10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(9):960-965. doi:10.7150/ijbs.6739 This issue Cite
Short Research Communication
Hoxc Gene Collinear Expression and Epigenetic Modifications Established during Embryogenesis Are Maintained until after Birth
Department of Anatomy, Embryology Laboratory, Brain Korea 21 project for Medical Science, Yonsei University College of Medicine, Seoul 120-752, Korea.
Received 2013-5-22; Accepted 2013-9-12; Published 2013-9-22
Abstract
The Hox genes, which are organized into clusters on different chromosomes, are key regulators of embryonic anterior-posterior (A-P) body pattern formation and are expressed at specific times and in specific positions in developing vertebrate embryos. Previously, we have shown that histone methylation patterns are closely correlated with collinear Hox gene expression patterns along the A-P axis of E14.5 mouse embryos. Since histone modification is thought to play a crucial mechanistic role in the highly coordinated pattern of collinear Hox gene expression, we examined the maintenance of the spatial collinear expression pattern of Hoxc genes and the corresponding histone modifications during embryogenesis and in early postnatal mice. Hox expression patterns and histone modifications were analyzed by semi-quantitative RT-PCR and chromatin immunoprecipitation (ChIP)-PCR analyses, respectively. The spatiotemporal expression patterns of Hoxc genes in a cluster were maintained until the early postnatal stage (from E8.5 through P5). Examination of histone modifications in E14.5 and P5 tissues revealed that level of H3K27me3 is only a weak correlation with collinear Hoxc gene expression in the trunk regions although diminished in general, however the enrichment of H3K4me3 is strongly correlated with the gene expression in both stages. In summary, the initial spatiotemporal collinear expression pattern of Hoxc genes and epigenetic modifications are maintained after birth, likely contributing to the establishment of the gene expression code for position in the anatomic body axis throughout the entire life of the organism.
Keywords: Hoxc cluster genes, collinear expression: anterior-posterior body axis, mouse embryogenesis, histone modification.
Introduction
Hox genes are transcription factors that play a pivotal role in patterning of the anterior-posterior (A-P) body axis during embryonic development. In mammals such as mouse and human, 39 Hox genes are organized into 4 clusters on 4 different chromosomes, with each cluster containing 9 to 11 genes [1-4]. The genomic locations of Hox genes within a cluster correlate with spatiotemporal expression during embryogenesis, a phenomenon termed collinearity [3, 5]. Hox genes located at the 3' end in a cluster are expressed more anteriorly and earlier in development than genes located at the 5' end of a cluster. Sequential initiation of Hox gene expression in a cluster is driven by dynamic regulatory events, including retinoic acid (RA) [6], FGF signaling [7], and regulatory interactions between the Hox and Cdx gene families [8]. In the developing mouse, Hox gene expression is initiated in the posterior region of the primitive streak during gastrulation [9]. The ontogenesis of Hox-mediated axial patterning during embryogenesis has been studied for decades. However, the molecular mechanisms underlying the collinear expression of Hox genes are not yet fully understood.
The Polycomb group (PcG) and trithorax group (TrxG) proteins, initially discovered in Drosophila, have been reported to be core epigenetic regulators of homeotic genes [10-12]. Polycomb Repressive Complex (PRC2), a class of PcG proteins, and tri-methylated H3K27 (H3K27me3) are essential for long-term repression of target genes [13, 14]. In contrast, TrxG protein complexes catalyze the tri-methylation of H3K4 (H3K4me3), which is generally associated with active transcription [15].
Recently, the antagonistic function of the PcG and TrxG complexes in regulating Hox gene expression during mammalian development has been actively investigated [16]. In vitro studies revealed that histone H3 modification is correlated with the maintenance phase of Hox gene transcriptional regulation in embryonic stem cells (ESC) and other types of cultured cells [17-20]. Soshnikova et al. (2009) reported the in vivo result that the organization of H3K4me3 and H3K27me3 chromatin marks was closely associated with the temporal activation of Hoxd genes in the developing mouse tail [16]. Through chromatin conformation capture (3C) and chromatin immunoprecipitation (ChIP) techniques, the loss of H3K27me3 and topological changes along the Hox cluster were shown to be prerequisites for collinear expression, and the concomitant acquisition of H3K4me3 was shown to be more closely associated with a collinear Hox gene expression pattern along the A-P body axis in E14.5 mice embryos [21].
As most spatial (A-P axis) collinear expression analyses have been performed in earlier stages including the mid-gastrulae stage of mouse embryos, Hox expression patterns in later embryonic stages are not fully understood. In this study, we analyzed Hoxc gene expression patterns during several embryonic stages (from E8.5 to E18.5) through the postnatal period (P5), as well as the dynamic changes in histone modifications along the A-P axis.
