in tooth germ development. Expression profiles of Dpysl4 mRNA in molar (A) and incisor (B) germs at ED12.5-18.5 using in situ hybridization analysis. Red dotted lines indicate the boundaries between the dental epithelial tissue and mesenchymal tissue. Scale bars, 100 μm.
A reduction in Dpysl4 expression causes an enamel defect
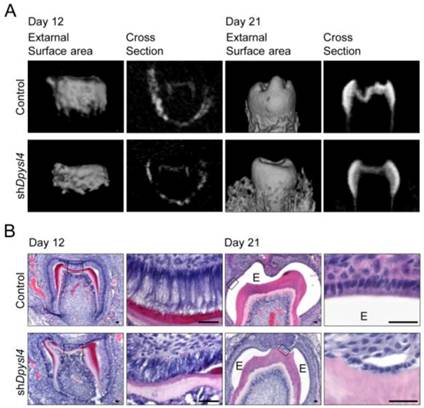
We next investigated whether Dpysl4 is involved in the regulation of ameloblast differentiation during tooth germ development. High-titer control or Dpysl4 shRNA adenovirus was injected into the ED13.5 molar dental epithelium to suppress the function of Dpysl4 during early tooth development. After two days of organ culture, shRNA infected molars were transplanted into the subrenal capsule to develop into a mature molar tooth. After 12 or 21 days post-transplantation into the subrenal capsule, the morphologies and hard tissue formation were analyzed by Micro-CT. Dpysl4 shRNA infected molars (shDpysl4 molars) developed to the secretory stage and formed dentin at day 12; however, abnormal cusp formation and reduced accumulation of enamel were observed at day 21 compared with control shRNA infected molars (control molars) (Fig. 2A). Histological analyses indicated that shDpysl4 molars showed an abnormal polarization of ameloblasts at day 12, and enamel formation was partially defective at day 21 (Fig. 2B). These results indicate that Dpysl4 is essential for enamel formation during amelogenesis and suggest that Dpysl4 may regulate ameloblast differentiation.
A reduction in Dpysl4 expression alters ameloblast morphology
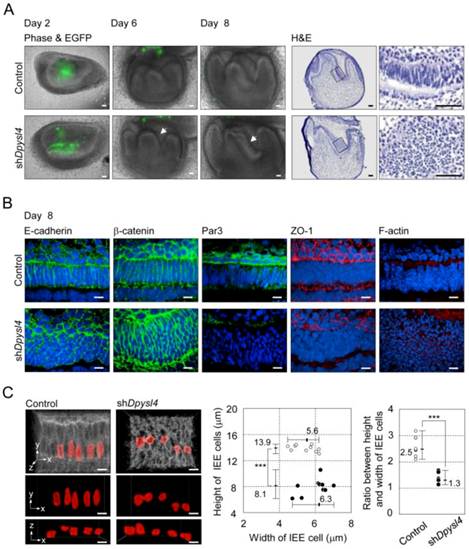
It is well known that ameloblast differentiation is strongly related to an alteration in cell morphology followed by the polarization of dental epithelial cells [8]. In the developing shDpysl4 molar, the abnormal multi-epithelial cell layers were observed after 6 to 8 days of organ culture (Fig. 3A arrows, Supplementary Material: Fig. S2B). Epithelial cells in these multi-layers presented globe-like shapes, while epithelial cells in the control molars exhibited a columnar shape (Fig. 3A). To investigate the effect of Dpysl4 on epithelial morphology, we examined the localization of cell adhesion molecules such as E-cadherin and β-catenin. Pre-ameloblasts in control molars showed a normal columnar shape, but the cells within shDpysl4 molars exhibited a miniaturized form as assessed by immunohistochemical analysis (Fig. 3B). It has been demonstrated that polarized ameloblasts have specific characteristics including a columnar cell shape and the localization of molecules including Par3, ZO-1 and F-actin at the apical edge [12]. Immunohistochemical analyses further showed that this localization of Par3, ZO-1 and F-actin was disrupted in the shDpysl4 molar epithelium as compared with control molars (Fig. 3B). We also analyzed the shape of cells using measurement and statistical analyses on 3D reconstructed images (Fig. 3C, left columns). The heights of the epithelial cells in the shDpysl4 molars (8.1 μm) were decreased as compared with epithelial cells in control molars (13.9 μm) (Fig. 3C center graph). In addition, the ratio between the height and width of the epithelial cells in shDpysl4 molars was also decreased as compared with epithelial cells in control molars (Fig. 3C, right graph). These results suggest that Dpysl4 plays an essential role in the formation of the columnar shape of pre-ameloblasts and in the localization of junction complex proteins at the apical edge.
