10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(9):989-1003. doi:10.7150/ijbs.6978 This issue Cite
Research Paper
Molecular Characterization and Alternative Splicing of a Sodium Channel and DSC1 Ortholog Genes in Liposcelis bostrychophila Badonnel (Psocoptera: Liposcelididae)
Key Laboratory of Entomology and Pest Control Engineering, College of Plant Protection, Southwest University, Chongqing 400716, P. R. China.
Received 2013-6-23; Accepted 2013-9-14; Published 2013-9-26
Abstract
Alternative splicing greatly contributes to the structural and functional diversity of voltage-gated sodium channels (VGSCs) by generating various isoforms with unique functional and pharmacological properties. Here, we identified a new optional exon 23 located in the linker between domains II and III, and four mutually exclusive exons (exons 27A, 27B, 27C, and 27D) in domains IIIS3 and IIIS4 of the sodium channel of Liposcelis bostrychophila (termed as LbVGSC). This suggested that more alternative splicing phenomena remained to be discovered in VGSCs. Inclusion of exon 27C might lead to generation of non-functional isoforms. Meanwhile, identification of three alternative exons (exons 11, 13A, and 13B), which were located in the linker between domains II and III, indicated that abundant splicing events occurred in the DSC1 ortholog channel of L. bostrychophila (termed as LbSC1). Exons 13A and 13B were generated by intron retention, and the presence of exon 13B relied on the inclusion of exon 13A. Exon 13B was specifically expressed in the embryonic stage and contained an in-frame stop codon, inclusion of which led to generation of truncated proteins with only the first two domains. Additionally, several co-occurring RNA editing events were identified in LbSC1. Furthermore, remarkable similarity between the structure and expression patterns of LbVGSC and LbSC1 were discovered, and a closer evolutionary relationship between VGSCs and DSC1 orthologs was verified. Taken together, the data provided abundant molecular information on VGSC and DSC1 orthologs in L. bostrychophila, a representative Psocoptera storage pest, and insights into the alternative splicing of these two channels.
Keywords: Liposcelis bostrychophila, VGSC, DSC1, qPCR, Alternative splicing, RNA editing.
Introduction
Voltage-gated sodium channels (VGSCs) are large integral transmembrane proteins that are responsible for the rapid rising phase of action potentials, and are essential for electrical signaling in neurons and other electrically excitable cells [1]. Initially, two homologs of sodium channels, para and DSC1, were identified from the genome of Drosophila [2-3]. Subsequent functional expression in Xenopus oocytes demonstrated that para was the only VGSC gene in Drosophila [4], and that DSC1 encoded voltage-gated calcium selective cation channels like BSC1 [5-6]. The VGSC and DSC1 orthologs had prominent structural similarity to mammalian sodium channels, characterized by four typical homologous structural domains (I-IV), each having six transmembrane α-helix segments (S1-S6), and connected by intracellular and extracellular loops [7]. The key structural features, including voltage sensors, an ion selective filter, and a fast inactivation gate, have been extensively compared between the orthologs [8].
Because of the crucial role of VGSCs in neuron signal transmission, insect VGSCs were targeted by a variety of neuron toxins and insecticides such as DDT, synthetic pyrethroids, and oxadiazines [9-11]. Since the genetic linkage between VGSCs and knockdown resistance (kdr) was confirmed in insects, quantities of resistance-associated mutations had been characterized in many agricultural and medically important pests [12-13]. Nevertheless, only a few mutations were confirmed to reduce the sensitivity to pyrethroids in Xenopus oocytes [13]. As for DSC1 orthologs, their identification only in some invertebrates implied their potential application in developing new insecticide targets. Previously, DSC1 was demonstrated to be involved in insect olfaction by p-element insertion [14]. A recent gene knock-out study on DSC1 confirmed its role in olfaction and suggested that DSC1 also had a role in regulating neuronal stability, particularly in extending the stability of neural circuits and behaviors under environmental stresses and insecticide exposure [15]. The work also suggested that DSC1 had a significant application in developing new and safer insecticides [15]. However, the specific biological function of DSC1 orthologs remains to be clarified in more species. Considering the high level of structural and sequence similarity between VGSCs and DSC1 orthologs, a comparative analysis will contribute further insights into their molecular characteristics and evolutionary relationship. The availability of full-length cDNA clones is the first crucial step for the functional expression in Xenopus oocytes, to elucidate their individual roles and gating properties. In 2007, the full-length cDNAs of VGSCs were only available in three insect species [8]. The last 5 years witnessed a rapid increase in the number of full-length cDNAs of VGSCs deposited in GenBank, and nearly 30 full-length cDNAs from different species in seven insect orders were available. Hence, cloning of cDNAs from more insect orders will advance studies on the evolutionary and functional characteristics of VGSCs, as well as contribute to the identification of conserved resistance-conferring mutations, which are crucial information for designing new insecticides.
Alternative splicing is the prominent mechanism for generating protein diversity, and RNA editing is an important post-transcriptional modification that can significantly change protein functions by introducing site-specific alterations in gene transcripts, including conversion of one base to another, or the insertion and/or deletion of nucleotides [16-17]. Alternative splicing and RNA editing are the main means of generating structural and functional diversity in voltage-gated ion channels of insects [18-20]. Alternative splicing variants were demonstrated to show different gating properties and generate a range of isoforms with distinctive sensitivities to pyrethroids [20-21]. With the increasing awareness of the importance of alternative splicing, alternative splicing events of VGSCs have been reported in many agricultural and medically-important pests [18, 21-27]. The majority of alternative exons were conserved in insects, while an optional exon 12 in the silkworm Bombyx mori sodium channel might be species-unique [25]. Intensive alternative splicing was also documented in DSC1 and BSC1 [5,28]. Although the precise biological functions of those splicing variants remain unknown, characterization of splicing events will greatly contribute to our understanding of the diversity of insect VGSCs, as well as facilitate the functional elucidation of certain isoforms.
