10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(5):481-495. doi:10.7150/ijbs.6109 This issue Cite
Research Paper
Candidate Chemosensory Genes in the Stemborer Sesamia nonagrioides
1. INRA, UMR 1272, Physiologie de l'Insecte : Signalisation et Communication, route de Saint-Cyr, F-78026 Versailles cedex, France;
2. IRD, UR 072, c/o CNRS, Laboratoire Evolution, Génomes et Spéciation, 91198 Gif-sur-Yvette Cedex, France;
3. IRISA, équipe GenScale, Campus universitaire de Beaulieu, 35042 Rennes cedex, France;
4. UPMC - Université Paris 6, UMR 1272 Physiologie de l'Insecte : Signalisation et Communication, 7 quai Saint-Bernard, F-75252 Paris cedex 05, France;
5. Université Paris-Sud 11, 91405 Orsay Cedex, France;
6. CNRS UPR9034, Laboratoire Evolution, Génomes et Spéciation, 91198 Gif-sur-Yvette Cedex, France.
7. IRD, UR 072, c/o ICIPE, NSBB Project, PO Box 30772-00100, Nairobi, Kenya.
Received 2013-3-20; Accepted 2013-5-2; Published 2013-5-21
Abstract
The stemborer Sesamia nonagrioides is an important pest of maize in the Mediterranean Basin. Like other moths, this noctuid uses its chemosensory system to efficiently interact with its environment. However, very little is known on the molecular mechanisms that underlie chemosensation in this species. Here, we used next-generation sequencing (454 and Illumina) on different tissues from adult and larvae, including chemosensory organs and female ovipositors, to describe the chemosensory transcriptome of S. nonagrioides and identify key molecular components of the pheromone production and detection systems. We identified a total of 68 candidate chemosensory genes in this species, including 31 candidate binding-proteins and 23 chemosensory receptors. In particular, we retrieved the three co-receptors Orco, IR25a and IR8a necessary for chemosensory receptor functioning. Focusing on the pheromonal communication system, we identified a new pheromone-binding protein in this species, four candidate pheromone receptors and 12 carboxylesterases as candidate acetate degrading enzymes. In addition, we identified enzymes putatively involved in S. nonagrioides pheromone biosynthesis, including a ∆11-desaturase and different acetyltransferases and reductases. RNAseq analyses and RT-PCR were combined to profile gene expression in different tissues. This study constitutes the first large scale description of chemosensory genes in S. nonagrioides.
Keywords: chemosensory receptors, pheromone biosynthesis, transcriptome, next-generation sequencing, Sesamia nonagrioides.
Introduction
The stemborer Sesamia nonagrioides Lefebvre (Lepidoptera: Noctuidae) is a polyphagous species with a fairly wide range of host plants. It is an important pest of maize in the Mediterranean Basin [1-3]. Pest management strategies are being developed against this species, including olfaction-mediated behaviour modification and the use of tachinids [4, 5]. A better knowledge of the molecular mechanisms of S. nonagrioides olfaction will contribute to the development of new tools for the control of this species since inhibition/artificial activation of the proteins implicated in the olfactory process would lead to the disruption of its chemical communication.
The molecular mechanisms of olfaction in insects are complex and rely on the intervention of a diversity of proteins expressed in the chemosensory sensilla that cover the sensory organs. These sensilla house olfactory sensory neurons (OSNs) whose dendrites are bathed in the sensillum lymph. Secreted proteins are found in abundance in the lymph, notably the odorant-binding proteins (OBPs) and the chemosensory proteins (CSPs). These soluble proteins are proposed to bind odorant molecules and to transport them to membrane bound receptors [6, 7]. Within moth OBPs, the pheromone-binding proteins (PBPs) are proposed to be specialized in binding pheromone components [8]. Although the role of OBPs and especially PBPs in olfaction is now well admitted, the exact function of CSPs remains unclear. Two families of volatile molecule receptors have been described in insects, the olfactory receptors (ORs) and the ionotropic receptors (IRs), these two types being involved in the recognition of different volatile families as demonstrated in Drosophila melanogaster [9]. Most OSNs express OR proteins, which have seven transmembrane domains with an inverted topology compared to vertebrate ORs: their N-terminus is located inside the cell [10]. A co-receptor highly conserved among species, named Orco [10-12], is required to form with ORs a complex proposed to function as an ion channel and which makes possible the detection of volatile compounds [13, 14]. A subset of OSNs expresses IRs, which may have evolved from ionotropic glutamate receptors to gain chemosensory function [15, 16]. Like ORs, IRs couple with obligate co-receptors that are highly conserved among insects [17]. Other protein families have been described in insect antennae, such as the sensory neuron membrane proteins (SNMP). One of these SNMPs, SNMP1, is located in the dendritic membrane of pheromone-specific OSNs and is thought to trigger ligand delivery to the receptor [18, 19]. Numerous enzymes are also found in antennae. Depending on their catalytic activities, they have been proposed to be involved in xenobiotic degradation and/or olfactory signal termination, via transformation of the odorant molecules [6].
Despite the economical importance of S. nonagrioides, only fragmentary information is available on the molecular actors used by this species for odorant detection. Only two PBPs and two other OBPs have been described by molecular cloning and proteomic analyses [20-22], one candidate pheromone-degrading esterase has been cloned [23] and no candidate ORs have been identified.
In this study, we applied a transcriptomic approach to identify a large array of candidate chemosensory genes in S. nonagrioides. Such transcriptomic approaches have been proven to be efficient in identifying large repertoires of chemosensory genes in insect species for which the genome is not sequenced, for example in the Lepidoptera Spodoptera littoralis [24, 25], Manduca sexta [26], Cydia Pomonella [27] and Helicoverpa armigera [28]. In particular, such approaches appeared to be efficient for the identification of candidate ORs. Indeed, the low level of sequence identity (20-40%) of ORs within insects precluded most attempts to identify new ORs by homology cloning, except for Orco [29] and for more conserved receptors involved in pheromone detection - the so-called pheromone receptors (PRs) [30-32]. Here, using next-generation sequencing technologies (NGS), we characterized transcripts produced in various tissues of S. nonagrioides, including the chemosensory organs of larvae and adults, and female ovipositors, these last being known to express some chemosensory genes [33]. Among the transcripts, we identified genes encoding binding proteins (OBPs & CSPs) and chemosensory receptors (IRs & ORs). Carboxylesterases were also annotated as candidate pheromone-degrading enzymes (PDEs) since the S. nonagrioides pheromone contains acetates [34, 35]. Focusing on the pheromonal detection system, we not only identified an additional pheromone-binding protein, but we also report the identification of four candidate pheromone receptors and two SNMPs. In addition, the inclusion of RNAs from female ovipositors that contain the pheromone glands allowed us to annotate candidate enzymes involved in pheromone biosynthesis, such as desaturases, acetyltransferases and reductases.
Materials and Methods
Insect rearing, tissue preparation, 454 and Illumina sequencing
S. nonagrioides were reared in the laboratory on a modified artificial diet from Poitout & Bues [36], containing agar, maize flour, wheat germ, dried yeast and a mixture of vitamins and antibiotics. The insects were kept in a controlled chamber at 24.4 ± 0.7°C, 54.4 ± 5.8% r.h. (means ± SD) and an L16:D8 reversed photoperiod. For transcriptome sequencing, antennae were dissected from 1-day-old adults (males and females) and antennae and maxillary palps were dissected from 4th instar larvae. Other tissues (adult brains and female ovipositors) were also prepared from the same animals to enrich the S. nonagrioides transcriptome. All dissected organs were immediately frozen in liquid nitrogen, and stored at -80°C until extraction. Total RNAs were extracted from each tissue using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA). RNA integrity was verified by gel electrophoresis and RNA quantity was determined on a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA). A pool of RNAs from each tissue (1 μg each) was used as a template for cDNA synthesis and 454 sequencing (454 Roche GS FLX Titanium, ½ Pico Titer Plate GATC Biotech SARL, Mulhouse, France). In parallel, RNAs from larvae antennae and palps, female ovipositors and female antennae were independently used as templates for Illumina sequencing (one channel for the two adult samples, one channel for the larvae sample, single read 51 pb lenght, HighSeq2000; GATC Biotech). All the data generated in this project have been deposited in LepidoDB (http://www.inra.fr/lepidodb/sesamia_nonagrioides), a centralized bioinformatic resource for the genomics of lepidopteran pests [37]. As a result, from the project page http://www.inra.fr/lepidodb/sesamia_nonagrioide, one can retrieve the whole sequence set.