Materials and methods
Animal preparation
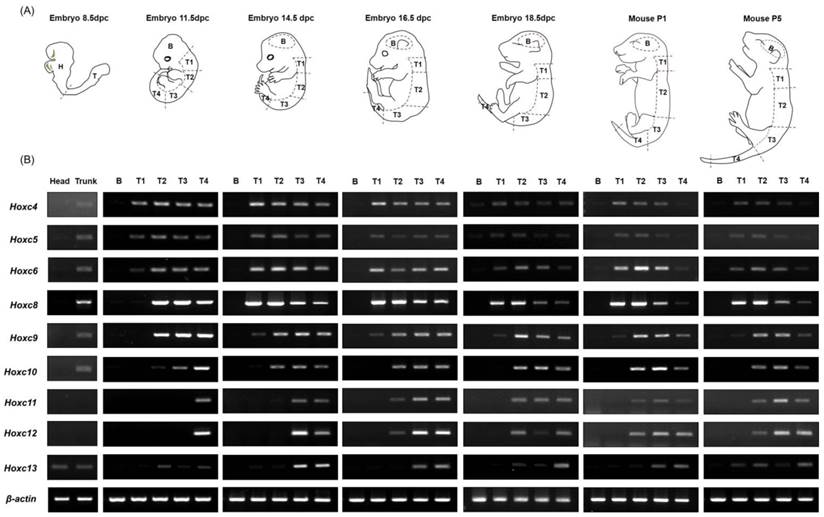
Pregnant Hsd:ICR (CD-1) mice were purchased from a pathogen-free laboratory animal company (Koatech, Gyeonggi-do, Korea). The embryos (E8.5, E11.5, E14.5, E16.5, and E18.5) and early postnatal mice (P1, P5) were dissected in PBS-containing plates on ice. After removing the internal organs, limbs and tail bud, each embryo or postnatal mouse was divided into five parts (B, brain; T1, somite number 5 to -14; T2, somite number 15 to -27; T3, somite number 28 to -32; T4, somite number 33 to the posterior end) along the A-P axis, except E8.5 embryo, which was divided into two parts, head (H, anterior end to somite number 6) and trunk (T, somite number 7 to the posterior end) as shown in Figure 1A. Experimental procedures were approved by the Animal Care and Use Committee of Yonsei University College of Medicine, Seoul, Korea.
RNA isolation and RT-PCR
Total RNA was isolated from freshly dissected samples using Trizol (Invitrogen, Carlsbad, CA, USA). Reverse transcription (RT) was performed with 1 μg of RNA using ImProm-llTM Reverse Transcriptase (Promega, Madison, WI, USA). A triplicate set of PCR was performed using Taq DNA polymerase (Bioneer, Daejeon, Korea). PCR amplification was performed under the following conditions: the initial denaturation for 3 minutes at 94°C, then 30 cycles of 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds. At least three independent biological replicates were analyzed. All PCR primer sequences used for detecting Hox gene expression and β-actin were as previously described [22].
Chromatin immunoprecipitation (ChIP)-PCR
ChIP analysis was performed following the X-ChIP protocol from Abcam (Abcam, Cambridge, UK), with minor modifications. ChIP-grade Abcam antibodies, Anti-Histone H3 (tri methyl K4) antibody (ab1012) and Anti-Histone H3 (tri methyl K27) antibody (ab6002), as well as Santa Cruz normal mouse IgG (sc-2025) were used. Embryonic samples were cross-linked with 1% formaldehyde diluted in serum-free Dulbecco's modified Eagle's medium (WelGENE Inc., Daegu, Korea) for 10 min at room temperature with gentle shaking. This reaction was stopped by adding 0.125 M glycine for 5 min, and the samples were then washed three times with PBS at room temperature. The resulting cells (approximately 4×107 cells) were lysed in 600 μl of sodium dodecyl sulfate (SDS) lysis buffer for 10 min on ice and sonicated with 30 sets of 10-second pulses with a VibracellTM sonicator (VCX130, Sonics & Materials Inc., Newton, CT, USA). The sonicated chromatin (500-1000 bp), salmon sperm DNA/protein coated A/G agarose beads (Santa Cruz, CA, USA), and appropriate antibodies were mixed and incubated overnight at 4°C with gentle shaking.
The following day, antibody-ChIP DNA complexes were washed, and the DNA was eluted and purified using a PCR purification kit (Cosmogenetech, Seoul, Korea). PCR amplification was performed under the following conditions: initial denaturation for 5 minutes at 95°C, and then 32 cycles of 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds. The PCR primer sequences used were as previously described [21, 23].