As yet, there were no reports on the VGSC and DSC1 orthologs of Psocoptera, a group that included many economically-important storage pests. In this study, we performed a comparative analysis between the VGSC and DSC1 orthologs first isolated from Liposcelis bostrychophila, one of the most economically-important pests in Psocoptera [29-30], and as a new risk for global food security and safety [31]. In addition, a series of alternative splicing events were characterized in both genes, and several potential RNA editing sites were revealed.
Materials and Methods
Insect culture
The stock colony of L. bostrychophila was originally collected from grain storage facilities in Beibei, Chongqing, China, in 2008. They were reared on an artificial diet composed of whole wheat flour, yeast powder, and skimmed milk (10:1:1) in air-conditioned rooms at a constant temperature of 27 ± 1°C and relative humidity of 75-80%, with a scotoperiod of 24 h. Plastic vials (1 cm high by 2.4 cm diameter) containing a small amount of diet were used to collect eggs, and about 20 adults were cultured in each of the vials. Four days later, the eggs were collected under a microscope (Olympus SZX12, Tokyo, Japan) using soft brushes. To collect the nymphs and adults, the adults were removed 4 days later, with only the eggs left behind. Consistent with the life history of L. bostrychophila at 27.5°C [32], we collected the nymphs during the period from 11-22 days, and the adults 26 days later. Body parts, including head (with antennae), thorax (with legs), and abdomen, were dissected from the adults under a microscope, and placed in a 1.5 mL centrifuge tube containing RNA store reagent (Tiangen, Beijing, China), respectively. All collected samples were stored at -80°C for future use.
RNA extraction and cDNA synthesis
Total RNA was extracted from 5 mg each of collected eggs, nymphs, adults, and the separated head (with antenna), thorax (with legs), and abdomen of adult L. bostrychophila, respectively, according to the manufacturer's protocols for the RNeasy plus Micro Kit (QIAGEN, Hilden, Germany). All RNA samples were treated with gDNA elimination columns supplied with the kit to remove genomic DNA. The quality, purity, and concentration of the RNA were measured at 260 nm and 280 nm using a NanoVue UV-Vis spectrophotometer (GE Healthcare Bio-sciences, Uppsala, Sweden), and the integrity was checked by 1.0% agarose gel electrophoresis. For first strand cDNA synthesis, 500 ng of total RNA was reverse transcribed using the PrimeScript 1st Strand cDNA Synthesis Kit (Takara, Dalian, China), with random hexamer and oligo (dT)18 primers, according to the manufacturer's instructions. The cDNA was stored at -20°C until future use. The cDNA templates used in full-length cloning were synthesized from a mixture of equal amounts of total RNA from eggs, nymphs, and adults.
Amplification of full-length cDNA of LbVGSC from L. bostrychophila
Two partial VGSC sequences were identified from transcriptome data of L. bostrychophila. The transcriptome was generated by Illumina sequencing of a mixture of RNA from eggs, nymphs, and adults (GenBank accession number: SRS390072). Specific primers were designed to fill the gaps based on the two sequences. Cloning strategies were shown in Fig. 1. To obtain the 3′ ends, we amplified fragment “e” using a set of specific primers, of which the antisense primer was derived from the cDNA sequence of a VGSC from Bactrocera dorsalis [33]. To obtain the complete open reading frame (ORF) by generating fragment “a”, rapid amplification of cDNA ends (RACE) was performed using the SMARTer RACE cDNA Amplification Kit (Clontech, Palo Alto, CA, USA) according to the manufacturer's instructions, in which only one round PCR was performed using a gene specific primer and universal primer NUP (5′-AAGCAGTGGTAACAACGCAGAGT-3′). Next, fragments “b” and “c” were isolated to fill the gaps. Five overlapping fragments corresponding to the cDNA sequence of VGSC from L. bostrychophila were cloned.
Degenerate primers were designed against the highly conserved domains from the sodium channels of other insects, and gene specific primers were designed based on the known sequences, using Primer Premier 5.0 software (Premier Biosoft International, CA, USA) (Supplementary Material: Table S1). All PCRs were performed using a C1000 Thermal Cycler (BIO-RAD, Hercules, CA, USA) in 25 μL reaction mixtures containing 1 μL template cDNA (500 ng), 2.5 μL 10 × La PCR buffer II (Mg2+ free), 2.5 μL Mg2+ (25 mM), 4.0 μL dNTPs (each dNTP 2.5 mM), 1 μL of each primer (10 μM), and 0.25 μL La Taq (5 U/μL) (Takara). Thermal cycling conditions were: one cycle of 94°C for 1 min; 30 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 1 min; one cycle of 72°C for 10 min, then holding at 16°C.
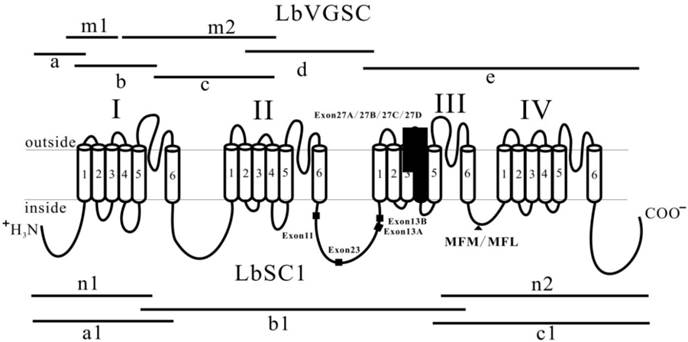
Strategies used in cloning LbVGSC and LbSC1 and schematic representation of topology of VGSCs and DSC1 orthologs. The four homologous domains (I-IV), each having six transmembrane fragments (S1-S6), were indicated. Fragments “m1”, “m2”, “n1”, and “n2” were derived from the transcriptome data of L. bostrychophila (SRS390072). Fragments “a”, “b”, “c”, “d”, and “e” were used to assemble LbVGSC, and “a1”, “b1”, and “c1” were used to assemble LbSC1. Lines indicated the length and relative location of amplicons in relation to the topological structure. The solid boxes indicated the alternatively spliced exons. The solid triangle showed the inactivation gate, labeled with MFM/MFL.