Sequence processing and assembly
454 and Illumina data were processed by removing adapters and by trimming low quality regions. Data were first analyzed with FastQC v. 0.10.0 (www.bioinformatics.babraham.ac.uk/projects/fastqc) that provided information on sequence quality and identified over-represented sequences within libraries. Over-represented sequences and low quality regions were removed with, respectively, Cutadapt [38] and PRINSEQ v 0.17.3. [39]. Sequences shorter than 20 bp long were also removed from all data sets. A first step of de novo assembly was performed on the Illumina reads with Trinity assembler (release 2012-01-25 [40]). Then, the processed sequences from 454 sequencing were added to the Trinity contig set and used as input in MIRA assembler v 3.2.1. using as parameters de novo assembly method, est assembly type, accurate quality, Sanger sequencing technology [41].
Transcriptome analyses and gene annotation
The contigs were compared to the NCBI non redundant protein database (NR, version January 20th 2013) using BLASTX, with a 1e-8 value threshold. BLAST2GO was used for the Gene Ontology (GO) annotation (GO association done by a BLAST against the NCBI NR database) [42]. Contigs were translated to peptides using FrameDP 1.2.0 [43] with three training iterations and using Swissprot (398.181, August 2009) as the reference protein database. GO annotation was then completed with Interproscan annotation of translated peptides. Olfactory gene transcripts were searched within the assembly with available lepidopteran OBP, CSP, OR, IR and SNMP amino acid sequences (see Phylogenetic analyses) as queries using TBLASTN. Enzyme-encoding genes were searched using carboxylesterase sequences (for candidate pheromone-degrading enzymes) and desaturase/acetyltransferase/reductase sequences (for pheromone biosynthesis enzymes) from Lepidoptera [44, 45]. The sequences that matched with the queries were further assembled using Cap3 [46], when possible, to obtain longer contigs. Resulting sequences were reversely compared to NCBI NR database using the BLASTX application to confirm annotation and their translation was manually verified or corrected. OBPs, CSPs and esterases were searched for the presence of a signal peptide using SignalP 4.0 [47], secondary structures were predicted using the Psipred server [48], and logos were generated using WebLogo [49]. Transmembrane domains of candidate ORs were predicted using the HMMTOP 2.0 [50].
Phylogenetic analyses
In addition to the 13 candidate S. nonagrioides OR sequences (SnonORs), the OR data set contained amino acid sequences from the moths Bombyx mori [51], Heliothis virescens [52, 53], S. littoralis [24, 25, 54] and M. sexta [26], and also from the butterflies Danaus plexippus [55] and Heliconius melpomene [56]. In total, the data set contained 248 sequences (343 amino acid positions for each). Only complete or nearly complete sequences were included in this data set, except some SnonOR sequences that were short but kept, which may affect the accuracy of the phylogenetic analysis. The OBP data set contained 12 amino acid sequences from S. nonagrioides, together with sequences from S. littoralis [24, 25, 54], B. mori [57], H. melpomene [56], H. virescens [45, 58, 59] and M. sexta [26, 60]. Signal peptide sequences were removed from the data set, which contained 182 sequences (256 amino acid positions for each). The CSP data set contained 19 sequences from S. nonagrioides and sequences from S. littoralis [24, 25, 54], B. mori [61], H. melpomene [56], H. virescens [62] and Papilio xuthus [63]. As for OBPs, signal peptide sequences were removed. The data set contained 124 sequences (103 amino acid positions for each). In the IR dataset, 10 S. nonagrioides candidate IR sequences were added to sequences identified in S. littoralis [54, 64], B. mori [16], and D. plexippus [55]. Since IRs are well conserved in insects, IR sequences from non-Lepidoptera species (D. melanogaster, Apis mellifera and Tribolium castaneum [16]) were also included in the data set. In addition, D. melanogaster iGluR sequences were included, and the final data set contained 179 sequences (618 amino acid positions for each). Amino acid sequences were aligned with MAFFT v.6 [65] using the FFT-NS-2 algorithm and default parameters, except for the OR sequences that were aligned using MUSCLE [66] as implemented in Seaview v.4 [67]. The alignments were manually curated to remove highly divergent regions. Phylogenetic reconstructions were carried out using maximum likelihood. For each data set, the LG+I+G substitution model [68] was determined as the best-fit model of protein evolution by ProtTest 1.3 [69] following Akaike information criterion. Rate heterogeneity was set at four categories, and the gamma distribution parameter and the proportion of invariable sites were estimated from the data set. Tree reconstruction was performed using PhyML 3.0 [70], with both SPR (Subtree Pruning and Regrafting) and NNI (Nearest Neighbour Interchange) methods for tree topology improvement. Branch support was estimated by approximate likelihood-ratio test (aLRT) [71]. We considered a branch was supported when the aLRT value was >0.95. Images were created using the iTOL web server [72].
Read mapping
All candidate chemosensory genes as well as the genes encoding candidate enzymes were used to perform unique read mapping of each Illumina library (female ovipositors, female antennae, larval palps and antennae). Each of the 85 individual gene mapping alignments was inspected for uniquely mapped reads. Read counts were normalized between libraries according to the size of the library with the DESeq package [73] implemented in R [74].
RT-PCR
Male and female antennae were dissected form of 1-3 day-old adults and total RNAs were extracted from both tissues using TRIzol® Reagent (Invitrogen). After a DNase I treatment (Promega, Madison, WI, USA), RNAs (0.5 to 1 μg) were used as templates for single stranded cDNA synthesis using the Advantage RT-for-PCR kit (Clontech, Mountain View, USA). PCRs were performed on the two tissues under the following conditions: 94 °C for 1 min, 35 cycles of (94 °C for 30 s, 57-67 °C - depending on primer pairs - for 30 s, 72 °C for 3 min) and 72 °C for 10 min as a final extension step, using Titanium Taq DNA polymerase (Clontech) and with specific primer pairs designed for the S. nonagrioides ORs (Table 1) using the Primer3+ software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). For each tissue, the ribosomal protein L8 gene (rpL8) was used as a positive control. Negative controls consisted of amplifications run on DNase-treated RNAs and water templates. The amplification products were loaded on 1.5% agarose gels and visualized using ethidium bromide. For each gene, at least one amplification product was verified by DNA sequencing (Biofidal, Vaulx-en-Velin, France) after gel extraction (Qiagen, Hilden, Germany).
Results and discussion
S. nonagrioides reference transcriptome and annotation
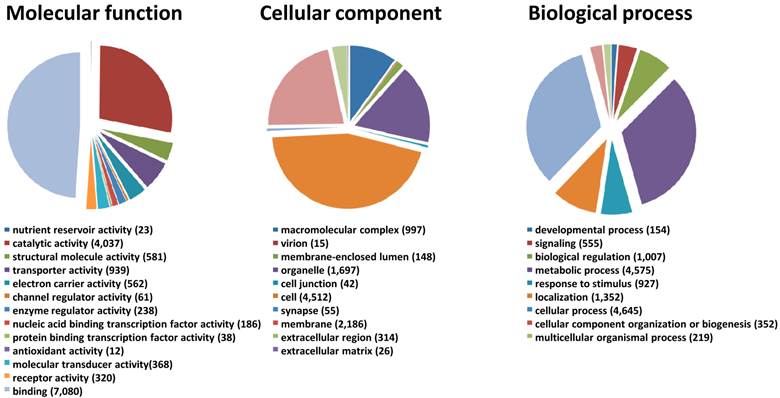
We generated a de novo transcriptome of S. nonagrioides using transcriptomic data sets obtained from 454 and Illumina sequencing. These sequencing platforms were combined since they lead to substantial differences in read length and they are supposed to recover different sequence types from a sample, thus we expected their combination to enhance the quality of the final assembly. In addition, the availability of Illumina libraries from isolated organs allows performing read mapping for expression studies. The 454 data set (995,424 processed sequences) was obtained from adult (male and female) and larvae antennae, larvae palps, adult brains and female ovipositors (1,004,420 raw reads) (Table 2). The Illumina data sets were obtained from female antennae (53,623,491 processed sequences), female ovipositors (71,544,332 processed sequences) and pooled larvae antennae and palps (190,673,453 processed sequences) (Table 2). We focused the Illumina sequencing on adult female and larval tissues to highlight genes putatively involved in host plant sensing, since male behaviors are mainly driven by the sex pheromone. The Illumina reads were assembled using Trinity, which generated a first assembly into 85,833 contigs. Then, the 995,424 processed sequences from 454 sequencing were added to the Trinity contigs to generate, using MIRA, a final assembly of 51,999 contigs (length from 40 to 29,697 bp, N50 : 1,741bp) which forms the final transcriptome of S. nonagrioides (Table 2). It has to be noticed that these contigs do not represent unigenes, since their assembly include possible splice variants, polymorphism or reverse transcriptase errors. Among these 51,999 contigs, a coding region could be predicted for 22,153 sequences (42.6%), and 16,280 predicted proteins (73.5%) translated from these regions showed similarity to known proteins when compared to the non-redundant protein database. Figure 1 represents the distribution of the S. nonagrioides contigs in GO terms. Among the 51,999 contigs, 11,369 (21.8%) corresponded to at least one GO term. As observed in other lepidopteran transcriptomes [25, 26], a large number of transcripts could not be associated with a GO term (78.2%). Among those associated to a GO-term, 9,961 were assigned to a molecular function (87.6%), 6,635 to a putative biological process (58.4%) and 4,836 to a cellular component (42.5%) (Figure 1). In the molecular function category, the terms “binding” and “catalytic activity” were the most represented (respectively 62.3% and 35.5%), as previously observed in the transcriptomes of S. littoralis [25] and M. sexta [26]. In the biological process category, the terms “metabolic process” and “cellular process” were the most represented (35.2 % and 40.9 % respectively). In the cellular component category, the terms “cell” and “membrane” were the most represented (39.7 % and 19.2 % respectively).
