10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(4):391-402. doi:10.7150/ijbs.4729 This issue Cite
Research Paper
Modulation of Cell Cycle Progression by 5-Azacytidine Is Associated with Early Myogenesis Induction in Murine Myoblasts
1. Department of Biomedical Sciences for Health, University of Milan, Milan, Italy.
2. Metabolism Research Centre, San Donato Hospital and Scientific Institute, Milan, Italy.
3. Division of Metabolic and Cardiovascular Science. Metabolism, Nutrigenomics and Cellular Differentiation Unit, DIBIT-San Raffaele Scientific Institute, Milan, Italy.
Received 2012-6-12; Accepted 2013-2-19; Published 2013-4-27
Abstract
Myogenesis is a multistep process, in which myoblasts withdraw from the cell cycle, cease to divide, elongate and fuse to form multinucleated myotubes. Cell cycle transition is controlled by a family of cyclin-dependent protein kinases (CDKs) regulated by association with cyclins, negative regulatory subunits and phosphorylation. Muscle differentiation is orchestrated by myogenic regulatory factors (MRFs), such as MyoD and Myf-5. DNA methylation is crucial in transcriptional control of genes involved in myogenesis. Previous work has indicated that treatment of fibroblasts with the DNA-demethylating agent 5-azacytidine (AZA) promotes MyoD expression. We studied the effects of AZA on cell cycle regulation and MRFs synthesis during myoblast proliferation and early myogenesis phases in C2C12 cells. During the proliferation phase, cells were incubated in growth medium with 5µM AZA (GMAZA) or without AZA (GM) for 24 hours. At 70% confluence, cells were kept in growth medium in order to spontaneously achieve differentiation or transferred to differentiation medium with 5μM AZA (DMAZA) or without AZA (DM) for 12 and 24 hours. Cells used as control were unstimulated.
In the proliferation phase, AZA-treated cells seemed to lose their characteristic circular shape and become elongated. The presence of AZA resulted in significant increases in the protein contents of Cyclin-D (FC:1.23 GMAZA vs GM p≤0.05), p21 (FC: 1.23 GMAZA vs GM p≤0.05), Myf-5 (FC: 1.21 GMAZA vs GM p≤0.05) and MyoD (FC: 1.20 GMAZA vs GM p≤0.05). These results propose that AZA could inhibit cell proliferation.
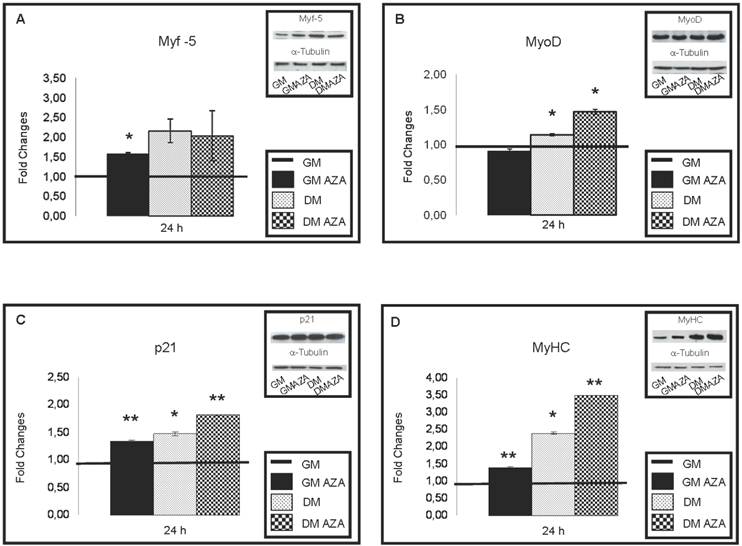
During 12 hours of differentiation, AZA decreased the downregulation of genes involved in cell cycle arrest and in restriction point (G1 and G1/S phase) and the expression of several cyclins, E2F Transcription Factors, cyclin-dependent kinase inhibitors, specific genes responsible of cell cycle negative regulation. During 24 hours of differentiation, AZA induced an increment in the protein expression of Myf-5 (FC: 1.57 GMAZA vs GM p≤0.05), MyoD (FC: 1.14 DM vs GM p≤0.05; FC: 1.47 DMAZA vs GM p≤0.05), p21 (FC: 1.36 GMAZA vs GM p≤0.01; FC: 1.49 DM vs GM p≤0.05; FC: 1.82 DMAZA vs GM p≤0.01) and MyHC (FC: 1.40 GMAZA vs GM p≤0.01; FC: 2.39 DM vs GM p≤0.05; FC: 3.51 DMAZA vs GM p≤0.01). Our results suggest that AZA-induced DNA demethylation can modulate cell cycle progression and enhance myogenesis. The effects of AZA may open novel clinical uses in the field of muscle injury research and treatment.
Keywords: cell cycle, DNA methylation, myogenic transcription factors, myogenic phenotype, myogenesis
Introduction
Skeletal muscle differentiation is a multistep process that involves two simultaneous phenomena. The first is the induction of muscle-specific gene expression by myogenic regulatory factors (MRFs) such as Myf-5, MyoD, MRF4 and Myogenin. The second is the commitment of myogenic cells to skeletal muscle through irreversible withdrawal from the cell cycle, inducing permanent G1 phase [1-4]. Achievement of these two pathways is required for the formation of multinucleated myotubes.
At the molecular level, several positive and negative cell cycle regulators have been identified [5]. Progression through the cell cycle phases is dependent on consecutive activation and inhibition of phosphoproteins by cyclin-dependent kinases (CDKs) complexed with their activatory cyclins. These complexes are in turn regulated by cyclin-dependent kinase inhibitors (CKIs) [5].
Moreover, the members of E2F family of transcription factors play a crucial role in the control of cell cycle progression and regulate the expression of genes required for the G1/S transition. These include genes encoding DNA replication proteins, enzymes involved in nucleotide synthesis and components of the origin recognition complex. E2F factors are controlled by a number of mechanisms including negative check of their activity by Retinoblastoma protein (Rb), an important negative regulator of G1 progression [6-8].
During muscle differentiation, MRFs are expressed in a defined sequence: MyoD and Myf-5 are primarily expressed in proliferating, undifferentiated cells, whereas Myogenin expression is only induced upon muscle differentiation [6,7,9,10].
MyoD can promote cell cycle arrest by activation of the cyclin-dependent kinase inhibitor p21 [11], which inhibits a wide range of CDKs essential for cell cycle progression [12]. It is generally accepted that activation of p21 follows Myogenin expression during myoblast differentiation and that a high level of p21 is required to maintain the postmitotic cellular state. MyoD can also induce expression of Rb [13-15].