Amplification of the full-length cDNA of LbSC1 from L. bostrychophila
Three overlapping PCR products, a1, b1, and c1, were generated (Fig. 1) using three sets of gene specific primers (Supplementary Material: Table S1) designed against the known sequences that were identified from the transcriptome data (SRS390072). The PCR reaction system and conditions were the same as mentioned above.
Cloning and sequencing
All PCR products were separated by 1.0% agarose gel electrophoresis and purified with a Gel Extraction Mini Kit (Watson Biotechnologies, Shanghai, China). The purified products were cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) and transformed into Trans5α chemically competent cells (Transgen, Beijing, China). Three to five positive clones were screened by ampicillin selection and verified by PCR using primers M13F and M13R. The inserts of cDNA clones were fully sequenced in both directions (BGI, Shenzhen, China).
Sequence analysis and phylogenetic tree reconstruction
Sequence assembly and similarity evaluation were performed using DNAMAN 5.2.2 (Lynnon BioSoft, Pointe-Claire, Canada). BLAST searches were performed online at NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignments were carried out in Clustal Omega [34]. The theoretic molecular weight and isoelectric point (pI) of deduced amino acid sequences were calculated by the ExPASy Proteomics Server (http://cn.expasy.org/tools/pi_tool.html). Protein kinase A (PKA) and protein kinase C (PKC) sites were predicted by KinasePhos [35]. N-Glycosylation sites were predicted by the NetNGlyc 1.0 server [36]. The Gblocks Sever was used to generate conserved blocks from multiple alignments for use in phylogenetic analysis [37]. A consensus tree was constructed using maximum likelihood (ML), by the program phyML 3.0, and with the neighbor-joining (NJ) method, by MEGA 5.1 [38]. The LG and Jones-Taylor-Thornton amino acid substitution models were used in ML and NJ, respectively. Phylogenies were tested by a bootstrap method, with 100 replicates for ML and 1000 replicates for NJ. The topology of the tree was refined using CorelDraw 12.0 (Corel Corporation, Ottawa, Canada), based on the higher bootstrap (Fig. 3).
Analysis of developmental and tissue-specific expression profiles of LbVGSC and LbSC1 by qPCR and RT-PCR
The mRNA expression patterns of LbVGSC and LbSC1 in embryos, nymphs, and adults were determined by the 2-ΔΔCT method using the StepOne Plus Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA), with the house-keeping gene α-tubulin as a reference. Primers for the α-tubulin gene were obtained from a previous study that validated it as a reference gene [39]. All gene specific primers were designed against regions where no alternative splicing exons were found (Supplementary Material: Table S1), using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome).The specificity of each set of primers was validated by melting profile. The efficiency and determination coefficient (R2) of PCR amplification were calculated from the slope of the standard curve constructed from five serial dilutions of one cDNA sample. According to the manufacturer's instructions, each qPCR was carried out in a 20 μL reaction volume containing 1 μL cDNA template (400 ng), 10 μL iQ SYBR Green Supermix (BIO-RAD), 1 μL of each primer (10 μM), and 7 μL of nuclease-free water. The reaction conditions were: one cycle at 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. Melting curve analysis from 60°C to 95°C was applied to all reactions to ensure specificity and consistency of all generated products. All PCR reactions were performed for each gene with three biological replicates. The data were analyzed statistically using one-way analysis of variance (ANOVA) by SPSS 20.0 (IBM Corp, Chicago, IL, USA).
For RT-PCR assays of transcripts abundance in the three developmental stages and three body parts of adults, each cDNA sample was diluted to 400 ng/μL. Each 25-μL reaction contained 1 μL template cDNA (400 ng), 2.5 μL 10 × PCR buffer (Mg2+-free), 2.5 μL Mg2+ (25 mM), 2.0 μL dNTPs (each dNTP 2.5 mM), 1 μL of each primer (10 μM), 0.25 μL rTaq (5U/μL) (Takara), and 14.75 μL nuclease-free water. Reactions were performed in a thermal cycler as follows: one cycle of 94°C for 1 min; 30 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 30 s; one cycle of 72°C for 5 min, and holding at 16°C. Each PCR product (10 μL) was analyzed by 2.0% agarose gel electrophoresis with GoldView II (Solarbio Co., Ltd., Shanghai, China) staining. All electrophoresis images were refined using Adobe Photoshop CS5 (Adobe System, San Jose, CA, USA). Equal amounts of RNA used in the cDNA synthesis were indicated by similar intensities of the α-tubulin gene bands. Three replicates were performed for each trial.
Genomic structure determination and diagnostic assay for the detection of alternatively spliced exons using RT-PCR
L. bostrychophila genomic DNA was extracted from 200 heads of adults, using a Tissue/cell gDNA Mini Kit (Watson Biotechnologies, Shanghai, China), following the manufacturer's protocols. To determine the genomic structure of each splicing site, primers flanking the alternative exons were designed to amplify the regions harboring all identified alternative splicing sites. PCR was carried out as mentioned above, using 500 ng of genomic DNA as a template for each reaction. Refer to Supplementary Material: Table S2 for detailed primer information.
To detect the presence of alternative exons in different developmental stages and body parts, cDNA templates were synthesized from the RNA pools of the three developmental stages and three body parts mentioned above. The cDNA templates were diluted to 400 ng/μL. One microliter of cDNA template was used in each PCR reaction. PCR products were separated by 2.0% agarose gel electrophoresis and visualized using a Gel Doc XR+System (BIO-RAD). The inclusion or exclusion of an alternative exon was indicated by the band patterns. Three replicates were carried out for each trial.