Primer pairs used in RT-PCR experiments, annealing temperature (T°), and amplification product sizes.
| Name | Forward primer | Reverse primer | T° | Size |
|---|---|---|---|---|
| SnonOrco | CATCACCGTGCTCTTCTTCA | GATGCTGCAGCTGTTCACAT | 60 | 467 |
| SnonOR6 | CTTACGTTTCACGCTGGTCA | TCGAGTTTTGGAGACCATCC | 60 | 479 |
| SnonOR10 | GGCCACATCCGAATAACTAC | GCTGATGTAGATGCTGACCA | 67 | 485 |
| SnonOR14 | TCCTGTGTTCGACGACTTTCT | CGTAAACGGCATCCTTCAAT | 60 | 471 |
| SnonOR15 | TTATTCAGCCGGGAACTACG | CGTCGTCATTTGTGAGCACT | 64 | 496 |
| SnonOR16 | ATATGGGCACGTTGAAGGAG | CAATCGCTTGATGGTGTTTG | 60 | 484 |
| SnonOR17 | CTGGTACCCCTTCGACAAGA | TCCCATTGTGCACTCAAAAA | 62 | 466 |
| SnonOR22 | CCACAGTTGCGGATTTTTCT | AATGGTCGCTTGGTGTTCTC | 60 | 473 |
| SnonOR33 | CAAGCTTTCCAGGAGATTCG | GGGAATCCACCAGATGAAGA | 60 | 484 |
| SnonOR45 | TCTACTGTCGAACGGAACCA | AGACGCGTATTCTCGACCAA | 60 | 461 |
Summary of data used for transcriptome assembly.
| Sequencing technology | 454 sequencing | Illumina sequencing | ||
|---|---|---|---|---|
| Tissues | Male and female antennae, adult and larval brains, female ovipositors, larval antennae and plaps | Larvae antennae and palps | Female antennae | Female ovipositors |
| Raw sequence number | 1,004,420 | 190,697,894 | 81,527,205 | 114,270,344 |
| Processed sequence number | 995,424 | 190,673,453 | 53,623,491 | 71,544,332 |
| Assembly | 51,999 contigs (N50: 1,741 bp) | |||
Distribution of S. nonagrioides contigs annotated at GO level 2.
Identification of putative S. nonagrioides odorant-binding proteins and chemosensory proteins
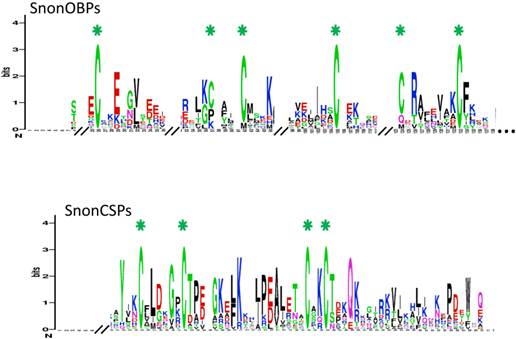
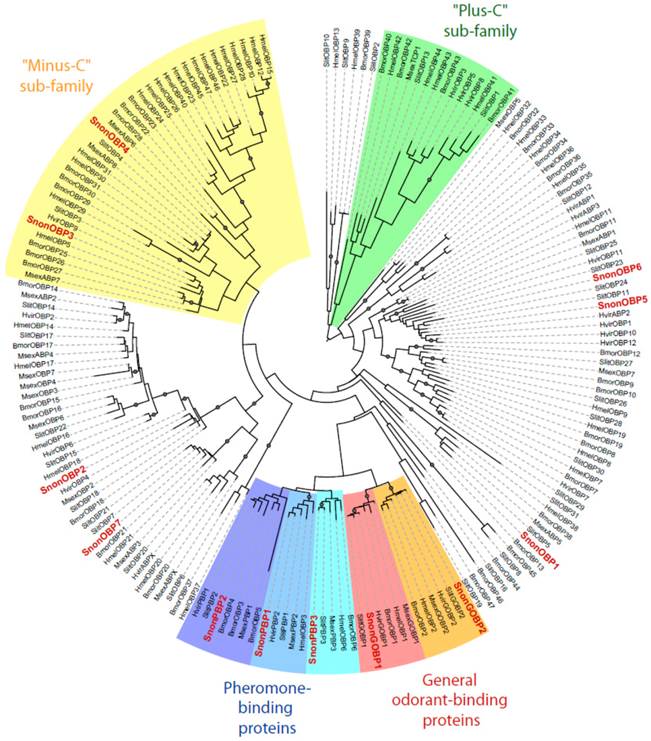
A total of 12 candidate OBPs and 19 candidate CSPs could be deduced from the analysis of the S. nonagrioides transcriptome. They are further referred to as SnonOBPs and SnonCSPs (Table 3, fasta format file in Supplementary material S1). Almost all the full-length predicted proteins have the characteristic hallmarks of the OBP and CSP protein families: the presence of a signal peptide and the highly conserved six (OBPs) and four (CSPs) cysteine profiles (Table 3, Figure 2). In Lepidoptera, the OBP family is characterized by the presence of three PBP lineages and two general odorant-binding protein (GOBP) lineages, GOBPs being proposed to bind “general” odours, such as plant odours [8]. Among the S. nonagrioides OBPs, we found the two previously cloned SnonPBP1 and SnonPBP2 [20] and we identified a third candidate PBP, named SnonPBP3, that clustered in the third lepidopteran PBP lineage (Figure 3). We also re-identified the previously described GOBP2 [21] and could extend the partial SnonGOBP1 sequence previously identified by proteomics [22]. Each of these SnonGOBPs clustered in one of the two lepidopteran GOBP lineages (Figure 3). Both GOBPs exhibited the highest numbers of mapped female antennal reads (Table 4), suggesting they are the most abundantly expressed OBPs in female antennae. They may thus be essential for female olfactory behaviours. In correlation with their cysteine number, some of the SnonOBPs clustered in the “minus-C” sub-family whereas no SnonOBP could be identified in the “plus-C” OBP sub-family (Figure 3). Some of the sequences were incomplete at their 5' ends and the corresponding proteins missed the signal peptide (Table 3). Although we likely identified the complete repertoires of PBPs and GOBPs in S. nonagrioides, the number of SnonOBPs is low compared to what has been identified in other species via genome or transcriptome analyses and additional sequencing would be needed to obtain the complete repertoire. For instance, 36 and 18 candidate OBPs have been identified in the S. littoralis [25, 54] and the M. sexta transcriptomes [26], respectively, and 44 OBPs were annotated in the genome of B. mori [57]. The 19 CSPs identified in this study may represent the nearly complete set of S. nonagrioides CSPs. For comparison, 18 putative CSPs have been annotated in B. mori [61], 21 in S. littoralis [25, 54] and 21 in M. sexta [26] (Phylogenetic analysis visible in Supporting information S2). These data confirm that Lepidoptera express a higher number of CSPs than other insect orders, such as Diptera [75]. The OBP and CSP transcripts showed diverse expression patterns, as revealed by Illumina read mapping (Table 4). The CSP family groups soluble proteins expressed in a diversity of tissues and whose function is unclear [7]. Here, the investigation of only a limited number of tissues does not allow us to propose possible functions for CSPs. However, one can note that a CSP, SnonCSP11, was highly expressed in the larval chemosensory organs and that some CSPs were clearly expressed in the ovipositors, as previously reported in another noctuid [76] (Table 4). More interesting features could be noticed for OBPs. Most of them, including the two GOBPs and the three PBPs, were observed to be highly expressed in female antennae. Some of them were also highly expressed in the larval chemosensory organs (Table 4), some others were not, and it could be speculated that these latter participate in adult specific behaviors. Interestingly, four OBPs were clearly expressed in the female ovipositors and 32 reads from the ovipositor library could be mapped on one PBP (Table 4).