Knockdown of Dpysl4 causes enamel defects. (A) Micro CT images of the external surface area (first or third columns from the left) and cross sections (second or fourth columns) of the control (upper row) and shDpysl4 (lower row) infected molars developed in a sub-renal capsule (SRC) transplantation for 12 days (left columns) or 21 days (right columns). (B) Histological analysis of the control and shDpysl4 infected molars at 12 (left columns) or 21 (right columns) days after SRC transplantation using hematoxylin and eosin (H&E) staining are also shown. Higher-magnification images of the crown area within the box are shown on the right. Scale bars, 25 μm. E, enamel.
Knockdown of Dpysl4 alters the morphology of the inner enamel epithelium. (A) EGFP fluorescence (green) are merged in phase-contrast images (first, second and third columns) of the control (upper row) and shDpysl4 (lower row) molar germs after 2, 6 and 8 days of adenovirus injection and histological analysis of the infected molar germs cultured for 8 days (fourth and fifth columns). Higher-magnification images of the inner enamel epithelium (IEE) area within the box are shown on the right. Scale bars, 50 μm. (B) Localization of E-cadherin (first column), β-catenin (second column), Par3 (third column), ZO-1 (fourth column) and F-actin (fifth column) in IEE cells of the infected molar germ at 8 days of organ culture were immunohistochemically analyzed using a specific antibody against each molecule. Nuclei were stained with Hoechst33342 (blue). E-cadherin, β-catenin and Par-3 are shown in green, ZO-1 and F-actin in red. Scale bars, 10 μm. (C) Three-dimensional images of the IEE cell structure of control (left) or shDpysl4 (right) molar germs are shown (upper). The lengths of IEE cells in the each of the infected molar germs cultured for 8 days were measured. The plasma membrane of IEE cells were immunohistochemically analyzed using a specific antibody against E-cadherin (gray). The measured cells are shown in red. And on the x-y plane (middle) and x-z plane (lower) images of the measured IEE cells are shown. Scatter diagrams showing the correlations between the IEE cell lengths, between the width and height of the control (white) or shDpysl4 (black) molar germs (left graph) and ratio between height and width of IEE cells (right graph) are shown. Median, +Max, -Min (n=10). ***p < 0.0005. Scale bars, 10 μm.
Dpysl4 regulates cell proliferation and differentiation in the developing dental epithelium
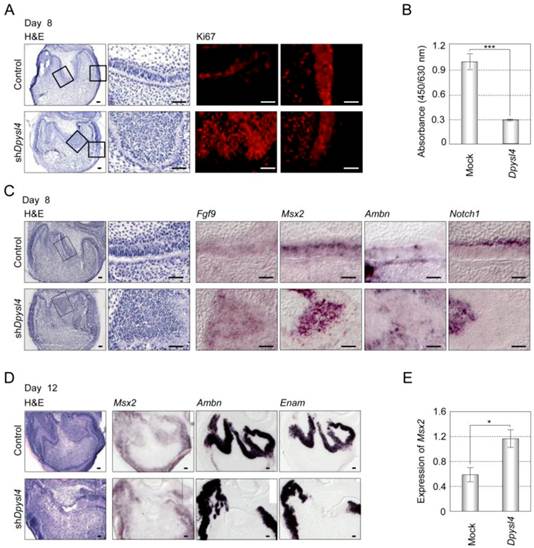
It has been suggested that dental epithelial cell growth is negatively regulated during the processes of ameloblast differentiation [26]. Cell proliferation analyses of shDpysl4 molars using an anti-Ki67 antibody showed that the area of multi-epithelial cell layers consisted of Ki67-positive epithelial cells; in contrast, no Ki67-positive cells were observed in control molars (Fig. 4A). We also examined whether Dpysl4 inhibits cell growth of the emtg2 mouse molar epithelial cell line. Overexpression of Dpysl4 in emtg2 cells induced a suppression of cell proliferation as compared with mock adenovirus-infected cells (Fig. 4B). These observations indicate that Dpysl4 regulates epithelial cell proliferation during ameloblast differentiation.