Results
Full-length cDNA cloning of LbVGSC and LbSC1
Five overlapping amplicons were generated by RT-PCR and RACE, and were assembled into a 6054 bp contiguous sequence, encoding 2014 amino acids and covering 98% of the predicted ORF (GenBank accession number: KC699919) (Fig. 1). Among them, fragment “e” harbored the stop codon. 5′-RACE was performed to complete the ORF, but after repeated attempts, only a 480 bp amplicon (fragment “a”) was amplified, which was short of the predicted start codon by 126 bp. Thus, we used the high homology between LbVGSC and VGSC of Pediculus humanus (89.5% amino acid sequence homology) and added 43 amino acid residues (MSDISDFHSEDEQRLFRPFTRESLAAIEQRIAQENEKFKELEK) from P. humanus VGSC to the N-terminus of LbVGSC to allow the following calculations. This hybrid product had a theoretical molecular weight of 232.8 kDa and pI of 5.12. To clone the full-length cDNA of LbSC1, we identified fragments “n1” and “n2” from the transcriptome of L. bostrychophila (SRS390072), and these two fragments harbored the start and stop codons, respectively. Based on these two sequences, we isolated three overlapping amplicons, “a1”, “b1”, and “c1” (Fig. 1), and assembled them into a contiguous 7618 bp sequence. The ORF of LbSC1 encoded 2535 amino acids (GenBank accession number: KC699920) (Fig. 1) with an evaluated theoretical molecular weight of 287.2 kDa and a pI of 8.43.
Comparative structure and homology analysis
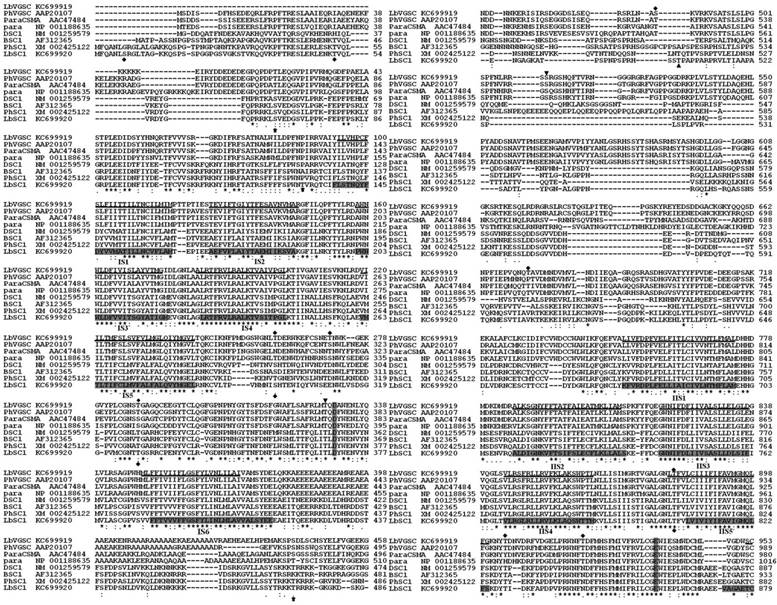
All of the typical structural features of VGSCs and DSC1 orthologs were highly conserved in LbVGSC and LbSC1. Amino acid sequence alignments with their counterparts in other insect species, para (NP_001188635), paraCSMA (AAC47484), P. humanus VGSC (AAP20107), DSC1 (NM_001259579), BSC1 (AF312365), and P. humanus DSC1 ortholog (XM_002425122), revealed that the canonical structural features of LbVGSC and LbSC1 were conserved (Fig. 2). Both LbVGSC and LbSC1 were composed of four putative homologous domains, each containing six transmembrane α-helical segments connected by intracellular and extracellular loops (Fig. 2). The four amino acid residues crucial for ion selectivity in the membrane reentrant loops, between S5 and S6 segments, were DEKA in LbVGSC and DEEA in LbSC1. The S4 segments responsible for voltage sensing had four to eight positively charged amino acids (arginine or lysine) at every third position. In LbVGSC, four, five, five, and eight positively charged amino acids were identified from domains IS4-IVS4 respectively, and four, four, six, and eight positively charged amino acids were identified in the corresponding domains of LbSC1. The three hydrophobic amino acid residue motif in the linker between domains III and IV, which served as the fast inactivation gate, was highly conserved in LbVGSC (MFM) and LbSC1 (MFL). Prediction of phosphorylation sites indicated that there were three and zero potential PKA and PKC sites, respectively, in LbVGSC, while LbSC1 possessed 10 and nine potential PKA and PKC sites, respectively (Fig. 2). In addition, 11 potential N-glycosylation sites were predicted in LbVGSC, and nine in LbSC1. These sites were mainly distributed in the linkers connecting S5 and S6, and some sites were shared by both channels.
Protein sequence analysis showed that the ORF of LbSC1 was longer, and its pI was much higher, than those of LbVGSC. This was the case for other DSC1 orthologs having longer ORF and higher pI than their corresponding VGSCs in the alignments. The longer ORFs of DSC1 orthologs were mainly attributed to the longer sequences in the linkers between domains II and III, and at the 3′ end (Fig. 1). To check whether this was the case for all the species, we investigated the length, pI, and theoretical molecular weight of all channels used in the phylogenetic analysis (Supplementary Material: Table S3, S4, and S5). In most cases, DSC1 orthologs had longer ORFs than VGSCs, and higher pIs (Supplementary Material: Table S3 and S4). The average lengths of DSC1 orthologs and VGSCs were about 2328 bp (1896-2961 bp) and 2093 bp (1830-2201 bp), respectively. The average pI was 7.00 (6.20-8.65) for DSC1 orthologs and 5.11 (4.95-5.41) for VGSCs.