Other molecular signaling pathways such as the Ca++/calmodulin-dependent transcriptional pathways should also be considered as potential mediators of muscle growth and muscle hypertrophy. In skeletal muscle, an increase in intracellular Ca++ levels not only results in contractile activity, but is also responsible for muscle-specific gene expression through activation of downstream transcriptional pathways [16].
Calmodulin is a ubiquitous Ca++-binding protein expressed in all eukaryotic cells where it is involved in a variety of signaling pathways in a Ca++-dependent manner. Changes in intracellular Ca++ concentrations regulate the physiological activities of calmodulin.
Ca++/calmodulin-dependent protein kinase II (CaMKII) is a multimeric serine/threonine kinase whose activity is detectable in skeletal muscle and requires binding of Ca++/calmodulin for its activation. Upon binding with Ca++/calmodulin, CaMKII is activated via autophosphorylation, then remains active independent of Ca++ levels. In the cytoplasm, where the activated CaMKII is localized, signaling pathways that are mainly involved in mitochondrial biogenesis and expression of contractile proteins are induced [16].
Increasing evidence indicates that DNA methylation is an important mechanism underlying the cascade of events that controls the process of muscle differentiation. The role of DNA methylation during myogenesis is not yet fully understood. Methylation of the 5´-position of cytosine in a CpG dinucleotide is an epigenetic mark commonly considered to mediate stable gene silencing [17-21], although the final effect strongly depends on CpG density.
The relationship between DNA methylation and the activation of the myogenic program was first established by the observation that treatment with the methyltransferase inhibitor, 5-aza-2´-deoxycytidine (AZA), converts C3H10T1/2 embryonic fibroblasts into muscle cells [18].
In particular, the majority of epigenetic studies have focused on the role of histone modifications in genome reprogramming during myogenesis [22], while the role of DNA methylation is less well established.
Data reported in literature are concerned with the role of DNA methylation in myogenic lineage acquisition [18-21,23,24] and MRFs expression [25-27].
Only in the last year, the study performed by Hupkes et al. [28] has demonstrated that genomic demethylation following treatment with AZA strongly enhances C2C12 maturity. This was the first work in which there was a global analysis of the effects of DNA demethylation during muscle differentiation (gene expression in myogenic differentiation, functional and contractile activity, calcium uptake, sarcomeric morphology and development). However, this work did not show any specific evidence of the role of DNA demethylation in cell cycle control, in myogenic regulator factors expression and in differentiation induction.
5-aza-2'-deoxycytidine (AZA) was originally developed as an antitumor agent. It has cytotoxic effects if used in pharmacological treatments but it can also induce differentiation in several experimental systems, most notably induction of muscle, adipocytes and chondrocytes in cultures of drug-treated mouse embryo fibroblasts [18,19]. The mechanisms underlying this drug-induced differentiation have been difficult to unravel [23]. As a demethylating agent, AZA has been extensively used in epigenetic research. It is incorporated into DNA where it binds DNA methyltransferase (DNMT) in an irreversible, covalent manner, thus sequestering the enzyme and preventing maintenance of the methylation state [29-31].
The immortalized murine C2C12 myoblastic cell line provides a good in vitro model for the major steps of myoblast proliferation and differentiation, being easily reproducible in cell culture [32-35].
In this cellular model, undifferentiated myoblasts are recognizable as flat, fusiform or star-shaped cells, which appear scattered on the substrate and are consistently mononucleated. After reaching confluence or 24 hours after serum removal, C2C12 cells are considered myoblasts in an early differentiation stage and they are characterized by a change in orientation, as well as by cell lengthening and thickening. Later, confluent mononucleated myocytes begin to fuse to form multinucleated myotubes (intermediate differentiation), which are positive for the characteristic muscle-specific protein MyHC (Myosin Heavy Chain). Myotubes become wider and longer over the next few days as additional myocytes fuse (late differentiation). Multinucleated and large myotubes appear to form a network with numerous nuclei arranged in multiple linear arrays (late differentiation). As occurs in vivo, C2C12 myoblastic differentiation is driven by activation of muscle-specific genes.
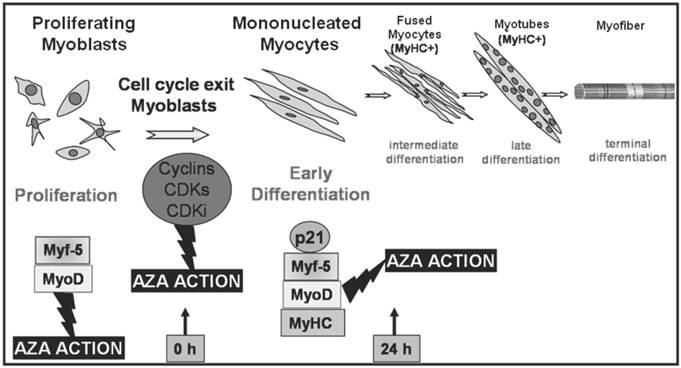
We used this cell line as a model for cell cycle withdrawal accompanying cellular differentiation. To gain further insight into the role of DNA methylation in cell cycle progression and in the induction of the process of muscle differentiation, we examined the action of AZA on the regulation of the cell cycle and MRFs synthesis during myoblast proliferation and early myogenesis phases in the C2C12 cell line (Fig. 1).
Materials and Methods
Materials
Mouse C2C12 myoblastic cells were purchased from the European Collection of Animal Cell Cultures (ECACC). Reagents were purchased from Sigma Chemical Co. (St. Louis, MO, U.S.A.) Primary antibodies : anti-MyoD (C-20), anti-Myf-5 (C-20), anti-MyHC (H-300), anti-p21 (C-19), anti-Cyclin B1 (H-433), anti-Cyclin D1 (C-20), anti α-Tubulin (TU-02) and anti-Calnexin (H-70). Monoclonal or polyclonal primary antibodies and peroxidase-conjugated secondary antibodies for Western Blot analysis and Rhodamine-conjugated antibodies for Immunofluorescence analysis were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.).
Experimental procedures
C2C12 skeletal myoblastic cells were routinely maintained at 37°C in a humidified atmosphere containing 5% CO2 in a growth medium comprising Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 20% (v/v) fetal bovine serum (FBS), 1% penicillin-streptomycin and 1% L-glutamine up to 70% of confluence. During this phase, representing in this study the proliferation phase, cells seeded at 6X102 cells/cm2 were maintained in mitogen-rich growth medium (GM) as single myoblasts. These proliferating cells were treated with AZA (5 μM) for 24 hours to induce DNA demethylation, or not treated with AZA.
The AZA concentration of 5 μM has been reported to be the optimal concentration to induce effects on the differentiation process without cytotoxicity [23,28], as validated in our dose-response experiments (data not shown).