We next analyzed the expression of ameloblast lineage markers in shDpysl4 molar germs. In situ hybridization analysis showed that the miniaturized cells within the shDpysl4 molar expressed Fgf9 and Msx2 mRNA, but no Ambn or Notch1 mRNA were detected (Fig. 4C). IEE cells in the control molar also expressed these genes, indicating that the miniaturized cells within shDpysl4 molar were committed to an IEE phenotype. Gene expression analysis of Ambn and Enam further showed that the expression of these genes was partially defected in the epithelial cells of the shDpysl4 molar, although these genes were fully detected in the ameloblasts of control molar after 12 days of organ culture (Fig. 4D right columns). We further investigated whether Dpysl4 promotes differentiation of primary molar epithelial cells. Overexpression of Dpysl4 induced Msx2 transcript expression in primary molar epithelial cells at a level that was increased approximately twofold in relation to mock transduced cells (Fig. 4E). These results suggest that Dpysl4 induces differentiation of the IEE to pre-ameloblast and regulates dental epithelial cell differentiation through the control of gene expression.
Dpysl4 is involved in the regulation of dental epithelial cell proliferation and differentiation. (A) Histological analysis using H&E staining and immunohistochemical analysis of Ki67 (red) expression of control (upper row) and shDpysl4 (lower row) molar germs cultured for 8 days. Higher magnification of the IEE area (second and third columns) and cervical loop area (fourth column) in the box is shown. Cell proliferation was analyzed immunohistochemically using a specific antibody against Ki67. Scale bars, 50 μm. (B) Cell proliferation of emtg2 cells after infection with Dpysl4 adenovirus for 3 days. Error bars show the standard deviation (n=3). ***p<0.0005. (C) In situ hybridization analysis of Fgf9 (third column), Msx2 (fourth column), Ambn (fifth column) and Notch1 (sixth column) expression in the IEE of infected molars at 8 days. Histological analysis of the infected molars is shown in the first column. Higher-magnification images of the IEE area (second columns) in the box are shown. Scale bar, 50 μm. (D) In situ hybridization analysis of Msx2 (second column), Ambn (third column) and Enam (fourth column) expression in the infected molar at 12 days. Histological analysis of the infected molars (first columns) is also shown. Scale bar, 50 μm. (E) Expression level of Msx2 mRNA was analyzed by real-time PCR. Error bars show the standard deviation (n=3). *p<0.05.
Discussion
In this study, we demonstrated that Dpysl4 is involved in enamel synthesis and secretion through the regulation of ameloblast differentiation. We also demonstrate that Dpysl4 regulates dental epithelial cell proliferation and is related to the regulation of pre-ameloblast differentiation of IEE cells. These findings suggest that Dpysl4 plays essential roles in dental epithelial cell differentiation through cytoskeletal rearrangement, and may be the regulator of cell polarization for epithelial differentiation.
It is thought that the negative regulation of epithelial cell proliferation plays important roles during tooth germ development in the formation of the enamel knot, crown morphogenesis and ameloblast differentiation [2, 27]. The inhibition of epithelial cell growth, which is regulated by p21 induced by mesenchymal BMP4 signaling, is essential for the formation of the enamel knot during the bud and cap stages [28]. Previously, we have shown that the spatiotemporal, negative regulation of the inner enamel epithelium within the prospective occlusal region is involved in determining the width of the molar crown [22]. Mathematical models that reproduce the morphology of mammalian teeth have also demonstrated that the inhibition of epithelial cell proliferation in the secondary enamel knots plays important roles in the regulation of cusp position and number [28, 29]. It has also been demonstrated that ameloblast differentiation is regulated by the suppression of ameloblast proliferation by Ambn through the expression of cyclin dependent kinase inhibitors such as p21 and p27 [30-32]. In the current study, we show that Dpysl4 is expressed during tooth germ development in growth inhibitory regions such as the enamel knot and the inner enamel epithelium. We also demonstrate that Dpysl4 is related to the negative regulation of dental epithelial cell growth. These results suggest that Dpysl4 is involved in a morpho-dynamic mechanism that regulates epithelial growth, suggesting a crucial role in tooth germ morphogenesis.