Homology analysis showed that LbVGSC shared the highest identity with P. humanus VGSC (89.5%), paraCSMA (82.7%), and para (76.7%). The similarity between DSC1 orthologs was relatively lower, as LbSC1 had 64.8% identity with that of P. humanus, 61.4% with BSC1, and 53.4% with DSC1. Amino acid sequence alignments between LbVGSC and LbSC1 showed only 28.5% overall similarity. Considering the high structural similarity, we performed partial sequence alignments of transmembrane segments and loops connecting domains between LbVGSC and LbSC1. The alignments showed that S4s of each domain forming the voltage sensor shared the highest homology with each other (50.0-85.0%), while other transmembrane fragments shared 25.0-73.3% homology. Only 10.0% and 6.7% homology was calculated in the linkers between domains I and II, but a predominantly higher homology (53.7%) was obtained between the linkers connecting domains III and IV.
Phylogenetic analysis among insect voltage-gated calcium channels (VGCCs), VGSCs, and DSC1 orthologs
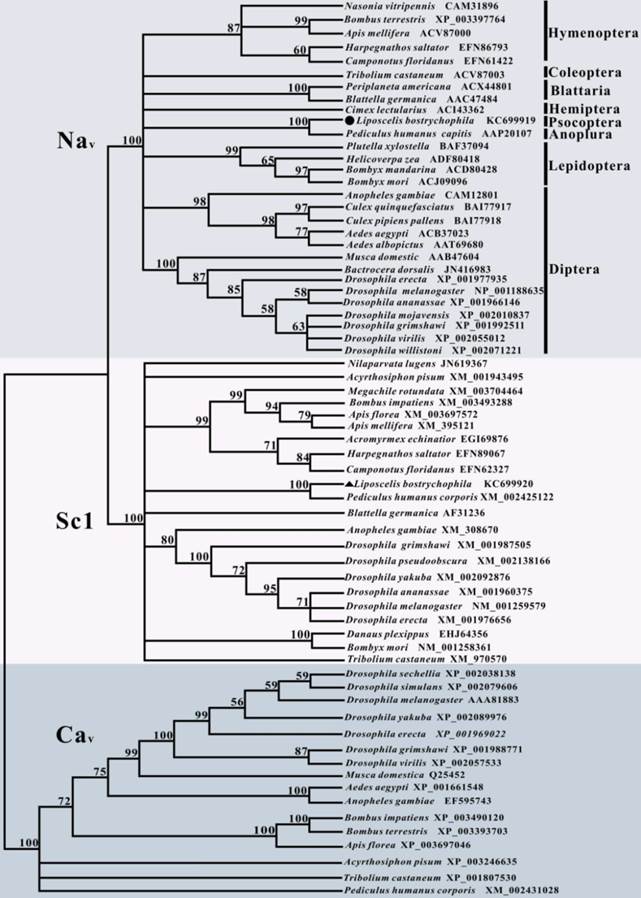
To clarify the evolutionary relationship of LbVGSC and LbSC1 with other insect orthologs, a phylogenetic tree was constructed based on the protein sequences of LbVGSC and LbSC1 and 67 other proteins sequences, including 28 insect VGSCs, 21 DSC1 orthologs, and 16 VGCCs (Fig. 3). This group included almost all insect orthologs of these three protein types with complete ORFs deposited in GenBank. The results revealed that all sequences clustered into three large clades (Fig. 3), which implicated that all three types were phylogenetically conserved among insects. VGSCs and DSC1 orthologs clustered into a larger clade separated from VGCCs, which suggested that VGSCs had a closer evolutionary relationship with DSC1 orthologs than with VGCCs, and VGCCs represented a more ancestral status.
Alignments of protein sequences of LbVGSC, LbSC1, para, DSC1, paraCSMA, BSC1, PhVGSC (para ortholog from Pediculus humanus), and PhSC1 (DSC1 ortholog from P. humanus). The GenBank accession numbers were indicated following the sequence names. The transmembrane segments (S1-S6) were underlined for LbVGSC and highlighted in black for LbSC1. The four-amino-acid DEKA motif in VGSCs and DEEA in DSC1 orthologs, for ion selection, in each linker between S5 and S6, were highlighted in black in each S4 region. The inactivation gates “MFM” and “MFL” between II and III were highlighted and boxed. All alternatively spliced exons included in the sequences were boxed, and thin arrows indicated the positions of those not included. The PKC sites were denoted with bold arrows. PKA sites were denoted with “▼” for LbVGSC and “▲” for LbSC1. The N-glycosylation sites were denoted with “◆”. “*” denoted identical residues; “:” denoted a conservative residue substitution; “.” denoted partial conservation of the residue.


Phylogenetic analysis of 67 insect amino acid sequences, including 29 VGSCs, 22 DSC1 orthologs, and 16 VGCCs. LbVGSC was denoted with a solid circle, and LbSC1was denoted with a solid triangle. Only bootstrap values exceeding 50% were shown at branch points. GenBank accession numbers of all sequences were listed following the species names and in Tables S3-S5. Members of VGSCs, DSC1 orthologs, and VGCCs were highlighted with different colors and denoted with corresponding names.
Expression profiles of LbVGSC and LbSC1 mRNA
The results of qPCR showed that the expression levels of both LbVGSC and LbSC1 were highest in the adult stage and lowest in the embryonic stage, in consistency with the results of RT-PCR (Fig. 4A and 5A). To determine the tissue-specific expression patterns, RT-PCR was performed to analyze the expression levels in adult head, thorax and abdomen (Fig. 4B and 5B). LbVGSC was highly expressed in the head and thorax, indicating that LbVGSC was mainly distributed in neurons and muscles. LbSC1 had a relatively higher expression in the head and abdomen than in the thorax.