Study plan. Simplified scheme of cell treatments and time points during the proliferation phase and early differentiation phase.
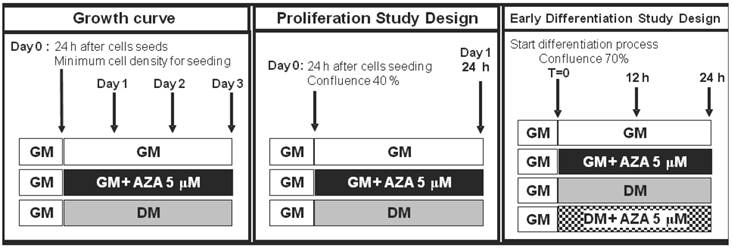
C2C12 myoblasts spontaneously fuse and differentiate into multinucleated myotubes as a result of both the achievement of confluence (as in the case of cells maintained in growth medium with 20% FBS: GM) and the removal of the serum growth factors (as in the case of cells transferred in DMEM supplemented with 1% Horse Serum: DM).
At 70% confluence cells were maintained in GM in order to spontaneously achieve differentiation or transferred in DM both and without AZA (GM,DM) or with 5 μM AZA (GMAZA, DMAZA) for 12 and 24 hours.
The medium was changed every day. Under these conditions myoblasts are known to exit the cell cycle and commit to the myogenic lineage [36].
To ensure reproducibility, for all experiments cultures were grown for a period no longer than 6 weeks after recovery from frozen stocks.
Growth curve
C2C12 myoblasts were plated on 60x15-mm culture dishes at 40% confluence and grown in the presence of GM supplemented with AZA (5 µM) or without AZA. The experiment was continued until subconfluence was achieved in the control cells (3 days), and the medium was changed every 24 hours. Daily during the culture period one cell plate per condition was trypsinized and the cells counted using a hemocytometer. The average number of cells for each day was then calculated and the values were used to plot growth curves for myoblasts treated with AZA and control cells without AZA (growth control GM and differentiation control DM). In addition, morphological changes were evaluated daily.
Electrophoretic techniques and immunoblotting analysis
C2C12 cells were homogenized in lysis buffer (50 mM Tris/HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM sodium orthovanadate (Na3VO4), 1 mM EDTA, 1 mM PMSF, 1 mg/ml aprotinin, 1 mg/ml leupeptin, 1 mg/ml pepstatin) and shaken for 1h at 4°C. Detergent-insoluble material was removed from the cell suspension by centrifugation at 12,000 x g for 30 min. Protein contents were determined using the Bradford method. The proteins in 30-µg aliquots of supernatant from the different samples were resolved by SDS-PAGE. Electrophoresed proteins were transferred to nitrocellulose membrane (Protran®, Whatman ® Schleicher & Schuell) as described previously [37]. The membranes were incubated with specific primary antibodies and then incubated with HRP-conjugated secondary antibodies. Immunoreactive bands were visualized by an enhanced chemiluminescence method (Amersham Pharmacia Biotech: Piscataway, NJ, USA). The membrane was stripped and reprobed with an antibody to calnexin or α-tubulin to confirm equal protein loading per sample. The immunoreactive bands were measured quantitatively by densitometric analysis using Scion Image Software (Scion Corporation, Frederick, MD, USA).
The data are presented as fold changes (FC) in relation to the controls.
Immunofluorescence analysis
For indirect Immunofluorescence, cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and blocked with phosphate-buffered saline containing 1% bovine serum albumin. Cells were then immunostained with Rhodamine-conjugated antibodies (Santa Cruz Biotechnology, California, USA) and the nuclei revealed with DAPI staining. Cells were viewed by fluorescence microscopy (Leica DM IRE2) and images of myotubes were captured using IM50 software (Leica Microsystems, Switzerland) for size comparison. Data were displayed and analyzed using Adobe® Photoshop® CS4.
RT-PCR array analysis
Gene expression levels were assessed by RT2 Profiler PCR Arrays RT-PCR system (SABiosciences Corporation, Frederick, MD 21703 USA). Cell Cycle RT2 Profiler PCR Arrays plates were used to analyze a panel of genes simultaneously. We studied the expression of the following genes during G1 phase and the G1/S transition: Calcium/Calmodulin-dependent protein kinase II alfa (Camk2a) and beta (Camk2b); Integrin beta 1 (Fibronectin receptor beta Itgb1); Nuclear Factor of activated-T cells, cytoplasmic, calcineurin-dependent 1 (Nfact1).
We also studied the expression of the following genes involved in the cell cycle checkpoint: Caspase 3 (Casp3); CDK5 regulatory subunit associated protein1 (Cdk5rap1); Cyclin-dependent kinase inhibitor 1a (Cdkn1a or p21) and Cyclin-dependent kinase inhibitor 1b (Cdkn1b); Growth arrest and DNA-damage-inducible 45 alpha (Gadd45a); Tumor susceptibility gene 101 (Tsg101); Cyclin A1 (Ccna1), Cyclin A2 (Ccna2), Cyclin B1 (Ccnb1) and Cyclin B2 (Ccnb2); E2F Transcription Factor E2f1, E2f2, E2f3, E2f4.
In addition, we evaluated the expression of the following genes implicated in negative regulation of the cell cycle: Retinoblastoma-like1 and Retinoblastoma-like 2 (Rbl1 and Rbl2); Transformation related protein 53 (Trp53).
GADPH and Actin housekeeping gene were included as endogenous normalization controls.
Total RNA was isolated from C2C12 cells using the RNeasy Plus Mini Qiagen kit (Qiagen, Germany) according to the manufacturer's instructions. Total RNA (1 µg) was reverse-transcribed using the RT2 First Strand kit (SABiosciences). The reverse transcripts were used as templates for analysis of gene expression level using Superarrays Cell Cycle RT2 - PCR Arrays RT2 - PCR Arrays plates according to the manufacturer's instructions. Each sample was run in triplicate. The expression level of the housekeeping genes chosen for normalization in the threshold cycle (Ct) for each experimental condition and then the fold change (ΔΔCt) for each gene for the treated group compared to the control group were calculated. If ΔΔCt is greater than 1, the result may be reported as a fold upregulation. If ΔΔCt is less than 1, the result may be reported as a fold downregulation.
Statistical analysis
All experiments were performed three times. For immunoblotting and immunofluorescence analysis, statistical evaluation was performed using an unpaired t-test. Data are presented as means ± SD. Results were considered statistically significant if p≤0.05.