List of S. nonagrioides contigs putatively involved in odorant binding. Signal peptides were determined using SignalP 4.0 [47] and α-helice structures were predicted using the Psipred server [48].
| Name | Length (amino acids) | Peptide signal | α-helice nb | C nb | BlastP hit | e-value |
|---|---|---|---|---|---|---|
| SnonGOBP1 | 163 | yes | 9 | 7 | gb|ABI24159.1| general odorant binding protein 1, partial [Agrotis segetum] | 1e-94 |
| SnonGOBP2 | 162 | yes | 8 | 6 | gb|AFM36760.1| general odorant-binding protein 2 [Agrotis ipsilon] | 2e-106 |
| SnonOBP1 | 145 | yes | 7 | 4 | gb|AEB54592.1| OBP9 [Helicoverpa armigera] | 9e-36 |
| SnonOBP2 | 139 | yes | 7 | 6 | gb|AEB54589.1| OBP8 [Helicoverpa armigera] | 6e-69 |
| SnonOBP3 | 139 | yes | 7 | 4 | gb|ACX53795.1| odorant binding protein [Heliothis virescens] | 3e-53 |
| SnonOBP4 | 133 | yes | 7 | 4 | gb|AFI57166.1| odorant-binding protein 17 [Helicoverpa armigera] | 6e-80 |
| SnonOBP5 | 147 | yes | 7 | 6 | gb|AAL66739.1|AF461143_1 pheromone binding protein 4 [Mamestra brassicae] | 6e-81 |
| SnonOBP6 | 150 | yes | 7 | 5 | gb|AEB54581.1| OBP5 [Helicoverpa armigera] | 8e-55 |
| SnonOBP7 | 141 | yes | 7 | 7 | gb|AFD34173.1| odorant binding protein 5 [Argyresthia conjugella] | 9e-62 |
| SnonPBP1 | 165 | yes | 8 | 6 | gb|AAS49922.1| pheromone binding protein 1 precursor [Sesamia nonagrioides] | 3e-117 |
| SnonPBP2 | 170 | yes | 8 | 6 | gb|AAS49923.1| pheromone binding protein 2 precursor [Sesamia nonagrioides] | 1e-116 |
| SnonPBP3 | 164 | yes | 8 | 6 | gb|AEQ30020.1| pheromone binding protein 3 [Sesamia inferens] | 1e-112 |
| SnonCSP1 | 128 | yes | 7 | 5 | gb|ACX53804.1| chemosensory protein [Heliothis virescens] | 2e-63 |
| SnonCSP2 | 120 | yes | 7 | 4 | gb|ACX53800.1| chemosensory protein [Heliothis virescens] | 1e-59 |
| SnonCSP3 | 122 | yes | 7 | 4 | gb|AFR92094.1| chemosensory protein 10 [Helicoverpa armigera] | 3e-76 |
| SnonCSP4 | 146 | yes | 8 | 4 | gb|ABM67686.1| chemosensory protein CSP1 [Plutella xylostella] | 2e-51 |
| SnonCSP5 | 127 | yes | 7 | 4 | gb|ABM67688.1| chemosensory protein CSP1 [Spodoptera exigua] | 9e-69 |
| SnonCSP6 | 123 | yes | 7 | 4 | gb|ACX53806.1| chemosensory protein [Heliothis virescens] | 2e-66 |
| SnonCSP7 | 121 | yes | 6 | 5 | gb|EHJ67380.1| chemosensory protein [Danaus plexippus] | 3e-48 |
| SnonCSP8 | 128 | yes | 7 | 4 | gb|AAF71290.2|AF255919_1 chemosensory protein [Mamestra brassicae] | 1e-72 |
| SnonCSP9 | 124 | yes | 7 | 4 | gb|ABM92663.1| chemosensory protein CSP3 [Plutella xylostella] | 8e-46 |
| SnonCSP10 | 235 | no | 6 | 4 | emb|CAJ01506.1| hypothetical protein [Manduca sexta] | 1e-69 |
| SnonCSP11 | 125 | yes | 7 | 4 | ref|NP_001037066.1| chemosensory protein precursor [Bombyx mori] | 7e-38 |
| SnonCSP12 | 120 | yes | 7 | 4 | gb|ACX53817.1| chemosensory protein [Heliothis virescens] | 6e-45 |
| SnonCSP13 | 122 | yes | 7 | 4 | gb|AEX07267.1| CSP6 [Helicoverpa armigera] | 4e-71 |
| SnonCSP14 | 127 | yes | 7 | 4 | gb|AAM77040.1| chemosensory protein 2 [Heliothis virescens] | 1e-69 |
| SnonCSP15 | 131 | no | 7 | 5 | ref|NP_001091781.1| chemosensory protein 15 [Bombyx mori] | 4e-41 |
| SnonCSP16 | 81 | no | 5 | 4 | gb|EHJ67380.1| chemosensory protein [Danaus plexippus] | 5e-39 |
| SnonCSP17 | 123 | yes | 7 | 4 | dbj|BAM20381.1| unknown secreted protein [Papilio polytes] | 5e-59 |
| SnonCSP18 | 122 | yes | 7 | 4 | dbj|BAG71920.1| chemosensory protein 12 [Papilio xuthus] | 7e-38 |
| SnonCSP19 | 124 | yes | 7 | 4 | dbj|BAF91716.1| chemosensory protein [Papilio xuthus] | 2e-50 |
Comparison of chemosensory gene expression in different tissues (female antennae, larval antennae and palps, female ovipositors) as revealed by Illumina read mapping. In each box, the number of uniquely mapped reads is indicated (read counts were normalized between libraries according to the size of the library with the DESeq package [75]). Color scales were established for each gene family using the conditional formatting option in Excel (dark red: max. value, blue: min. value).

SnonOBP and CSP sequence logos. Degree of amino acid sequence conservation [49] along the primary sequence axis of odorant-binding proteins (OBPs) and the chemosensory proteins (CSPs) of S. nonagrioides. Depicted amino acid character size correlates to relative conservation across aligned sequences. Green asterisks indicate the conserved six and four cysteine motifs of OBPs and CSPs, respectively.
Maximum likelihood tree of candidate odorant-binding proteins (OBPs) from S. nonagrioides and other Lepidoptera. Sequences used were from B. mori [57], S. littoralis [24, 25, 54], H. melpomene [56], H. virescens [45, 58, 59] and M. sexta [26, 60]. Signal peptide sequences were removed from the data set. Branch support was estimated by approximate likelihood-ratio test (aLRT) (circles: >0.95) [71]. Images were created using the iTOL web server [85]. The SnonOBPs identified in this study are in red.