Developmental and tissue-specific expression of LbVGSC. (A) RT-qPCR analysis of developmental expression patterns from the embryonic to adult stages. Different letters above the bar indicated significant differences in the expression level of LbVGSC (p < 0.05, one-way ANOVA). (B) RT-PCR analysis of LbVGSC in head, thorax, and abdomen of adult insects. Each PCR product (10 μL) was separated on a 2.0% agarose gel.
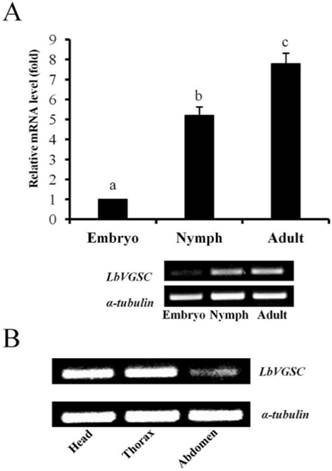
Developmental and tissue-specific expression of LbSC1. (A) RT-qPCR analysis of developmental expression patterns from embryonic to adult stages. Different letters above the bar indicated significant differences in the expression level of LbSC1 (p < 0.05, one-way ANOVA). (B) RT-PCR analysis of LbSC1 in head, thorax, and abdomen of adult insects. Each PCR product (10 μL) was separated on a 2.0% agarose gel.
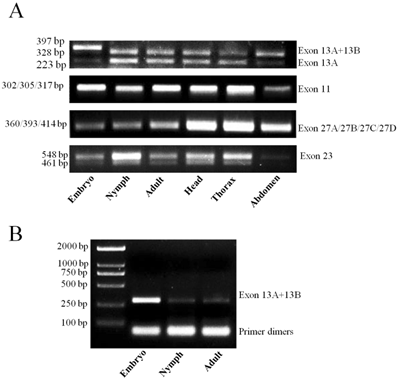
Identification of alternative exons and detection in developmental stages and body parts
Comparison of multiple cDNA clones revealed that there were several alternative splicing variants in LbVGSC and LbSC1 transcripts, leading to the discovery of two variable regions in LbVGSC and two in LbSC1 (Fig. 1). Three alternatively spliced exons in LbVGSC were discovered, including exon 23 (87 bp) located in the linker connecting domains II and III, and exons 27A (102 bp) and 27B (123 bp) spanning IIIS3 and IIIS4. The alternative exons were named based on the numeral of the silkworm B. mori sodium channel (BmNav) and Culex mosquito sodium channel [25-26]. Exon 23 in LbVGSC was located next to the counterpart of exon 22ii in BmNav and partially overlapped with the counterpart of exon h in para [18]. Exon 11 and exon 13A located in the linker connecting domains II and III, were identified in LbSC1 and named based on the numeral from DSC1 [28].
RT-PCR detected exon 13A in all stages and body parts. In addition, a larger than expected band was observed in samples corresponding to the embryonic stage, indicating another alternatively spliced exon existed in this region, and variants containing this exon might be embryo-specific (Fig. 6A). Hence, we cloned and sequenced the band, which identified it as a new exon, named exon 13B. This exon was 69 bp long and located downstream from exon 13A. Retention of exon 13B generated an in-frame stop codon, which might lead to formation of truncated proteins with only the first two domains. In addition, exon 13B had relatively high expression in the embryonic stage, but a very low expression level in nymphal and adult stages (Fig. 6B). Interestingly, only a band containing both exon 13A and 13B was detected; exon 13B was never detected alone. This suggested that variants with inclusion of exon 13B could be produced only if exon 13A were also included. RT-PCR assay against exon 23 showed that it was present in all tested samples.
RT-PCR was unable to generate distinguishing bands for exons 27A, 27B, and 11 (Fig. 6A). Therefore, we sequenced five randomly-selected clones from each cDNA pool, for a total of 30 clones for the two regions containing exon 27A and 27B, and exon 11. Multiple sequence alignments were performed for all obtained sequences (Supplementary Material: Fig. S1 and S2). Two new exons, designated as exons 27C and 27D, were identified at the same sites as exons 27A and 27B. In all sequenced clones, only one copy of exon 27A, 27B, 27C, or 27D was present in a single transcript (Supplementary Material: Fig. S1), indicating they might be mutually exclusive. Variants containing exon 27B were the main transcripts in all developmental stages and body parts. Exon 27A was only detected in adults, and exon 27C was only detected in the heads of adults. Exon 27D was detected in five clones, four from adult thorax and one from nymph. Exon 11 was only detected in three clones from adult samples (Supplementary Material: Fig. S2), suggesting that variants excluding exon 11 were the main transcripts of LbSC1.
Inclusion or exclusion of certain exons influenced the phosphorylation level of LbVGSC. Inclusion of exon 23 generated no additional PKA or PKC sites, and only one N-glycosylation was detected in exon 23. One PKA site was present within IIIS4 in exon 27B. One more cGMP-dependent protein kinase PKG site was predicted in exon 27D than in 27B. Inclusion of 27A generated a PKC site, at the position corresponding to the PKA site in 27B. No extra PKC or PKA sites were detected after inclusion or exclusion of exons 13A and 13B.
Detection of alternatively spliced exons in different developmental stages and tissues by RT-PCR. (A) PCR detection for exons in different developmental stages. Primers were designed to flank the splicing sites. Different bands indicated various products by inclusion or exclusion of specific exons. Each PCR product (10 μL) was separated on a 2.0% agarose gel. (B) PCR detection for exons 13A and 13B. The amplified region included exon 13A and part of exon 13B. The antisense primer (13A/13B-R1) was designed within exon 13B. 20 μL of the product was separated on a 2.0% agarose gel to detect any low-abundance transcripts.