Results
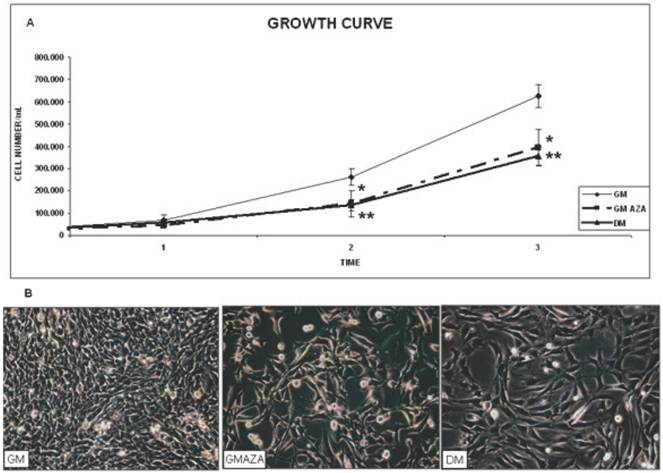
We first evaluated the effect of AZA-induced demethylation on the proliferation phase (Fig. 2A). The cell growth curves showed that C2C12 cells grew slowly during the first 2 days of culture, while cell numbers increased more rapidly during the third day. AZA treatment induced a decrease in cell division in relation to the untreated control (GM) cells (Day 2: GMAZA vs GM p≤0.05; DM vs GM p≤0.01. Day 3: GMAZA vs GM p≤0.05; DM vs GM p≤0.01). Cells treated with GMAZA showed a growth trend similar to cells maintained in DM medium, in respect to GM control cells, indicating the potential ability of AZA to modify cell growth, promoting proliferation arrest.
Very important support for these findings was the morphological change observed in AZA-treated cells during the proliferation phase: the cells appeared to lose their characteristic circular shape (Fig. 2B, GM) and become elongated (Fig. 2B, DM and GMAZA). Phase contrast images obtained on day 3 of proliferation confirmed comparable morphological changes in the DM and GMAZA conditions.
Taken together, these results suggest that AZA could be able to promote the induction of differentiation by decreasing the rate of cell growth and causing the acquisition of a specific myocyte phenotype.
The expression of Cyclin D is induced during differentiation [7,38,39], in contrast to most cyclins [7,40].
Growth curve and morphological study. A) C2C12 proliferation rate through kinetics of cell growth curve analysis. Proliferation curve shows that AZA treatment induces a decrease of cell division in respect to untreated control cells. (Day 2: GMAZA p≤0.05; DM p≤0.01; Day 3: GMAZA p≤0.05; DM p≤0.01 ). GMAZA cells reveal a growth trend similar to that of DM cells. Significance: * p≤0.05, ** p≤0.01. B) Evaluation of cells morphology. Morphological changes are apparent in DM cells and AZA-treated cells on day 3 during the proliferation phase. The cells have lost their characteristic circular shape and have become elongated in contrast to control cells (GM).
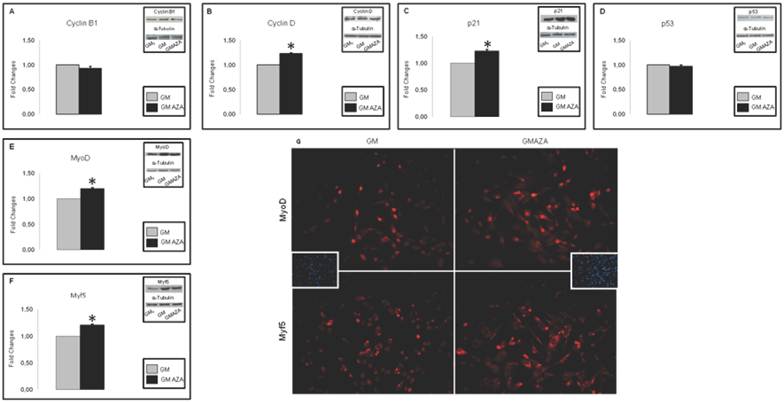
To determine AZA effects on regulators of cell cycle withdrawal during proliferation, we measured the protein contents of cell cycle regulators (Cyclin B1, Cyclin D, p21) and early myogenic regulatory factors (Myf-5 and MyoD). AZA stimuli resulted in significant increase in the protein amount of: Cyclin D (FC: 1.23 GMAZA vs GM p≤0.05), p21 (FC: 1.23 GMAZA vs GM p≤0.05), Myf-5 (FC: 1.21 GMAZA vs GM p≤0.05) and MyoD (FC: 1.20 GMAZA vs GM p≤0.05). However, in the presence of AZA there was no significant detectable rise in the levels of Cyclin B (Fig. 3A,B,C,E,F).
To confirm the absence of an AZA-mediated apoptotic action on proliferating C2C12 cells, we measured p53 protein levels. The p53 regulator is known to play a key role in growth arrest and apoptosis after cell stress [41]. AZA-treated cells showed a p53 protein content comparable to that of GM cells (Fig. 3D).
As shown in DAPI Immunofluorescence images (Fig. 3G), obtained during the first day of growth curve, after 24 hours of proliferation, cells densities in the GM and GMAZA conditions were superimposable. This finding is additional proof of the absence of an AZA-mediated apoptotic action.
Fig. 3G shows Immunofluorescence images of Myf-5 and MyoD expression in AZA-treated cells obtained after 24 h of proliferation during the first day of growth curve and at 40% cell confluence.
The AZA-treated cells validate the morphological changes previously described.
The GMAZA treatment revealed a major number of cell positive for MyoD and Myf-5 in respect to the GM control, and this could indicate that the differentiation process began earlier than in the untreated cells (GM).
Given the effect of AZA on the proliferation phase, we wanted to study its effects on the subsequent early stage of differentiation (12-24 hours after differentiation induction, Fig. 1).
To verify the hypothetical role of AZA on global cell cycle gene expression, we performed quantitative RT-PCR 12 hours after the start of differentiation in C2C12 cells (Fig. 1), in both GM and DM conditions. We wanted to analyze the action of AZA in terms of the number of cell cycle genes regulated over time. Fig. 4A shows that in AZA-treated cells a larger number of cell cycle genes were regulated than in untreated cells.