It is well known that epithelial cell polarity is essential for the intracellular trafficking and secretion of molecules such as signaling molecules or extracellular matrices [9, 11]. During tooth development, polarization of dental epithelial cells is crucial for the formation of the columnar structure of ameloblasts and secretion of the enamel matrix [8]. Sonic hedgehog has been shown to be involved in ameloblast polarization and organization according to the result of conditional inactivation in dental epithelial cells [33]. Smoothened (Smo), which is essential for all hedgehog-signaling and is a Frizzled family member, also is crucial for regulation of ameloblast polarization and differentiation as Smo mutants lose the accumulation of junctional complex protein, ZO-1 and cytoskeletal complex protein, β-tubulin and the expression of ameloblast marker genes [34]. It has also been reported that Msx2 controls terminal ameloblast differentiation through the maintenance of cell adhesion complexes of ameloblast by the induction of Laminin α5 expression in secretary ameloblast [6]. In the present study, we show that down-regulation of Dpysl4 results in inhibition of ameloblast differentiation and polarization. A previous study indicated that Dpysl4 plays a critical role in neuronal polarity during the formation of dendrite arborization and spine morphology in the hippocampus [19]. Furthermore, Dpysls regulated by Rho kinase can affect microtubule dimerization to induce neuronal polarity [15]. Thus, the present results suggest that promotion of ameloblast polarization via a Dpysl4 cascade plays an important role in tooth development.
Enamel matrix secretion has also been reported to be vital in amelogenesis, and the impairment of this ability causes amelogenesis imperfecta [8]. It has been reported that oculodentodigital dysplasia (ODDD) patients harboring autosomal-dominant mutations in Connexin43 exhibit hypoplastic enamel due to a severely disorganized ameloblast layer [35]. The genetic mutations of Family with sequence similarity 83, member H (Fam83h) and WD repeat domain 72 (Wdr72) are well known to cause enamel defects [36, 37]. Functional mutation of amelogenin (Amelx) has shown to inhibit the co-assembly of cytokeratin14 and AMELX, which promotes the translocation of AMELX to the apical region of ameloblasts at the site of secretion [38]. It has been suggested that tubulin rearrangement is essential for differentiation of ameloblasts and enamel formation during the maturation stage of ameloblasts [13, 14]. In the current study, abnormal cusp formation and the reduced accumulation of enamel were observed when shDpysl4 molars were ectopically transplanted into the subrenal capsule, and this effect is thought to act through the inhibition of ameloblast differentiation. These results suggest that the dysfunction of Dpysl4 may be a candidate for the cause of amelogenesis imperfecta.
In conclusion, this study demonstrates that Dpysl4 is involved in growth regulation, differentiation and polarization of dental IEE cells during tooth germ morphogenesis. Further studies of molecules that induce Dpysl4 expression or the molecular mechanisms such as signaling cascades and tubulin polymerization as a target of Dpysl4 and their associations with amelogenesis imperfecta will contribute to our understanding of amelogenesis during tooth germ morphogenesis.
Supplementary Material
Fig.S1 - S2.