Identification of putative S. nonagrioides chemosensory membrane proteins
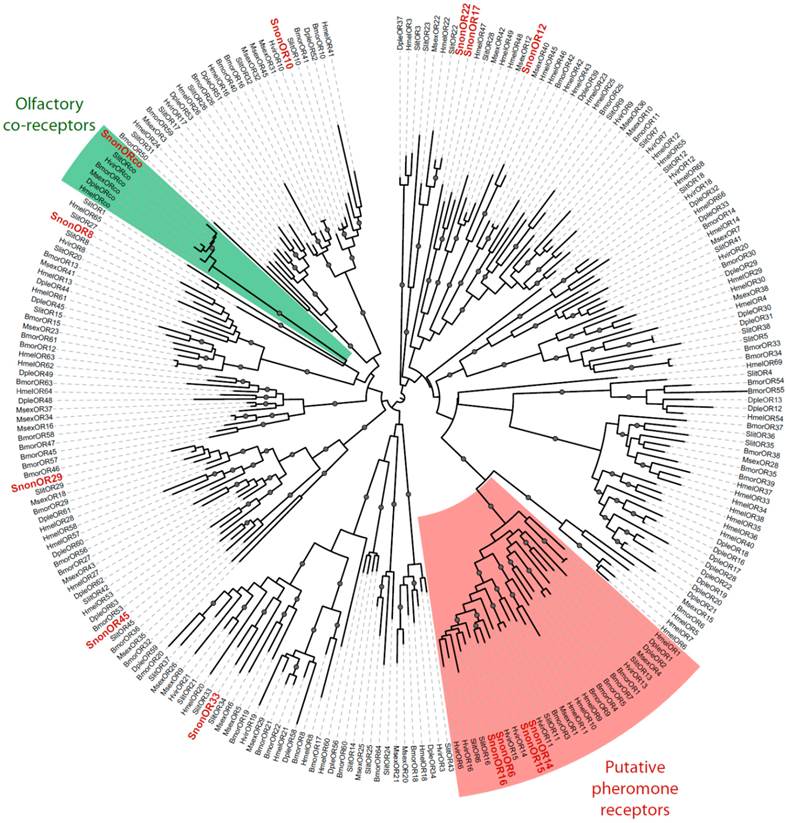
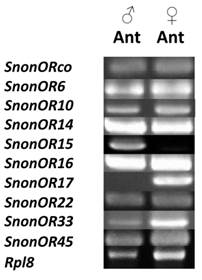
A total of 13 putative OR-encoding genes, named SnonORs, were identified in the S. nonagrioides transcriptome (Table 5, fasta format file in Supplementary material S1). For convenience and when possible, these ORs were numbered according to their S. littoralis homologs found in the phylogenetic analyses (Figure 4). In other moths, 43 to 47 candidate ORs have been annotated via similar transcriptomic strategies in S. littoralis [25, 54], M. sexta [26] and C. pomonella [27], and 66 ORs were annotated in the genome of B. mori. By comparison, the number of SnonORs identified here is quite small. It is probable that the SnonORs are expressed at a very low level, which makes additional ORs difficult to identify. Seven sequences encoded complete proteins (SnonOrco, OR10, OR12, OR15, OR17, OR22, OR33) predicted to contain between 6 and 8 transmembrane domains, as usually observed for insect ORs [10]. Depending on the size of the fragments, the other SnonORs were predicted to contain between 3 and 6 transmembrane domains (Table 5). Among the SnonORs, we identified the S. nonagrioides Orco, and four SnonORs (OR6, OR14, OR15 and OR16) clustered in the sex pheromone receptor sub-family (Figure 4), a number that fits well with the number of components described in the S. nonagrioides sex pheromone blend: (Z)-11-hexadecenyl acetate (Z11-16:Ac) -the main component-, (Z)-11-hexadecen-1-ol (Z11-16:OH), (Z)-11-hexadecenal (Z11-16:Ald), and dodecyl acetate (12:Ac) [34, 35]. As revealed by Illumina read mapping analyses (Table 4), some SnonORs appeared to be expressed in both adult and larvae antennae. It was the case of SnonOrco, as previously observed in other noctuid species [29]. Some other SnonORs had a limited number of larval mapped reads, suggesting a role restricted to adult chemosensation. Interestingly, reads from the ovipositor library could be mapped on some of the SnonORs we identified, including Orco (Table 4). Expression of OBPs (see upper), ORs and Orco in this organ suggest that the ovipositors could detect volatile molecules, as suggested in H. virescens [33]. In this latter species, PBP and PR but not Orco expressions could be evidenced in the female ovipositors. Here, the presence of Orco suggests that the ORs are functional in these organs. In addition to read mapping, RT-PCR was performed on male and female antennae to reveal sex-biased ORs. Using RT-PCR, we were able to retrieve 10 OR transcripts (Figure 5), and three transcripts were not amplified in spite of numerous attempts. Seven SnonORs could be amplified in both male and female antennae, including three of the candidate PRs, and only one PR, SnonOR15, was male-specific (SnonOR15, Figure 5). Interestingly, S. nonagrioides female antennae do not respond to the sex pheromone blend, as previously revealed by electroantennography [22]. Taken together, these observations either suggest that PRs alone may not be sufficient to trigger an electrical response to the pheromone, or that actual male-specific SnonPRs remain to be identified, apart SnonOR15. In H. virescens, PRs responding to Z11-16:Ald (HvirOR13), Z11-16:Ac (HvirOR14) and Z11-16:OH (HvirOR16) have been previously characterized [77, 78], these three components being also found in the S. nonagrioides sex pheromone blend. The SnonPRs presenting the higher percentage of identity with these HvirPRs were, respectively, SnonOR14 (46.3%), SnonOR15-14 (65.4%- 63.3%) and SnonOR6-16 (64.6%-61.7%). Further functional studies would be required to verify whether these SnonORs recognize the same ligands as their H. virescens counterparts or not. One SnonOR (SnonOR17) appeared to be female-specific in the RT-PCR experiment (Figure 5). Female-specific and female-enriched ORs have been reported in diverse Lepidoptera species, as potentially involved female-specific behaviours (localization of oviposition sites, responses to the male pheromone [24, 79, 80]). However, this female-specific SnonOR is unrelated to these Lepidoptera ORs (for instance MsexOR3, BmorOR19 and SlitOR37 in Figure 4). Another SnonOR (SnonOR33) grouped in the clade of female-specific/female-enriched ORs, although it is expressed in both male and female antennae in the RT-PCR analysis (Figure 5). Interestingly, SnonOR33 exhibited the highest number of mapped female antennal reads (Table 4), suggesting that it is one of the most abundantly expressed ORs in female antennae. It may thus be essential for female-specific behaviours.
We identified 10 putative IRs, named SnonIRs, and 5 ionotropic glutamate receptors (iGluR) in the S. nonagrioides transcriptome (Table 5), based on the phylogenetic analysis (Figure 6). This analysis notably indicates that we identified the S. nonagrioides IR8a and IR25a, which are both supposed to encode co-receptors that couple with other IRs [17]. These two co-receptors were highly expressed in adult antennae, as expected for co-receptors, but were also found to be expressed in the larval chemosensory organs and the ovipositors (Table 4). We also identified members belonging to 5 of the 9 other conserved IR sub-families (highlighted in colors in Figure 6). No SnonIR candidate clustered in the divergent IR clade, whose members are not expressed in antennae and likely not involved in olfaction in D. melanogaster [16]. In agreement, most of the SnonIRs were highly expressed in adult (at least female) antennae, a limited number of which was also expressed in the larval chemosensory organs (Table 4). One IR, SnonIR2, was highly expressed in the ovipositors. This IR appeared as atypical since it was expressed at a low level in antennae (Table 4) and was unrelated to previously described insect IRs (Figure 6). Its presence in the ovipositors together with the two co-receptors IR25a and IR8a suggests that it is functional and it may thus be used by the ovipositing females to select an adequate host plant. SnonIR1 appeared in a group that included only lepidopteran IR proteins (Figure 6), supporting our previous hypothesis of the occurrence of a lepidopteran specific IR sub-group [27, 64]. Among the tissues we sequenced, this IR was only observed to be expressed in adult antennae.
In Lepidoptera, two SNMPs have been described. In accordance with their best hit, we annotated in the S. nonagrioides transcriptome two putative SNMPs, defined as SnonSNMP1 and SnonSNMP2. SNMPs were first identified in pheromone-sensitive neurons of Lepidoptera [81, 82] and are thought to play an important role in pheromone detection, as demonstrated for the D. melanogaster SNMP1 homolog [19]. Both were abundantly expressed in female antennae but reads were also detected from the other libraries, especially for SnonSNMP2 (Table 4).