Results and Discussion
Maintenance of the spatial collinear expression pattern of Hox genes during embryogenesis and in early postnatal period in mice
In order to investigate whether the collinear expression patterns of Hox genes are maintained throughout embryogenesis as well as after birth, tissues from embryos (E8.5, E11.5, E14.5, E16.5, and E18.5) and postnatal mice (P1 and P5) were used for RT-PCR analysis after isolating RNA from each segment along the A-P axis, as described in the Materials and Methods. In general, there were no Hoxc genes expressed in brain tissues in any of the developmental stages examined (E8.5 through P5), which was expected based on previous results [21, 24]. In trunk tissue, the spatial collinear expression patterns of Hoxc genes along the A-P axis were detected throughout embryogenesis, and this expression pattern was maintained until the postnatal period P5 (Fig. 1).
In E11.5 embryos, T1 (somite 5-14) and T2 (somite 15-27) tissues expressed only the 3' anterior Hoxc genes (Hoxc4 to Hoxc6 in T1, Hoxc4 to Hoxc10 in T2), but posterior gene expression was seen as development continued. In the T1 tissue of E14.5 embryos, expression of Hoxc4 through Hoxc9 was seen, and this expression pattern was retained until postnatal stage P5. In the case of T2 tissue, sequentially expanded Hoxc genes to the 5' side were detected, up to Hoxc11 at E14.5, Hoxc12 at E16.5, and Hoxc13 at E18.5 and after. This expanded expression pattern could be due to boundary elements and/or insulator-binding proteins implicated in the establishment of gene expression boundaries being driven to move to the 5' position within a locus, or the changing of the interaction/binding affinities of transcription factors. However, tissue (segment) contamination during the dissection process could be also possible. Posterior tissues, such as T3 and T4, showed expression of the entire Hoxc genes at stage E14.5 and maintained this pattern until P5. In early embryos such as E11.5, however, only the most posterior tissue, T4, showed expression of the entire Hoxc genes. Additionally, T3, which is anterior to T4, did not show expression of posterior genes; only the 3' genes Hoxc4 to Hoxc10 were expressed. In the case of E8.5 mouse embryos, anterior (H) and posterior (T) tissues were generated by dissecting tissues between somite number 6 and 7. The anterior future brain region (H) did not express any Hoxc genes, whereas the posterior tissue (T) expressed the anterior Hoxc genes (Hoxc4 to Hoxc10). Even though E8.5 usually has 8 -12 somites, T tissues expressed more posterior 5' Hoxc genes, than expected based on somite level, although very weakly, indicating that Hox genes seemed to be expressed prior to somite formation.
Expression levels of each Hoxc gene throughout development show that gene expression is relatively low at E8.5, increases to a peak between E11.5 and E14.5, and then decreases. Unusual expression of Hoxc13, a deviation from the collinear expression pattern of Hox cluster genes, was found in several tissues at different stages, as has been previously reported [23, 25, 26]. Although the mechanisms are not known, Hoxc13 seems to escape the mechanisms regulating the collinear expression of Hoxc genes.
Interestingly, Hox genes are not only expressed during embryogenesis, but also widely expressed in adult human organ tissues, providing regional identities via differential and collinear expression along the A-P and proximal-distal axes of the body [27]. Even though there was a small extension of Hoxc gene expression in each trunk region following the mid-gastrula stage (E11.5), our results indicate that the spatial collinearity of the Hoxc gene expression pattern along the A-P body axis is established during early gastrulation and persists until after birth.