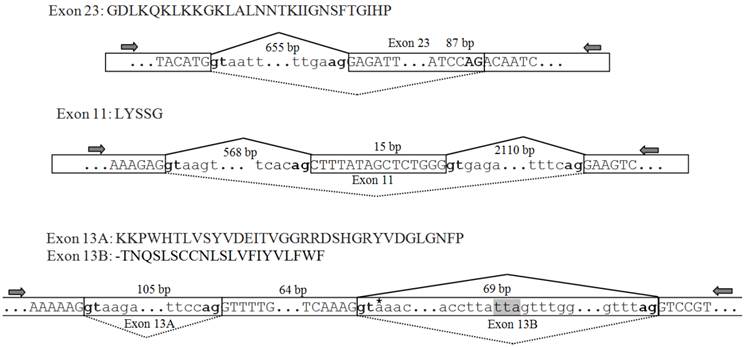
Genomic structure and unique splicing patterns
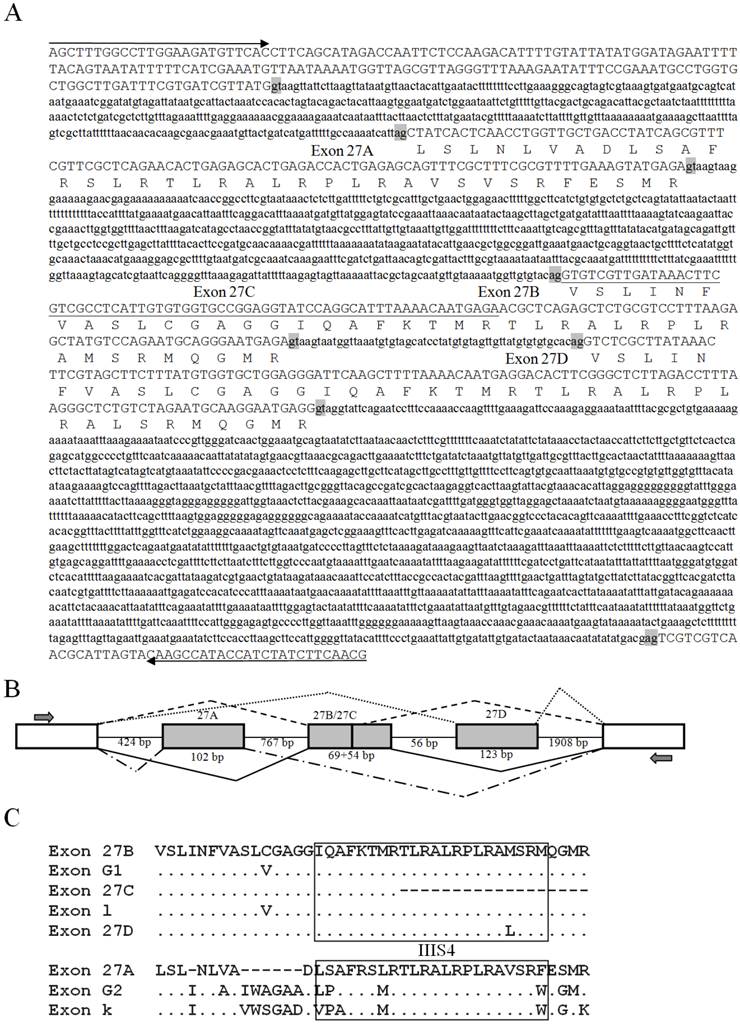
The genomic sequences corresponding to the splicing sites were cloned to clarify the splicing patterns, and all sequences were deposited in GenBank [accession numbers: KF059949 (exon 23), KF059950 (exons 27A/27B/27C/27D), KF059951 (exon 11), and KF059951 (exons 13A and 13B)]. The genomic structure and splicing mechanism of the observed splicing sites were presented in Figs. 7 and 8B. Consensus splice donor and acceptor sequences, gt/ag, at each intron/exon border, were conserved in all splice variants (Figs. 7 and 8A). Exon 23 located in the linker between domains II and III, was an internally optional spliced exon that were generated using an alternative 3′ splicing site in an adjacent downstream constitutive exon, as shown in Fig. 7. Complicated splicing patterns were found in the sites corresponding to exons 27A/27B/27C/27D, which spanned IIIS3 and IIIS4. Exons 27A, 27B, and 27D were cassette exons, separated by three introns, while 27C was part of 27B, resulting from alternative 3′ splice site selection (Fig. 8A, B). Exons 27B and 27D differed at 25 nucleotides, but still shared extremely high identity, with only one amino acid difference between them. Exons 27B and 27D each had only one amino acid difference from exon l in para and exon G1 in paraCSMA (Fig. 8C). Exon 27A shared 68.3% identity with exon G2 and 53.7% with exon k in para, much higher than with exon 27B or 27D. Exon 27C shared no significant amino acid homology with exon G3 and harbored no in-frame stop codons. Additionally, a conserved sequence of ATGAGA/G, at the end of exons 27A/27B/27C/27D, was identified (Fig. 8A). In LbSC1, the short exon 11 (LYSSG) was confirmed to be a cassette exon bordered by two relatively long introns (Fig. 7). Exon 11 was generated by the same splicing sites as exons 11A, 11B, and 11C in DSC1, but shared no significant sequence similarity with them. Both exons 13A and 13B were classified as retained introns for their unique splicing mechanism, as shown in Fig. 7. Exons 13A and 13B were located within constitutive exon 13 in DSC1, but whether DSC1 underwent such splicing events in exon 13 remained to be determined. In addition, compared with the genomic DNA sequence, the cDNA sequence of exon 13B lacked a TTA sequence (Fig. 7).