List of candidate S. nonagrioides ORs, IRs and ionotropic glutamate receptors. Transmembrane domains (Tm) were predicted using HMMTOP 2.0. [50].
| Name | Length (amino acids) | Tm nb | Blast P hit | e-value |
|---|---|---|---|---|
| SnonOrco | 474 | 7 | dbj|BAG71415.1| olfactory receptor-2 [Mythimna separata] | 0 |
| SnonOR6 | 345 | 6 | emb|CAG38117.1| putative chemosensory receptor 16 [Heliothis virescens] | 5e-168 |
| SnonOR8 | 231 | 3 | emb|CAD31949.1| putative chemosensory receptor 8 [Heliothis virescens] | 8e-84 |
| SnonOR10 | 390 | 7 | emb|CAG38111.1| putative chemosensory receptor 10 [Heliothis virescens] | 0 |
| SnonOR12 | 399 | 7 | gb|AFC91721.1| putative odorant receptor OR12 [Cydia pomonella] | 0 |
| SnonOR14 | 218 | 4 | gb|ACF32964.1| olfactory receptor 14 [Helicoverpa armigera] | 1e-91 |
| SnonOR15 | 442 | 8 | dbj|BAG71414.1| olfactory receptor-1 [Mythimna separata] | 0 |
| SnonOR16 | 107 | 3 | emb|CAG38117.1| putative chemosensory receptor 16 [Heliothis virescens] | 8e-37 |
| SnonOR17 | 411 | 5 | gb|AFC91725.1| putative odorant receptor OR17 [Cydia pomonella] | 5e-84 |
| SnonOR22 | 429 | 7 | gb|AFC91732.1| putative odorant receptor OR24 [Cydia pomonella] | 2e-142 |
| SnonOR29 | 374 | 6 | ref|NP_001166894.1| olfactory receptor 29 [Bombyx mori] | 8e-161 |
| SnonOR33 | 403 | 8 | gb|ADM32898.1| odorant receptor OR-5 [Manduca sexta] | 6e-85 |
| SnonOR45 | 304 | 5 | ref|NP_001166892.1| olfactory receptor 36 [Bombyx mori] | 5e-115 |
| SnonIR1 | 196 | 1 | gb|EHJ76709.1| ionotropic glutamate receptor-invertebrate [Danaus plexippus] | 2e-36 |
| SnonIR2 | 352 | 0 | gb|EHJ72235.1| hypothetical protein KGM_01297 [Danaus plexippus] | 9e-145 |
| SnonIR8a | 192 | 1 | gb|AFC91764.1| putative ionotropic receptor IR8a, partial [Cydia pomonella] | 2e-125 |
| SnonIR21a | 380 | 2 | gb|ADR64678.1| putative chemosensory ionotropic receptor IR21a [Spodoptera littoralis] | 0 |
| SnonIR25a | 630 | 3 | gb|EHJ78658.1| hypothetical protein KGM_04141 [Danaus plexippus] | 0 |
| SnonIR41a | 508 | 3 | gb|ADR64681.1| putative chemosensory ionotropic receptor IR41a [Spodoptera littoralis] | 0 |
| SnonIR75p | 518 | 3 | gb|ADR64684.1| putative chemosensory ionotropic receptor IR75p [Spodoptera littoralis] | 0 |
| SnonIR75q | 622 | 3 | gb|ADR64685.1| putative chemosensory ionotropic receptor IR75q.2 [Spodoptera littoralis] | 0 |
| SnonIR76b | 340 | 3 | gb|ADR64687.1| putative chemosensory ionotropic receptor IR76b [Spodoptera littoralis] | 0 |
| SnonIR93a | 389 | 7 | gb|EAT43564.1| AAEL005012-PA [Aedes aegypti] | 8e-100 |
| SnonGluR1 | 475 | 1 | gb|EHJ66743.1| hypothetical protein KGM_16050 [Danaus plexippus] | 0 |
| SnonGluR2 | 904 | 5 | gb|EHJ66742.1| hypothetical protein KGM_16053 [Danaus plexippus] | 2e-169 |
| SnonGluR3 | 434 | 5 | ref|XP_001655460.1| ionotropic glutamate receptor subunit ia [Aedes aegypti] | 0 |
| SnonNmdaR1 | 465 | 0 | gb|EHJ78211.1| putative NMDA-type glutamate receptor 1 [Danaus plexippus] | 0 |
| SnonNmdaR2 | 485 | 6 | gb|EHJ66761.1| putative glutamate receptor, ionotropic, n-methyl d-aspartate epsilon [Danaus plexippus] | 0 |
Maximum likelihood tree of candidate ORs from S. nonagrioides and other Lepidoptera. Sequences used were from B. mori [51], S. littoralis [24, 25, 54], H. virescens [52, 53], M. sexta [26], D. plexippus [55] and H. melpomene [56]. Branch support was estimated by approximate likelihood-ratio test (aLRT) (circles: >0.95) [71]. Images were created using the iTOL web server [85]. The SnonORs identified in this study are in red.
RT-PCRs of S. nonagrioides OR transcripts (SnonORs) in male and female antennae.
Maximum likelihood tree of candidate ionotropic receptors (IRs) from S. nonagrioides and other insects. Sequences used were from B. mori [16], S. littoralis [24, 25, 54], D. plexippus [55], D. melanogaster, Apis mellifera and Tribolium castaneum [16]. Branch support was estimated by approximate likelihood-ratio test (aLRT) (circles: >0.95) [71]. Images were created using the iTOL web server [85]. SlitIRs are in bold and the new SlitIRs identified in this study are in red.

Identification of candidate S. nonagrioides enzymes involved in pheromone clearance and biosynthesis
As demonstrated in the GO analysis (see above), the transcriptome appeared to be enriched in genes involved in catalytic activity and we thus particularly focused on candidate enzymes involved in pheromone clearance and pheromone biosynthesis. Twelve carboxylesterases could be annotated, as potentially involved in the degradation of acetate pheromone components within the sensillum lymph (Table 6). Among these carboxylesterases, we could identify a previously cloned sequence [23] that we named SnonCXE1. The others were named according to their S. littoralis carboxylesterase best hit. All but two of the deduced SnonCXE proteins displayed the Ser-active site included in the conserved pentapeptide Gly-X-Ser-X-Gly common in enzymes of the α-/β hydrolase family [83] (amino acid sequences available in Supplementary material S1) and eight sequences displayed a signal peptide. These SnonCXEs presented diverse expression patterns, some being highly expressed in female antennae, and one (SnonCXE5) being highly expressed in the female ovipositor (Table 4). The contribution of esterases in the biosynthesis of pheromone components has not yet been reported. However, since the S. nonagrioides pheromone blend contains both an acetate (Z11-16:OAc) and its corresponding alcohol (Z11-16:OH), such enzymatic activities could participate in the generation of the alcohol component from its acetate precursor. Seventeen other enzymes putatively involved in pheromone biosynthesis were also annotated, among them a Δ11-desaturase and different acetyltransferases and reductases (Table 6), that may be involved in the biosynthesis of Z11-16:Ac, the main component of S. nonagrioides sex pheromone. Indeed, it has been previously demonstrated that Z11-16:Ac is biosynthesized from palmitic acid by Δ11-desaturation followed by reduction and acetylation [84]. Reduction of the intermediate Z11-16:COOH has also been proposed to generate the minor component Z11-16:Ald [84]. Most of the transcripts were expressed in the female ovipositors and, noticeably, the Δ11-desaturase transcript was highly expressed in this organ (Table 4), supporting its function in introducing the double bound in a specific position in the acid precursor of the S. nonagrioides pheromone components.
List of S. nonagrioides contigs encoding enzymes putatively involved in pheromone degradation and pheromone biosynthesis. CXE: carboxylesterases.