Maintenance of epigenetic modifications at Hoxc loci along the A-P axis of postnatal mice
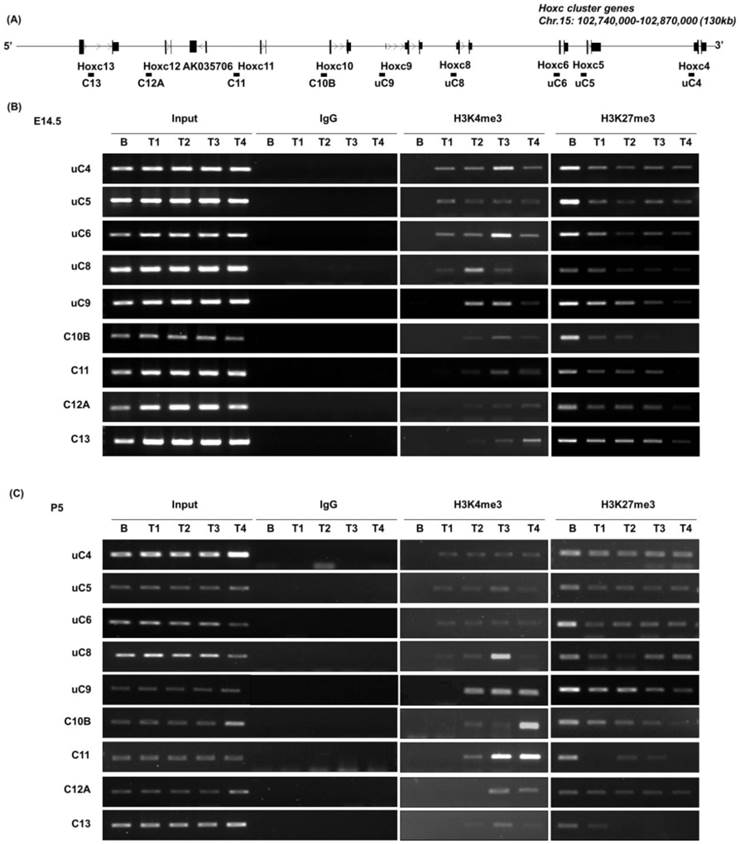
Although there have been several reports describing the functional relationship between histone marks such as H3K27me3 and H3K4me3 with the spatial collinear expression of Hox genes in embryos as well as in ES cells [19, 21, 24], little information is available regarding postnatal mice. To understand whether the maintenance of the collinear Hoxc gene expression pattern is accompanied by histone modifications up until the postnatal stage, we analyzed epigenetic modifications along the Hoxc cluster in P5 mice and the E14.5 embryo.
In E14.5 embryos, high levels of the repressive marker H3K27me3 were observed over the entire Hoxc locus in brain tissues that do not express Hox. However, the level of H3K27me3 was significantly reduced in all trunk (T1 to T4) tissues (Fig. 2A, B). There is only a weak correlation between the enrichment profile of H3K27me3 and the onset of collinear Hox gene expression in the trunk regions, which is in agreement with our previous report that the diminishment of H3K27me3 seemed to predate and was insufficient for the initiation of Hox gene expression [21]. However, the enrichment of H3K27me3 was significantly reduced in T3 compared to that in T1 and T2, and furthermore was almost completely diminished in T4, especially in the genomic region of the posterior Hoxc genes. This could be due to the composition of T4 tissues, which have more or less homogenous populations of Hox-expressing cells, and thus the majority of cells are not enriched with H3K27me3. In the case of the active expression marker H3K4me3, the whole cluster in the brain tissue was unmarked with H3K4me3, whereas H3K4me3 modification was closely correlated with the collinear expression pattern of Hoxc genes in the trunk regions of E14.5 embryos (Fig. 2B), as described previously [21].
The epigenetic modifications in P5 mice were similar to those detected in E14.5 (Fig. 2C). A strong H3K27me3 mark was detected in the brain tissue, which lacks Hoxc gene expression, whereas less strong in the trunk tissues of postnatal P5 mice. In the case of H3K4me3 modification, a strong positive correlation was detected with the expression of Hoxc genes, as seen in E14.5 embryos. Compared to the Hox-silent brain tissue, H3K4me3 mark was detected in the 3' Hox genes (uC4 through uC8-uC9, Figure 2) in the anterior trunk tissue (T1), where Hoxc4 through Hoxc8 were expressed (Fig. 1). This positive H3K4me3 mark was detected in the entire cluster in the posterior trunk tissues, as the entire Hoxc cluster genes were expressed like in the E14.5 trunk tissues. This result indicates that positive and negative histone modifications, once established, seemed to be maintained throughout development and even after birth. We conclude that the basic mode of epigenetic mechanisms regulating Hox gene expression are intrinsically maintained throughout development and following birth, and probably over the lifetime of the organism, as previously implicated [27].
Expression pattern of Hoxc cluster genes during embryogenesis and in the early postnatal period along the anterior-posterior (A-P) body axis. (A) A diagram depicting mouse embryos of different developmental stages and the scheme of the embryo dissection. The dashed lines indicate the areas where each tissue was dissected. Each trunk region was carefully isolated on the basis of somite numbers (See Materials and Methods). (B) Expression patterns of Hoxc genes (Hoxc4 through Hoxc13) in different tissues along the A-P axis. RT-PCR was performed using the primers specific to Hoxc genes. A pair of β-actin primers was used as a positive control for RNA integrity and cDNA synthesis. Selected data from three replicate experiments is shown.