Key cyclins and myogenic protein levels in proliferation phase (Day 1, 24 h proliferation, 40% cell confluence). A) Protein levels of cell cycle regulators during proliferation phase. In GMAZA conditions the graphs show a negligible effect on Cyclin B protein levels. B) Protein levels of cell cycle regulators during proliferation phase. In GMAZA conditions the graphs illustrate a significant increase in Cyclin D protein amount (p≤0,05) in respect to control (GM). C) Protein levels of cell cycle regulators during proliferation phase. GMAZA conditions shows a significant rise in p21 protein content in respect to GM (p≤0,05). D) Apoptotic marker p53 protein levels during proliferation phase. The graph shows no differences between GMAZA treatment and control. E) Protein levels of early MRFs during proliferation phase. AZA treatment seems to cause a significant raise in MyoD protein levels in respect to control (p≤0,05). F) Protein levels of early MRFs during proliferation phase. AZA significant increases Myf5 protein levels in respect to control (p≤0,05). G) Myf-5 and MyoD immunofluorescence images in proliferation phase. The images show a major density of MyoD (up panel) and Myf-5 (down panel) positive cells (red) in GMAZA condition respect to control (GM). Representative DAPI images of GM and GMAZA conditions during the proliferation phase (fist day of growth curve, 24 h proliferation) are also shown. A-F Insets representative immunoblots of analyzed proteins.
Cell cycle RT-PCR ARRAY analysis (Early Differentiation, 12 h after the start of differentiation). A) Global gene expression analysis in early differentiation. Each area in the graphic represents the number of genes regulated in time after 12 hours of differentiation induction. In AZA-treated cells several cell cycle genes are regulated compared to untreated cells. B) Expression analysis of cell cycle regulating genes in early differentiation (12 hours). The figure shows the expression analysis of genes involved in cell cycle and arrest restriction point. AZA treatment decreases the downregulation of these genes promoting myoblast differentiation. C) Expression analysis of Cyclins and E2F Transcription Factors genes in early differentiation (12 hours). The figure shows the expression analysis of Cyclins and E2F Transcription Factors genes. AZA treatment slightly decreases the downregulation of these important regulators of cell cycle progression. D) Expression analysis of cell cycle negative regulators genes in early differentiation (12 hours). Array panel shows the significant AZA-induced downregulation decrease in the expression of specific genes responsible of negative regulation of cell cycle.
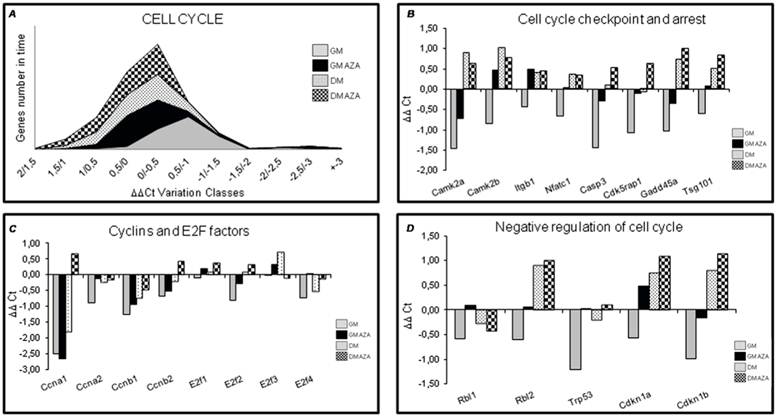
In detail, RT-PCR illustrated that AZA decreased the downregulation of genes involved in cell cycle arrest including Caspase 3 (Casp3), Cdk5 regulatory subunit associated protein1 (Cdk5rap1), Growth arrest and DNA-damage inducible 45 alpha (Gadd45a), Tumor susceptibility gene 101 (Tsg101). This effect was particularly relevant for genes modulating the cell cycle restriction point (G1 and G1/S phase) when cells are committed to continue in the cell cycle (proliferate) or exit from the cell cycle (differentiate) [41]. AZA also decreased the downregulation of Calcium/Calmodulin-dependent protein kinase 2 alpha (CamK2a), Calcium/Calmodulin-dependent protein kinase 2 beta (CamK2b), Integrin beta 1 (Itgb1), Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (Nfact 1) genes too (Fig. 4B).
In order to obtain supplementary information relating to cell cycle progression, we analyzed gene expression of some important cyclins and E2F Transcription Factors.
The expression of most cyclins appeared to be rapidly suppressed with the onset of differentiation when cells are blocked in G1 phase [7,40]. Fig. 4C shows that AZA induced a slight reduction in the downregulation of the expression of several cyclins (Ccna1, Ccna2, Ccnb1, Ccnb2) and E2f 1,2,3,4 Transcription Factors.
The expression of positive regulators of cell cycle progression, Cyclins and Cyclin-dependent kinase (CDKs), is regulated at several levels during early differentiation. In particular, expression of some CDKs drops during differentiation [7,42,43].
In contrast, the expression levels of Cyclin-dependent kinase inhibitors (CKIs) important for in vitro and in vivo myoblast differentiation, increase during differentiation [7,44,45].
AZA decreased the downregulation of Cyclin-dependent kinase inhibitor 1A (Cdkn1A) also known as p21, and of Cyclin-dependent kinase inhibitor 1B (Cdkn1b) (Fig. 4D).
A close association between cell cycle arrest and induction of myogenic differentiation has also been shown by the activity of proliferative negative regulators. In particular, data in the literature emphasize the mutual activity of p21 and Rb in early differentiation that maintains a postmitotic state by ensuring the absence of DNA replication [46].
Furthermore, Fig. 4D shows that AZA treatment decreased the downregulation of specific genes that negatively regulate the cell cycle including Retinoblastoma-like 1 (Rbl1), Retinoblastoma-like 2 (Rbl2), Transformation related protein 53 (Trp53).
These data suggest that AZA could have a negative effect on cell cycle progression promoting cell cycle arrest.
In order to further validate our hypothesis, we performed Western Blot analysis in C2C12 cells after 24 hours of differentiation, in both GM and DM (Fig. 1). We evaluated the expression levels of key proteins involved in cell cycle progression and early differentiation.
Differentiation process requires the upregulation of MyoD and Myf-5 [2,6,7,9,10].
As shown in Fig. 5A, only GMAZA condition showed a significant increase in Myf-5 protein content (FC: 1.57 GMAZA vs GM p≤0.05). DM and DMAZA conditions did not show significant increases in Myf-5 expression. In contrast to the expression of Myf-5, significantly higher levels of MyoD expression were seen in DM and DMAZA conditions in respect to GM (FC: 1.14 DM vs GM p≤0.05; FC: 1.47 DMAZA vs GM p≤0.05). In fact at 24 hours the level of expression of MyoD was delayed compared to that of Myf-5 and was not significantly different in GM and GMAZA, but occurred earlier in DM and DMAZA conditions in which differentiation was accelerated.
Increases in p21 protein levels were seen after 24 hours of differentiation (Fig. 5C) (FC: 1.36 GMAZA vs GM p≤0.01; FC: 1.49 DM vs GM p≤0.05; FC: 1.82 DMAZA vs GM p≤0.01).