| Names | Length (amino acids) | BlastP hit | e-value |
|---|---|---|---|
| SnonCXE1 | 532 | gb|ABH01082.1| esterase [Sesamia nonagrioides] | 0 |
| SnonCXE2 | 465 | gb|AFO65061.1| esterase [Helicoverpa armigera] | 0 |
| SnonCXE3 | 177 | gb|ACV60230.1| antennal esterase CXE3 [Spodoptera littoralis] | 2e-99 |
| SnonCXE4 | 666 | gb|AAR26516.1| antennal esterase [Mamestra brassicae] | 0 |
| SnonCXE5 | 577 | gb|ADR64702.1| antennal esterase CXE5 [Spodoptera exigua] | 0 |
| SnonCXE9 | 555 | gb|ACV60236.1| antennal esterase CXE9 [Spodoptera littoralis] | 0 |
| SnonCXE11 | 537 | gb|ACV60238.1| antennal esterase CXE11 [Spodoptera littoralis] | 0 |
| SnonCXE13 | 560 | gb|ACV60240.1| antennal esterase CXE13 [Spodoptera littoralis] | 0 |
| SnonCXE14 | 560 | gb|ACV60241.1| antennal esterase CXE14 [Spodoptera littoralis] | 0 |
| SnonCXE16 | 472 | gb|ACV60243.1| antennal esterase CXE16 [Spodoptera littoralis] | 0 |
| SnonCXE17 | 551 | gb|ACV60244.1| antennal esterase CXE17 [Spodoptera littoralis] | 0 |
| SnonCXE19 | 617 | gb|ACV60246.1| antennal esterase CXE19 [Spodoptera littoralis] | 0 |
| Snon-Acyl-CoA Δ9 desaturase | 355 | gb|AAF81788.1|AF272343_1 acyl-CoA delta-9 desaturase [Helicoverpa zea] | 0 |
| Snon-Acyl-CoA Δ 11 desaturase | 332 | gb|ACX53794.1| desaturase [Heliothis virescens] | 0 |
| Snon-N-acetyltransferase | 178 | gb|EHJ73917.1| N-acetyltransferase [Danaus plexippus | 1e-113 |
| Snon-acyl-CoA Δ9 desaturase | 356 | gb|AAF81790.2|AF272345_1 acyl-CoA delta-9 desaturase [Helicoverpa zea] | 0 |
| Snon-acetyltransferase 1 | 405 | gb|EHJ65205.1| acetyltransferase 1 [Danaus plexippus] | 0 |
| Snon-desaturase | 374 | gb|AAQ74260.1| desaturase [Spodoptera littoralis] | 0 |
| Snon-putative acetyl transferase | 231 | dbj|BAH96561.1| putative acetyl transferase [Bombyx mori] | 8e-151 |
| Snon-N-acetyltransferase 3 | 104 | gb|EHJ68864.1| N-acetyltransferase [Danaus plexippus] | 4e-66 |
| Snon-fatty acid synthase | 2380 | ref|XP_970417.2| PREDICTED: similar to fatty acid synthase [Tribolium castaneum] | 0 |
| Snon_fatty-acyl CoA reductase 1 | 514 | gb|ADI82774.1| fatty-acyl CoA reductase 1 [Ostrinia nubilalis] | 0 |
| Snon_fatty-acyl CoA reductase 2 | 624 | gb|ADI82775.1| fatty-acyl CoA reductase 2 [Ostrinia nubilalis] | 0 |
| Snon_fatty-acyl CoA reductase 3 | 104 | gb|ADI82776.1| fatty-acyl CoA reductase 3 [Ostrinia nubilalis] | 4,00e-41 |
| Snon_fatty-acyl CoA reductase 4 | 498 | gb|ADI82777.1| fatty-acyl CoA reductase 4 [Ostrinia nubilalis] | 0 |
| Snon_fatty-acyl CoA reductase 5 | 534 | gb|EHJ72233.1| fatty-acyl CoA reductase 5 [Danaus plexippus] | 0 |
| Snon_fatty-acyl CoA reductase 6 | 525 | gb|EHJ76493.1| fatty-acyl CoA reductase 6 [Danaus plexippus] | 0 |
| Snon_fatty-acyl CoA reductase b | 480 | gb|ADI82779.1| fatty-acyl CoA reductase 6 [Ostrinia nubilalis] | 0 |
| Snon_fatty-acyl CoA reductase II | 450 | gb|ADD62441.1| fatty-acyl CoA reductase II [Yponomeuta rorrellus] | 3,00e-131 |
Conclusion
Through sequencing of the transcriptome, we identified a variety of genes potentially involved in olfactory signal detection and pheromone biosynthesis in an important pest of maize in the Mediterranean Area. We annotated a total of 68 contigs encoding putative proteins involved in all the steps of odorant detection - transport, docking, recognition and degradation - and 17 enzymes potentially involved in pheromone biosynthesis. Concerning the pheromone detection process, we identified in this species three PBPs, two SNMPs, four candidate pheromone receptors and many carboxylesterases as putative pheromone-degrading enzymes. This study constitutes the first large scale description of chemosensory genes in S. nonagrioides.
Supplementary Material
Supplementary Material S1 and S2.
Acknowledgements
We thank Ferial Kaoula (LEGS, Gif-sur-Yvette, France) for insect rearing and the BioGenouest platform for their bioinformatics support. This work was supported by funding from the French ANR (Agence Nationale de la Recherche) (Adaptanthrop project ANR-09-PEXT-009).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Anglade P. Les Sésamia. Entomologie appliquée à l'agriculture Tome II, Lépidoptères, II Paris, France: Masson et Cie. 1972:1389-401
2. Melamed-Madjar V, Tam S. A field survey of changes in the composition of corn borer population in Israel. Phytoparasitica. 1980;8:201-4
3. Rousseau D. Maïs: la sésamie progresse cap au nord, dans l'air marin et la douceur angevine: Progression de la sésamie dans les Pays-de-la-Loire: à gérer dans la région, à méditer ailleurs en France. Phytoma-La Défense des Végétaux. 2009;622:38-41
4. Frérot B, Guillon M, Bernard P, Madrennes L, De Schepper B, Mathieu F. et al. Mating disruption on pink corn borer, Sesamia nonagrioïdes Lef. (Lep, Noctuidae). Technology Transfer in Mating Disruption. IOBC wprs Bulletin. 1997;20:119-28
5. Larue P. La Sésamie (Sesamia nonagrioïdes Lef.): dégâts et actualisation de la lutte. Phytoma - La défense des végétaux. 1984;277:163-71
6. Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373-91
7. Pelosi P, Zhou JJ, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cell Mol Life Sci. 2006;63:1658-76
8. Vogt RG, Prestwich GD, Lerner MR. Odorant-binding-protein subfamilies associate with distinct classes of olfactory receptor neurons in insects. J Neurobiol. 1991;22:74-84
9. Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GS. et al. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J Neurosci. 2011;31:13357-75
10. Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20
11. Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703-14
12. Vosshall LB, Hansson BS. A Unified Nomenclature System for the Insect Olfactory Coreceptor. Chem Senses. 2011;36:497-8
13. Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH. et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007-11
14. Sato K, Pellegrino M, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002-6
15. Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149-62
16. Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H. et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010 6
17. Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44-60
18. Nichols Z, Vogt RG. The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem Mol Biol. 2008;38:398-415
19. Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450:289-93
20. de Santis F, Francois MC, Merlin C, Pelletier J, Maibeche-Coisne M, Conti E. et al. Molecular cloning and in Situ expression patterns of two new pheromone-binding proteins from the corn stemborer Sesamia nonagrioides. J Chem Ecol. 2006;32:1703-17
21. Konstantopoulou M, Pratsinis H, Kletsas D, Mazomenos BE. Pheromone-binding protein and general odorant-binding protein of Sesamia nonagrioides: sex- and diel-dependent expression. Ent Exp Appl. 2006;119:129-36
22. Acin P, Carrascal M, Abian J, Guerrero A, Quero C. Expression of differential antennal proteins in males and females of an important crop pest, Sesamia nonagrioides. Insect Biochem Mol Biol. 2009;39:11-9
23. Merlin C, Rosell G, Carot-Sans G, Francois MC, Bozzolan F, Pelletier J. et al. Antennal esterase cDNAs from two pest moths, Spodoptera littoralis and Sesamia nonagrioides, potentially involved in odourant degradation. Insect Mol Biol. 2007;16:73-81
24. Legeai F, Malpel S, Montagne N, Monsempes C, Cousserans F, Merlin C. et al. An Expressed Sequence Tag collection from the male antennae of the Noctuid moth Spodoptera littoralis: a resource for olfactory and pheromone detection research. BMC Genomics. 2011;12:86
25. Jacquin-Joly E, Legeai F, Montagné N, Monsempes C, François MC, Poulain J. et al. Candidate chemosensory Genes In Female Antennae Of The Noctuid Moth Spodoptera littoralis. Int J Biol Sci. 2012;8:1036
26. Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, Hansson BS. Antennal transcriptome of Manduca sexta. Proc Natl Acad Sci U S A. 2011;108:7449-54
27. Bengtsson JM, Trona F, Montagné N, Anfora G, Ignell R, Witzgall P. et al. Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS ONE. 2012;7(2):e31620
28. Liu Y, Gu S, Zhang Y, Guo Y, Wang G. Candidate olfaction genes identified within the Helicoverpa armigera Antennal Transcriptome. PLoS One. 2012;7:e48260
29. Malpel S, Merlin C, Francois MC, Jacquin-Joly E. Molecular identification and characterization of two new Lepidoptera chemoreceptors belonging to the Drosophila melanogaster OR83b family. Insect Mol Biol. 2008;17:587-96
30. Mitsuno H, Sakurai T, Murai M, Yasuda T, Kugimiya S, Ozawa R. et al. Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur J Neurosci. 2008;28:893-902