Acknowledgements
This work was partially supported by a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Culture, Sports and Technology, Japan (T.T.).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev Biol. 2003;262:195-205
2. Thesleff I. Epithelial-mesenchymal signalling regulating tooth morphogenesis. J Cell Sci. 2003;116:1647-8
3. Tucker A, Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nature reviews Genetics. 2004;5:499-508 doi:10.1038/nrg1380
4. Mitsiadis TA, Graf D. Cell fate determination during tooth development and regeneration. Birth defects research Part C, Embryo today: reviews. 2009;87:199-211 doi:10.1002/bdrc.20160
5. Kettunen P, Thesleff I. Expression and function of FGFs-4, -8, and -9 suggest functional redundancy and repetitive use as epithelial signals during tooth morphogenesis. Dev Dyn. 1998;211:256-68 doi:10.1002/(SICI)1097-0177(199803)211:3<256::AID-AJA7>3.0.CO;2-G
6. Bei M, Stowell S, Maas R. Msx2 controls ameloblast terminal differentiation. Dev Dyn. 2004;231:758-65 doi:10.1002/dvdy.20182
7. Harada H, Ichimori Y, Yokohama-Tamaki T, Ohshima H, Kawano S, Katsube K. et al. Stratum intermedium lineage diverges from ameloblast lineage via Notch signaling. Biochem Biophys Res Commun. 2006;340:611-6 doi:10.1016/j.bbrc.2005.12.053
8. Bei M. Molecular genetics of ameloblast cell lineage. Journal of experimental zoology Part B, Molecular and developmental evolution. 2009;312B:437-44 doi:10.1002/jez.b.21261
9. Nelson WJ. Remodeling epithelial cell organization: transitions between front-rear and apical-basal polarity. Cold Spring Harbor perspectives in biology. 2009;1:a000513. doi:10.1101/cshperspect.a000513
10. Baum B, Georgiou M. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J Cell Biol. 2011;192:907-17 doi:10.1083/jcb.201009141
11. Datta A, Bryant DM, Mostov KE. Molecular regulation of lumen morphogenesis. Current biology: CB. 2011;21:R126-36 doi:10.1016/j.cub.2010.12.003
12. Inai T, Sengoku A, Hirose E, Iida H, Shibata Y. Differential expression of the tight junction proteins, claudin-1, claudin-4, occludin, ZO-1, and PAR3, in the ameloblasts of rat upper incisors. Anatomical record. 2008;291:577-85 doi:10.1002/ar.20683
13. Biz MT, Marques MR, Crema VO, Moriscot AS, dos Santos MF. GTPases RhoA and Rac1 are important for amelogenin and DSPP expression during differentiation of ameloblasts and odontoblasts. Cell Tissue Res. 2010;340:459-70 doi:10.1007/s00441-010-0961-0
14. Otsu K, Kishigami R, Fujiwara N, Ishizeki K, Harada H. Functional role of Rho-kinase in ameloblast differentiation. Journal of cellular physiology. 2011;226:2527-34 doi:10.1002/jcp.22597
15. Yamashita N, Goshima Y. Collapsin response mediator proteins regulate neuronal development and plasticity by switching their phosphorylation status. Molecular neurobiology. 2012;45:234-46 doi:10.1007/s12035-012-8242-4
16. Yoshimura T, Arimura N, Kaibuchi K. Molecular mechanisms of axon specification and neuronal disorders. Annals of the New York Academy of Sciences. 2006;1086:116-25 doi:10.1196/annals.1377.013
17. Arimura N, Menager C, Fukata Y, Kaibuchi K. Role of CRMP-2 in neuronal polarity. Journal of neurobiology. 2004;58:34-47 doi:10.1002/neu.10269
18. Fei T, Xia K, Li Z, Zhou B, Zhu S, Chen H. et al. Genome-wide mapping of SMAD target genes reveals the role of BMP signaling in embryonic stem cell fate determination. Genome research. 2010;20:36-44 doi:10.1101/gr.092114.109
19. Quach TT, Massicotte G, Belin MF, Honnorat J, Glasper ER, Devries AC. et al. CRMP3 is required for hippocampal CA1 dendritic organization and plasticity. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2008;22:401-9 doi:10.1096/fj.07-9012com
20. Ito T, Kagoshima M, Sasaki Y, Li C, Udaka N, Kitsukawa T. et al. Repulsive axon guidance molecule Sema3A inhibits branching morphogenesis of fetal mouse lung. Mech Dev. 2000;97:35-45
21. Yoneda A, Morgan-Fisher M, Wait R, Couchman JR, Wewer UM. A collapsin response mediator protein 2 isoform controls myosin II-mediated cell migration and matrix assembly by trapping ROCK II. Molecular and cellular biology. 2012;32:1788-804 doi:10.1128/MCB.06235-11