As expected, a substantial raise in the amount of MyHC was seen in AZA-treated cells (FC: 1.40 GMAZA vs GM p≤0.01; FC: 2.39 DM vs GM p≤0.05; FC: 3.51 DMAZA vs GM p≤0.01) confirming the role of AZA on the myogenic process.
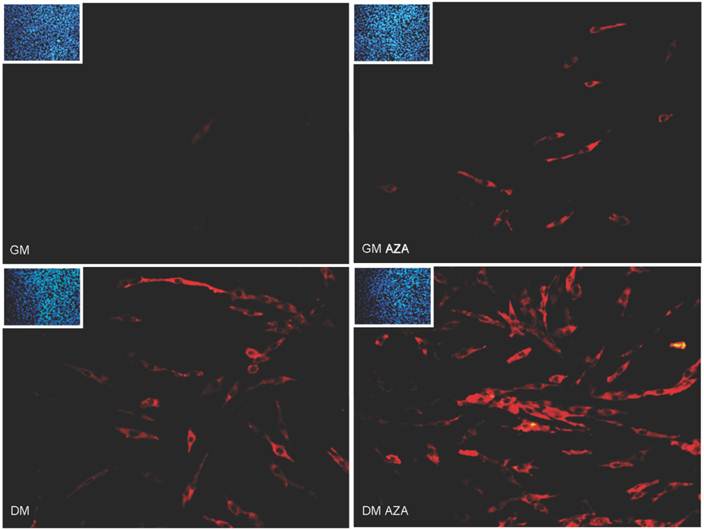
Finally, Immunofluorescence analysis after 24 hours of differentiation (Fig. 6) revealed the presence of a major number of MyHC-positive cells in GMAZA and DMAZA conditions in contrast to the number in untreated cells (GM and DM). This effect was not caused by differences in cell density, since DAPI imaging showed that cell numbers under all conditions were superimposable.
Morphological changes in AZA-treated C2C12 cells were observed (Fig. 6).
Key myogenic protein expression in early differentiation (24 h after differentiation induction). A) Immunoblot shows that only GMAZA condition showed a significant increase in Myf-5 protein content (GMAZA vs GM p≤0.05). DM and DMAZA conditions did not show a significant increment of Myf-5 expression. B) The bar graph shows a significant higher level of MyoD expression in DM and DMAZA conditions respect to GM (DM vs GM p≤0.05; DMAZA vs GM p≤0.05). C) Immunoblot showed the increase of p21 protein level at 24 hours of differentiation (GMAZA vs GM p≤0.01; DM vs GM p≤0.05; DMAZA vs GM p≤0.01). D) Immunoblot showed an important increment in MyHC content in AZA-treated cells (GMAZA vs GM p≤0.01; DM vs GM p≤0.05; DMAZA vs GM p≤0.01). Representative immunoblots of analyzed proteins are shown on the graphic. Significance: * p≤0.05, ** p≤0.01.
MyHC immunofluorescence analysis in early differentiation (24 hours after differentiation start). Immunofluorescence analysis at 24 hours of differentiation reveals the presence of a major number of MyHC positive cells (red) in GMAZA and DMAZA conditions in respect to untreated cells (DM and GM). Moreover, in GMAZA and DM conditions myoblasts are becoming elongated in respect to GM. This behavior is particularly noticeable in DMAZA condition, in which confluent mononucleated myocites began to fuse to form multinucleated myotubes. Insets representative DAPI images of all conditions collected at 24 hours of early differentiation were added to confirm comparable cell densities.
Transcription factors regulating different stages of myogenesis and AZA action. Schematic illustration of skeletal muscle differentiation and AZA effects. The committed myoblasts exit cell cycle, cease to divide and begin to elongate (early differentiation). In a second stage mononucleated myocytes begin to fuse forming multinucleated myocites (intermediate differentiation) which become wider and longer and start to organize into multinucleate myotubes (late differentiation). Cell cycle withdrawal and each phase of differentiation is orchestrated by activation of specific Cyclins, CDKs, CDKi and muscle regulatory factors which lead to the induction of specific muscle proteins as shown in the lower portion of the figure. During the proliferation phase, AZA could cause a cut in cell division and a decreased growth trend. Furthermore, AZA seems to anticipate early MRFs protein expression. During early differentiation AZA may also be able to induce protein expression of early MRFs, p21 and muscle specific protein MyHC, marker of differentiation.
In the AZA and DM conditions myoblasts started to become elongated in contrast to their shape in GM. This was particularly evident in DMAZA condition, in which confluent mononucleated myocytes began to fuse to form multinucleated myotubes.
Taken together, these results appear to support our hypothesis that, in vitro, AZA causes cell cycle withdrawal and enhances myogenic progression in C2C12 cells.
Discussion
Myogenesis is a multistep process orchestrated by an ordered and sequential expression of muscle regulatory protein and transcription factors. The muscle-specific gene program is governed by a combination of genetic and epigenetic events that affect the satellite cells switching them from a quiescent to an activated state [1-4,6,7,9,10].
An epigenetic mechanism is DNA methylation, considered a mark leading to gene silencing and acting as a switch capable of turning on or off the expression of genes regulating all cellular processes including development and differentiation [17,47,48].
It is well known that DNA methylation and myogenesis are linked [22,27,28].
The first evidence that DNA demethylation plays a role in myogenesis regulation was the finding that muscle differentiation is induced in fibroblast treated with AZA, a potent demethylating agent [18,49].
Our previous results indicate that DNA hypomethylation induces the activation of factors promoting myoblast proliferation and differentiation and then muscle growth [50].
The effects of AZA on changes in gene expression patterns are due to its ability to inhibit DNA methyltransferases. For this reason we chose to study the effects of AZA on C2C12 myoblasts that are a good model of muscle differentiation.
Proliferation and differentiation are mutually exclusive processes in myogenesis.
Proliferation of undifferentiated myoblasts ceases in response to the downregulation of Cyclin D1 expression, thus promoting cell cycle exit and allowing muscle differentiation to start [7].
Investigating the effect of AZA on the growth capacity of C2C12 myoblasts, we observed the ability of this DNA-demethylating agent to reduce cell proliferation leading to cell cycle arrest, as occurs with growth factors removal (DM condition). In support of this, cell growth curves showed the ability of AZA to interrupt proliferation. The demethylation effects were visible not only in the kinetics of cell growth, but also in the morphological analysis: AZA-treated cells appeared to lose their original circular shape to achieve a new, specific, elongate morphology, typical of muscle phenotype. Morphological changes observed in DM and GMAZA treated cells were superimposable showing that, under these two conditions, differentiation was faster than in the control condition, in which differentiation was determined only by cell contact. This suggests that the presence of AZA may favor more rapid differentiation.