31. Miura N, Nakagawa T, Tatsuki S, Touhara K, Ishikawa Y. A male-specific odorant receptor conserved through the evolution of sex pheromones in Ostrinia moth species. Int J Biol Sci. 2009;5:319-30
32. Wanner KW, Nichols AS, Allen JE, Bunger PL, Garczynski SF, Linn CE. et al. Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS One. 2010;5:e8685
33. Widmayer P, Heifetz Y, Breer H. Expression of a pheromone receptor in ovipositor sensilla of the female moth (Heliothis virescens). Insect Mol Biol. 2009;18:541-7
34. Sreng I, Maume B, Frérot B. Analyse de la secretion phéromonale produite par les femelles vierges de Sesamia nonagrioides (Lépidoptère, Noctuidae). CR Acad Sc Paris. 1985;301:439-42
35. Mazomenos BE. Sex pheromone components of corn stalk borer, Sesamia nonagrioides (Lef.). Partial chemical purification and its biological activity under laboratory conditions. J Chem Ecol. 1989;6:565-72
36. Poitout S, Buès R. Elevage de chenilles de vingt-huit espèces de Lépidoptères Noctuidae et de deux espèces d'Arctiidae sur milieu artificiel simple. Particularités de l'élevage selon les espèces. Ann Zool Ecol anim. 1974;6:431-41
37. d'Alençon E, Sezutsu H, Legeai F, Permal E, Bernard-Samain S, Gimenez S. et al. Extensive synteny conservation of holocentric chromosomes in Lepidoptera despite high rates of local genome rearrangements. Proc Natl Acad Sci U S A. 2010;107:7680-5
38. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011 17
39. Schmieder R, Edwards R. Fast Identification and Removal of Sequence Contamination from Genomic and Metagenomic Datasets. PLoS ONE. 2011;6:e17288
40. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotech. 2011;29:644-52
41. Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WEG, Wetter T. et al. Using the miraEST Assembler for Reliable and Automated mRNA Transcript Assembly and SNP Detection in Sequenced ESTs. Genome Research. 2004;14:1147-59
42. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674-6
43. Gouzy J, Carrere S, Schiex T. FrameDP: sensitive peptide detection on noisy matured sequences. Bioinformatics. 2009;25:670-1
44. Durand N, Carot-Sans G, Chertemps T, Montagne N, Jacquin-Joly E, Debernard S. et al. A diversity of putative carboxylesterases are expressed in the antennae of the noctuid moth Spodoptera littoralis. Insect Mol Biol. 2010;19:87-97
45. Vogel H, Heidel AJ, Heckel DG, Groot AT. Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens. BMC Genomics. 2010;11:29
46. Huang X, Madan A. CAP3: A DNA Sequence Assembly Program. Genome Research. 1999;9:868-77
47. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785-6
48. Buchan DW, Ward SM, Lobley AE, Nugent TC, Bryson K, Jones DT. Protein annotation and modelling servers at University College London. Nucleic Acids Res. 2010;38:W563-8
49. Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188-90
50. Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849-50
51. Tanaka K, Uda Y, Ono Y, Nakagawa T, Suwa M, Yamaoka R. et al. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol. 2009;19:881-90
52. Krieger J, Raming K, Dewer YM, Bette S, Conzelmann S, Breer H. A divergent gene family encoding candidate olfactory receptors of the moth Heliothis virescens. Eur J Neurosci. 2002;16:619-28
53. Krieger J, Grosse-Wilde E, Gohl T, Dewer YM, Raming K, Breer H. Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc Natl Acad Sci USA. 2004;101:11845-50
54. Poivet E, Gallot A, Montagné N, Glaser N, Legeai F, Jacquin-Joly E. A comparison of the olfactory gene repertoires of adults and larvae in the noctuid moth Spodoptera littoralis. PLoS ONE. 2013;8:e60263
55. Zhan S, Merlin C, Boore JL, Reppert SM. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011;147:1171-85
56. The Heliconius Genome Consortium. Islands of divergence underlie adaptive radiation in a butterfly genome. Nature. 2012;487:94-8
57. Gong DP, Zhang HJ, Zhao P, Xia QY, Xiang ZH. The Odorant Binding Protein gene family from the genome of silkworm, Bombyx mori. BMC Genomics. 2009;10:332
58. Krieger J, Ganssle H, Raming K, Breer H. Odorant binding proteins of Heliothis virescens. Insect Biochem Mol Biol. 1993;23:449-56
59. Krieger J, Mameli M, Breer H. Elements of the olfactory signaling pathways in insect antennae. Invertebrate Neuroscience. 1997;3:137-44
60. Robertson HM, Martos R, Sears CR, Todres EZ, Walden KK, Nardi JB. Diversity of odourant binding proteins revealed by an expressed sequence tag project on male Manduca sexta moth antennae. Insect Mol Biol. 1999;8:501-18
61. Gong DP, Zhang HJ, Zhao P, Lin Y, Xia QY, Xiang ZH. Identification and expression pattern of the chemosensory protein gene family in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2007;37:266-77
62. Picimbon J-F, Dietrich K, Krieger J, Breer H. Identity and expression pattern of chemosensory proteins in Heliothis virescens (Lepidoptera, Noctuidae). Insect Biochemistry and Molecular Biology. 2001;31:1173-81
63. Ozaki K, Utoguchi A, Yamada A, Yoshikawa H. Identification and genomic structure of chemosensory proteins (CSP) and odorant binding proteins (OBP) genes expressed in foreleg tarsi of the swallowtail butterfly Papilio xuthus. Insect Biochem Mol Biol. 2008;38:969-76
64. Olivier V, Monsempes C, Francois MC, Poivet E, Jacquin-Joly E. Candidate chemosensory ionotropic receptors in a Lepidoptera. Insect Mol Biol. 2011;20:189-99
65. Katoh K, Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 2010;26:1899-900
66. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792-7
67. Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221-4
68. Le SQ, Lartillot N, Gascuel O. Phylogenetic mixture models for proteins. Philosophical Transactions of the Royal Society B: Biological Sciences. 2008;363:3965-76
69. Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104-5
70. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307-21
71. Anisimova M, Gascuel O. Approximate Likelihood-Ratio Test for Branches: A Fast, Accurate, and Powerful Alternative. Systematic Biology. 2006;55:539-52
72. Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Research. 2011;39:W475-W8
73. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106
74. R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria http: //wwwR-projectorg. 2011
75. Vieira FG, Rozas J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol Evol. 2011;3:476-90
76. Jacquin-Joly E, Vogt RG, Francois MC, Nagnan-Le Meillour P. Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem Senses. 2001;26:833-44
77. Grosse-Wilde E, Gohl T, Bouche E, Breer H, Krieger J. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur J Neurosci. 2007;25:2364-73
78. Wang G, Vasquez GM, Schal C, Zwiebel LJ, Gould F. Functional characterization of pheromone receptors in the tobacco budworm Heliothis virescens. Insect Mol Biol. 2011;20:125-33
79. Patch HM, Velarde RA, Walden KK, Robertson HM. A candidate pheromone receptor and two odorant receptors of the hawkmoth Manduca sexta. Chem Senses. 2009;34:305-16
80. Wanner KW, Anderson AR, Trowell SC, Theilmann DA, Robertson HM, Newcomb RD. Female-biased expression of odourant receptor genes in the adult antennae of the silkworm, Bombyx mori. Insect Mol Biol. 2007;16:107-19
81. Rogers ME, Sun M, Lerner MR, Vogt RG. Snmp-1, a novel membrane protein of olfactory neurons of the silk moth Antheraea polyphemus with homology to the CD36 family of membrane proteins. J biol Chem. 1997;272:14792-9
82. Rogers ME, Krieger J, Vogt RG. Antennal SNMPs (sensory neuron membrane proteins) of Lepidoptera define a unique family of invertebrate CD36-like proteins. J Neurobiol. 2001;49:47-61
83. Oakeshott JG, Claudianos C, Russell RJ, Robin GC. Carboxyl/cholinesterases: a case study of the evolution of a successful multigene family. Bioessays. 1999;21:1031-42
84. Mas E, Lloria J, Quero C, Camps F, Fabrias G. Control of the biosynthetic pathway of Sesamia nonagrioides sex pheromone by the pheromone biosynthesis activating neuropeptide. Insect Biochem Mol Biol. 2000;30:455-9
85. Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127-8
Author contact
![]() Corresponding author: Dr Emmanuelle Jacquin-Joly, INRA, UMR 1272 INRA-UPMC Physiologie de l'Insecte : Signalisation et Communication, route de Saint-Cyr, F-78026 Versailles cedex, France. Tel: (33) 1 30 83 32 12; fax: (33) 1 30 83 31 19; E-mail: emmanuelle.jacquininra.fr.
Corresponding author: Dr Emmanuelle Jacquin-Joly, INRA, UMR 1272 INRA-UPMC Physiologie de l'Insecte : Signalisation et Communication, route de Saint-Cyr, F-78026 Versailles cedex, France. Tel: (33) 1 30 83 32 12; fax: (33) 1 30 83 31 19; E-mail: emmanuelle.jacquininra.fr.

 Global reach, higher impact
Global reach, higher impact