Histone modifications at Hoxc genomic loci in E14.5 mouse embryos and P5 mice. (A) A schematic representation of the Hoxc locus indicating the genomic position of each Hoxc gene and the amplicons used for ChIP-PCR. (B, C) The results of ChIP-PCR to detect histone modifications in the genomic region of the Hoxc cluster. Immunoprecipitated DNAs and input DNAs were derived from E14.5 (B) and P5 (C) brain and trunk (T1-T5) tissues using anti-H3K4me3 and anti-H3K27me3 antibodies. IgG ChIP DNA samples were amplified using the same primers as the negative control. The data is representative of three independent experiments on triplicate samples.
Acknowledgements
Ms. H. Min is a graduate student receiving a Brain Korea 21 Scholarship and Hi Seoul science fellowship from Seoul Scholarship Foundation (SSF). This research was supported by the Basic Science Research Program through the National Research Foundation (NRF-2010-0026759 and -2010-0025149) and partly by PJ00905601 from the BioGreen21 Program, Rural Development Administration (RDA), Korea.
Competing Interests
The authors have declared that no competing interest exists.
References
1. McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283-302
2. Kim MH, Kessel M. Homeobox genes as regulators of vertebrate development. AgBiotech News and Information. 1993;5:189-194
3. Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358-364
4. Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191-201
5. Kmita MF, van Der Hoeven F, Zakany J. et al. Mechanisms of Hox gene colinearity: transposition of the anterior Hoxb1 gene into the posterior HoxD complex. Genes Dev. 2000;14:198-211
6. Simeone A, Acampora D, Arcioni L. et al. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature. 1990;346:763-766
7. Partanen J, Schwartz L, Rossant J. Opposite phenotypes of hypomorphic and Y766 phosphorylation site mutations reveal a function for Fgfr1 in anteroposterior patterning of mouse embryos. Genes Dev. 1998;12:2332-2344
8. Young T, Deschamps J. Hox, Cdx, and anteroposterior patterning in the mouse embryo. Curr Top Dev Biol. 2009;88:235-255
9. Duboule D. Temporal colinearity and the phylotypic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Dev Suppl. 1994:135-142
10. Francis NJ, Kingston RE. Mechanisms of transcriptional memory. Nat Rev Mol Cell Biol. 2001;2:409-421
11. Simon JA, Tamkun JW. Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr Opin Genet Dev. 2002;12:210-218
12. Orlando V. Polycomb, epigenomes, and control of cell identity. Cell. 2003;112:599-606
13. Czermin B, Melfi R, McCabe D. et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185-196
14. Ebert A, Lein S, Schotta G. et al. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377-392
15. Bernstein BE, Mikkelsen TS, Xie X. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315-326
16. Soshnikova N, Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science. 2009;324:1320-1323
17. Bernstein BE, Kamal M, Lindblad-Toh K. et al. Genomic Maps and Comparative Analysis of Histone Modifications in Human and Mouse. Cell. 2005;120:169-181
18. Bracken AP, Dietrich N, Pasini D. et al. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & Development. 2006;20:1123-1136
19. Lee TI, Jenner RG, Boyer LA. et al. Control of Developmental Regulators by Polycomb in Human Embryonic Stem Cells. Cell. 2006;125:301-313
20. Barski A, Cuddapah S, Cui k. et al. High-resolution profiling of histone methylation in the human genome. Cell. 2007;129:823-837
21. Min H, Lee JY, Kim MH. Structural Dynamics and Epigenetic Modifications of Hoxc Loci along the Anteroposterior Body Axis in Developing Mouse Embryos. Int J Biol Sci. 2012;8:802-810
22. Lee JY, Min H, Wang X, Khan AA, Kim MH. Chromatin organization and transcriptional activation of Hox genes. Anat Cell Biol. 2010;43:78-85
23. Kong KA, Yoon H, Kim MH. Akt1 as a putative regulator of Hox genes. Gene. 2013;513:287-291
24. Noordermeer D, Leleu M, Splinter E. et al. The dynamic architecture of Hox gene clusters. Science. 2011;334:222-225
25. Cao R, Wang L, Wang H. et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039-1043
26. Negishi M, Saraya A, Miyagi S. et al. Bmi1 cooperates with Dnmt1-associated protein 1 in gene silencing. Biochem Biophys Res Commun. 2007;353:992-998
27. Rinn JL, Bondre C, Gladstone HB. et al. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006;2:e119
Author contact
![]() Corresponding author: E-mail: mhkim1ac. Tel: +82-2-2228-1647, Fax: +82-2-365-0700.
Corresponding author: E-mail: mhkim1ac. Tel: +82-2-2228-1647, Fax: +82-2-365-0700.

 Global reach, higher impact
Global reach, higher impact