Since activation of the muscle differentiation program requires irreversible cell cycle withdrawal and tissue-specific gene expression, we then investigated the effect of AZA on the exit of C2C12 myoblast from the cycle. Our results confirmed that the expression level of genes including p21, required to maintain the postmitotic cellular state, and Cyclin D, whose expression is induced during differentiation, increased after AZA treatment.
Similarly, the expression levels of MyoD and Myf-5 transcription factors, myogenic markers already expressed in undifferentiated proliferating myoblasts, were increased after AZA treatment.
Also during the early phase of differentiation, AZA was found to have a negative effect on cell cycle progression, promoting the arrest of cell proliferation. In fact, AZA stimuli seem to cause a considerable downregulation of the expression of genes implicated in cell cycle checkpoint, arrest and negative control.
The effects of AZA on mRNA expression of genes involved in cell cycle arrest was also confirmed by determining the protein content of cell cycle regulators and early myogenic regulatory factors.
Our results demonstrated that only GMAZA condition showed a significant increase in Myf-5 protein content. DM and DMAZA conditions did not show a significant increase in Myf-5 expression probably because the removal of serum growth factors and AZA treatment, which accelerated the differentiation process, led to Myf-5 expression earlier than in GM. There is also some evidence that Myf-5 expression upregulates the expression of MyoD, driving exit from the cell cycle towards myogenic differentiation.
This could explain the significantly higher level of MyoD expression, in contrast to Myf-5 expression, in DM and DMAZA conditions in respect to GM. In fact at 24 h its expression, which was delayed compared to that of Myf-5, was not significantly different in GM and GMAZA, but occurred earlier in DM and DMAZA, in which differentiation is accelerated.
These data support our hypothesis that AZA induces the expression of genes related to muscle differentiation at an early time point, when cell fusion has not yet taken place.
Surprisingly p21 protein levels and MyHC content showed an important rise after 24 hours of differentiation, occurring earlier than MyoD expression.
Such an event may be explained by the possibility that AZA treatment induces the expression of genes essential to cell cycle arrest and the drive towards differentiation, without following the ordered and sequential expression of the muscle-specific gene program. In relation to our findings, this could mean that p21, principal factor responsible for cell cycle withdrawal, and MyHC, the first marker of muscle phenotypic differentiation, are AZA-specific targets. If this hypothesis will be confirmed, AZA would represent a topic drug for the treatment of disease in which p21 or MyHC expression are directly compromised (some forms of myopathy and hypertrophic cardiomyopathy).
Moreover, AZA seemed to induce cell morphological changes during early differentiation phases and demethylation could have positive effects on MRFs and muscle-specific MyHC protein expression during acquisition of the commitment to the myogenic lineage.
These results suggested that AZA not only suppresses proliferation but also causes elongation and fusion of myoblasts.
The study of MRFs activity regulation may have important implications in understanding the mechanisms underlying several myopathies and regulating muscle growth and tissue regeneration. Our results seem to confirm that AZA-induced demethylation is involved in myoblast determination after specific control of cell cycle progression.
In summary, our findings suggest that the sequence of events following treatment with AZA results in improved maturity of C2C12 myotubes. First, this DNA methylation inhibitor promotes an increased expression of checkpoint genes involved in cell cycle progression and arrest, and upregulation of key myogenic transcription factors.
The present results may propose novel clinical uses for AZA which is presently used in the clinic for its antineoplastic effects only. Our in vitro studies may constitute novel proof of principle for the potential use of the compound in the treatment of serious clinical conditions characterized by reduced or functionally impaired muscle mass for its ability to promote the formation of new muscle.
Author contributions
Conception and design of the experiments: AM, LL, PS, IT. Performance of the experiments: AM, LL, PS, IT. Analysis of the data: AM, LL, PS, IT. Writing of the paper: AM, LL, PS, IT.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Stockdale FE. Myogenic cell lineage. Dev Biol. Dec. 1992;154(2):284-98
2. Perry RL, Rudnick MA. Molecular mechanism regulating myogenic determination and differentiation. Front Biosci Sep. 2000;5:D750-67
3. Rescan PY. Regulation and functions of myogenic regulatory factors in lower vertebrates. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2001;130(1):1-12
4. McKinsey TA, Zhang CL, Olson EN. Control of muscle development by duelling HATs and HDACs. Curr Opin Genet Dev. 2001;11(5):497-504
5. Kitzmann M. and Fernandez A. Crosstalk between cell cycle regulators and the myogenic factor MyoD in skeletal myoblasts. Cellular and Molecular Life Sciences. 2001;58:571-579
6. Joulia D, Bernardi H, Garandel V, Rabenoelina F, Vernus B, Cabello G. Mechanisms involved in the inhibition of myoblasts proliferation and differentiation by myostatin. Experimental Cell Research. 2003;286:263-275
7. James DR Knight and Rashmi Kothary. The myogenic kinome: protein kinases critical to mammalian skeletal myogenesis. Skeletal Muscle. 2011;1:29
8. Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr. Opin. Gent. 1998;8(1):28-35
9. Hans-Henning Arnold and Barbara Winter. Muscle differentiation: more complexity to the network of myogenic regulators. Current Opinion in Genetics e Development. 1998;8:539-544
10. Shen X, Collier JM, Hlaing M, Zhang L, Delshad EH. et al. Genome-wide examination of myoblast cell cycle withdrawal during differentiation. Developmental dynamics. 2003;226:128-138
11. Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J. et al. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267(5200):1018-21
12. Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501-12
13. Ostrovsky O, Bengal E. The mitogen-activated protein kinase cascade promotes myoblast cell survival by stabilizing the cyclin-dependent kinase inhibitor, p21WAF1 protein. J Biol Chem. 2003;278(23):21221-31