22. Ishida K, Murofushi M, Nakao K, Morita R, Ogawa M, Tsuji T. The regulation of tooth morphogenesis is associated with epithelial cell proliferation and the expression of Sonic hedgehog through epithelial-mesenchymal interactions. Biochem Biophys Res Commun. 2011;405:455-61 doi:10.1016/j.bbrc.2011.01.052
23. Oshima M, Mizuno M, Imamura A, Ogawa M, Yasukawa M, Yamazaki H. et al. Functional tooth regeneration using a bioengineered tooth unit as a mature organ replacement regenerative therapy. PloS one. 2011;6:e21531. doi:10.1371/journal.pone.0021531
24. Komine A, Suenaga M, Nakao K, Tsuji T, Tomooka Y. Tooth regeneration from newly established cell lines from a molar tooth germ epithelium. Biochem Biophys Res Commun. 2007;355:758-63 doi:10.1016/j.bbrc.2007.02.039
25. Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K. et al. Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Developmental cell. 2012;23:317-28 doi:10.1016/j.devcel.2012.05.012
26. Smith CE. Cellular and chemical events during enamel maturation. Critical reviews in oral biology and medicine: an official publication of the American Association of Oral Biologists. 1998;9:128-61
27. Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 2000;92:19-29
28. Jernvall J, Aberg T, Kettunen P, Keranen S, Thesleff I. The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development. 1998;125:161-9
29. Salazar-Ciudad I, Jernvall J. A gene network model accounting for development and evolution of mammalian teeth. Proc Natl Acad Sci U S A. 2002;99:8116-20 doi:10.1073/pnas.132069499
30. Casasco A, Calligaro A, Casasco M. Proliferative and functional stages of rat ameloblast differentiation as revealed by combined immunocytochemistry against enamel matrix proteins and bromodeoxyuridine. Cell Tissue Res. 1992;270:415-23
31. Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G. et al. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973-83 doi:10.1083/jcb.200409077
32. Sonoda A, Iwamoto T, Nakamura T, Fukumoto E, Yoshizaki K, Yamada A. et al. Critical role of heparin binding domains of ameloblastin for dental epithelium cell adhesion and ameloblastoma proliferation. The Journal of biological chemistry. 2009;284:27176-84 doi:10.1074/jbc.M109.033464
33. Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775-85
34. Gritli-Linde A, Bei M, Maas R, Zhang XM, Linde A, McMahon AP. Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development. 2002;129:5323-37
35. Toth K, Shao Q, Lorentz R, Laird DW. Decreased levels of Cx43 gap junctions result in ameloblast dysregulation and enamel hypoplasia in Gja1Jrt/+ mice. Journal of cellular physiology. 2010;223:601-9 doi:10.1002/jcp.22046
36. El-Sayed W, Parry DA, Shore RC, Ahmed M, Jafri H, Rashid Y. et al. Mutations in the beta propeller WDR72 cause autosomal-recessive hypomaturation amelogenesis imperfecta. American journal of human genetics. 2009;85:699-705 doi:10.1016/j.ajhg.2009.09.014
37. El-Sayed W, Shore RC, Parry DA, Inglehearn CF, Mighell AJ. Ultrastructural analyses of deciduous teeth affected by hypocalcified amelogenesis imperfecta from a family with a novel Y458X FAM83H nonsense mutation. Cells, tissues, organs. 2010;191:235-9 doi:10.1159/000252801
38. Ravindranath RM, Tam WY, Bringas P Jr, Santos V, Fincham AG. Amelogenin-cytokeratin 14 interaction in ameloblasts during enamel formation. The Journal of biological chemistry. 2001;276:36586-97 doi:10.1074/jbc.M104656200
Author contact
![]() Corresponding author: Research Institute for Science and Technology, Tokyo University of Science, Noda, Chiba, 278-8510, Japan. TEL: +81-4-7122-9711, FAX: +81-4-7122-1499 E-Mail: t-tsujinoda.tus.ac.jp.
Corresponding author: Research Institute for Science and Technology, Tokyo University of Science, Noda, Chiba, 278-8510, Japan. TEL: +81-4-7122-9711, FAX: +81-4-7122-1499 E-Mail: t-tsujinoda.tus.ac.jp.

 Global reach, higher impact
Global reach, higher impact