14. Martelli F, Cenciarelli C, Santarelli G, Polikar B, Felsani A, Caruso M. MyoD induces retinoblastoma gene expression during myogenic differentiation. Oncogene. 1994;9(12):3579-90
15. Kitzmann M, Carnac G, Vandromme M, Primig M, Lamb NJ, Fernandez A. The muscle regulatory factors MyoD and Myf-5 undergo distinct cell cycle-specific expression in muscle cells. J Cell Biol. 1998;142:1447-1459
16. Nasser Al-Shanti, Claire E. Stewart. Ca2+/Calmodulin-dependent transcriptional pathways: potential mediators of skeletal muscle growth and development. Biological Reviews. 2009;84:637-652
17. Klose R.J. and Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31(2):89-97
18. Taylor S. M. and Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17(4):771-9
19. Nakatsuka R, Nozaki T, Uemura Y, Matsuoka Y, Sasaki Y. et al. 5-Aza-2'-deoxycytidine treatment induces skeletal myogenic differentiation of mouse dental pulp stem cells. Arch Oral Biol. 2010;55(5):350-7
20. Jost JP, Oakeley EJ, Zhu B, Benjamin D, Thiry S. et al. 5-Methylcytosine DNA glycosylase participates in the genome-wide loss of DNA methylation occurring during mouse myoblast differentiation. Nucleic Acids Res. 2001;29(21):4452-61
21. Aguirre-Arteta A. M, Grunewald I, Cardoso MC, Leonhardt H. Expression of an alternative Dnm1 isoform during muscle differentiation. Cell Growth Diff. 2000;11(10):551-9
22. Perdiguero E, Sousa-Victor P, Ballestar E, Muñoz-Cánoves P. Epigenetic regulation of myogenesis. Epigenetics. 2009;4(8):541-50
23. Taylor SM. 5-Aza-2'-deoxycytidine: cell differentiation and DNA methylation. Leukemia. 1993;7(Suppl 1):3-8
24. Palacios D, Puri PL. The epigenetic network regulating muscle development and regeneration. J Cell Physiol. 2006;207(1):1-11
25. Lucarelli M, Fuso A, Strom R, Scarpa S. The dynamics of myogenin site-specific demethylation is strongly correlated with its expression and with muscle differentiation. J Biol Chem. 2001;276(10):7500-6
26. Wakitani S, Saito T, Caplan AI. Myogenic cells derived from rat bone marrow mesenchymal stem cells exposed to 5-azacytidine. Muscle & Nerve. 1995;18(12):1417-26
27. Scarpa S, Lucarelli M, Palitti F, Carotti D. Strom R. Simultaneous myogenin expression and overall DNA hypomethylation promote in vitro myoblast differentiation. Cell Growth Differ. 1996;7(8):1051-8
28. Hupkes M, Jonsson MK, Scheenen WJ, Van Rotterdam W, Sotoca AM. et al. Epigenetics: DNA demethylation promotes skeletal myotube maturation. FASEB J. 2011;25(11):3861-72
29. Fruhwald M. C. and Plass. Global and gene-specific methylation patterns in cancer: aspects of tumor biology and clinical potential. Mol. Genet Metab. 2002;75:1-16
30. Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res. 1998;72:141-196
31. Haaf T. The effects of 5-azacytidine and 5-azadeoxycytidine on chromosome structure and function: implications for methylation-associated cellular processes. Pharmacol. Ther. 1995;65:19-46
32. Yamaguchi A. Regulation of differentiation pathway of skeletal mesenchymal cells in cell lines by transforming growth factor-beta superfamily. Semin Cell Biol. 1995;6(3):165-73
33. Cossins J, Vernon AE, Zhang Y, Philpott A, Jones PH. Hes6 regulates myogenic differentiation. Development. 2002;129(9):2195-207
34. Burattini S, Ferri P, Battistelli M, Curci R, Luchetti F, Falcieri E. C2C12 murine myoblasts as a model of skeletal muscle development: morpho-functional characterization. Eur J Histochem. 2004;48(3):223-33
35. Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725-727
36. S.B.P. Chargè and M.A. Rudnicki. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209-238
37. Terruzzi I, Senesi P, Magni C, Montesano A, Scarafoni A. et al. Insulin-mimetic action of Conglutin-γ, a lupin seed protein, in mouse myoblasts. Nutrition, Metabolism and Cardiovascular Disease. 2010;21(3):197-205
38. Rao SS. And Kohtz DS. Positive and negative regulation of D-type cyclin expression in skeletal myoblasts by basic fibroblast growth factor and transforming growth factor beta. A role for cyclin D1 in control of myoblast differentiation. J Biol Chem. 1995;270(8):4093-100
39. Bartkova J, Lukas J, Strauss M. and Bartek J. Cyclin D3: requirement for G1/S transition and high abundance in quiescent tissues suggest a dual role in proliferation and differentiation. Oncogene. 1998;17(8):1027-37
40. Wang J. and Nadal-Girard B. Regulation of cyclins and p34CDC2 expression during terminal differentiation of C2C12 myocytes. Biochem Biophys Res Commun. 1995;206(1):82-8
41. Pietenpol JA, Stewart ZA. Cell cycle checkpoint signaling: Cell cycle arrest versus apoptosis. Toxicology. 2002;181-182:475-481
42. Kitzmann M, Vandromme M, Schaeffer V, Carnac G, Labbè JC. et al. cdk1-and cdk2-mediated phosphorylation of MyoD Ser200 in growing C2 myoblasts: role in modulating MyoD half-life and myogenic activity. Mol Cell Biol. 1999;19(4):3167-76
43. Guo K. and Walsh K. Inhibition of myogenesis by multiple cyclin-Ckd complexes. Coordinate regulation of myogenesis and cell cycle activity at the level of E2F. J Biol Chem. 1997;272(2):791-7
44. Reynaud EG, Pelpel K, Guiller M, Leibovitch MP, Leibovitch SA. p57 (Kip2) stabilizes the MyoD protein by inhbiting cyclin E-Cdk2 kinase activity in growing myoblasts. Mol Cell Biol. 1999;19(11):7621-9
45. Chu CY, Lim RW. Involvement of p27 (Kip1) and cyclin D3 in the regulation of cdk2 activity during skeletal muscle differentiation. Biochim Biophys Acta. 2000;1497(2):175-85
46. Mal A, Chattopadhyay D, Ghosh MK, Poon RY, Hunter T. and Harter ML. p21 and retinoblastoma protein control the absence of DNA replication in terminally differentiated muscle cells. J Cell Biol. 2000;149(2):281-92
47. Geiman TM, Muegge K. DNA methylation in early development. Mol Reprod Dev. 2010;77(2):105-113
48. Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55(3):451-8
49. Taylor SM. and Jones PA. Changes in phenotypic expression in embryonic and adult cells treated with 5-azacytidine. J Cell Physiol. 1982;111(2):187-94
50. Terruzzi I, Senesi P, Montesano A, La Torre A, Alberti G. et al. Genetic polymorphisms of the enzymes involved in DNA methylation and synthesis in elite athletes. Physiol Genomics. 2011;43(16):965-73
Author contact
![]() Corresponding author: Ileana Terruzzi. Division of Metabolic and Cardiovascular Science. Metabolism, Nutrigenomics and Cellular Differentiation Unit. DIBIT-San Raffaele Scientific Institute, Milan, Italy. e-mail: terruzzi.ileanait
Corresponding author: Ileana Terruzzi. Division of Metabolic and Cardiovascular Science. Metabolism, Nutrigenomics and Cellular Differentiation Unit. DIBIT-San Raffaele Scientific Institute, Milan, Italy. e-mail: terruzzi.ileanait

 Global reach, higher impact
Global reach, higher impact