By comparing the cDNA sequence with the corresponding genomic DNA sequence, a series of RNA editing events were identified in the region containing exon 11 (Supplementary Material: Fig. S2). Some of the nucleotide changes might result from PCR errors, therefore only nucleotide changes detected in at least two variants were defined as RNA editing sites. Nine clones had a deletion of three bases (CAG), resulting in loss of one amino acid (Gln) but no frame-shift of the ORF. This deletion event was mainly observed in the adult stage, but also in the embryonic and nymphal stages. Four single nucleotide changes were also observed, including one A>C, one A>G, and two T>C changes (Supplementary Material: Fig. S2). The first C>T change was only detected in two clones, one from whole adults and another from the adult abdomen, and led to an Ala>Val substitution. The second C>T change was a synonymous editing event, resulting in no amino acid change. The A>C and A>G changes caused Ile>Leu and Lys>Arg changes, respectively. The latter three editing sites co-occurred and were detected in seven clones from whole adults or the three body parts of adult, and were accompanied by deletion of CAG. Variants without alterations in these sites were the most common transcripts in all developmental stages and body parts.
Discussion
VGSCs, as the target of pyrethroids, have been studied in insects for decades. Here, we characterized the first VGSC gene in L. bostrychophila of Psocoptera, and clarified its expression profile and unique alternative splicing phenomena. DSC1 orthologs were reported in several invertebrates but absent in vertebrates, and this ancient gene family was lost in several groups of invertebrates [40]. In this study, we confirmed the presence of DSC1 orthologs in Psocoptera by cloning the full-length cDNA of LbSC1 from L. bostrychophila. The isolation of LbSC1 would allow further comparative analysis and studies of the biological function of DSC1 orthologs in L. bostrychophila.
It was confirmed that the four amino acids, which determined ion selectivity were DEKA in VGSCs, DEEA in DSC1 orthologs, and EEEE or EEDD in VGCCs [1,6,41]. Previous studies showed that substitution of the third amino acid could alter the ion selectivity, indicating its important role in modulating this process [6,42]. In this study, LbVGSC encoded a VGSC that possessed the conserved DEKA motif. The DEEA motif of LbSC1 implied that it encoded a voltage-gated calcium selective cation channel, like DSC1 and BSC1. It was well known that the S4 regions served as voltage sensors, and were characterized by four to eight positively charged arginine or lysine residues, each separated by two neutral amino acids [7]. Most of the positively charged amino acids were conserved between VGSCs and DSC1 orthologs in the alignments, except for minor differences between LbVGSC and LbSC1in IIS4 and IIIS4. LbVGSC had four and six positively charged amino acids in the IIS4 and IIIS4, respectively, which were conserved in other DSC1 orthologs except BSC1 [28]. Whether such differences have a functional effect on gating properties remains to be elucidated.
Phosphorylation is involved in down-regulating sodium currents and slowing Na+ channel inactivation in mammalian sodium channels [43]. Furthermore, phosphorylation sites in alternative exons could have a role in regulating sodium channel expression [44]. A PKA/PKC site in variants containing exon 27A/27B was located in IIIS4 of LbVGSC, which might be involved in voltage sensing. Interestingly, a potential PKC site in IIIS4 in LbSC1 corresponded to a site in LbVGSC (Fig. 1), indicating that phosphorylation at this position might be relatively important. Glycosylation is necessary for maintenance of the normal steady state of biosynthesis and degradation of sodium channels [45]. Several glycosylation sites were predicted in both channels; however, whether they have functional consequences remains to be investigated.
Amino acid sequences, schematic of the genomic structure, and splicing patterns of exon 23 in LbVGSC, and exons 11, 13A and 13B in LbSC1. Amino acid sequences of alternative exons were shown above the nucleotide sequence. The genomic structure of the corresponding exon was indicated below. All exons and introns were indicated with six bases at the junction. The exons were represented by boxed uppercase letters, and retained introns 13A and 13B were indicated by boxed lowercase letters. The adjacent introns were indicated using lowercase letters without boxes. The alternative exons were labeled with corresponding names. Sizes of the bordering introns and alternative exons were indicated. The junctions of exons were indicated with bridge lines. The consensus splice donor and acceptor sequences, gt-ag, were in bold. The arrows showed the relative positions of primers used in PCR. The three nucleotides, TTA, highlighted in gray, were present in the genomic sequence but not in exon 13A. The premature stop codon TAA was denoted with an asterisk in exon 13B.
Genomic sequences, splicing patterns, and amino acid sequence alignments of exons 27A, 27B, 27C, and 27D. (A) The amplified genomic sequences containing exon 27A, 27B, 27C, and 27D. Introns were indicated with lowercase letters. The deduced amino acid sequences of the alternative exons were indicated below the nucleotides. The splicing consensus sequences were highlighted in gray. Exon 27C, located within exon 27B, was underlined. Arrows indicated primers used in PCR. (B) Schematic of splicing patterns. Constitutive exons were indicated by blank boxes, alternative exons were indicated by gray boxes, and introns were indicated by straight lines. The junctions of exons were indicated by bridge lines. The scale was not proportional. (C) Alignments of exons 27A/27B/27C/27D with k/l in para and G1/G2 in paraCSMA. The transmembrane fragment IIIS4 was boxed.
Interestingly, LbVGSC and LbSC1 shared low overall amino acid identity but significantly higher identity within specific domains. Alignment of protein sequences showed that the highest identity was observed among S4s, which formed the voltage sensors. The linkers between domains III and IV, where the fast inactivation motif was located, also shared high identity. These results showed that VGSCs and DSC1 orthologs shared significant similarity in structure and function, which had been evolutionarily conserved.
Recently, DSC1 orthologs have been found to mediate Ca2+, but clustered with VGSCs in phylogenetic analysis [46]. In the present study, phylogenetic analysis showed similar results that VGSCs and DSC1 orthologs clustered in one clade. Moreover, investigation of basic parameters showed that both the length and pI of VGSCs were less variable than in DSC1 orthologs and VGCCs. We postulated DSC1 orthologs might be much more diversified, and a more ancient member of the voltage-gated ion channels superfamily, like VGCCs.
In Blattella germanica, RT-PCR analysis showed that there were distinct differences in the developmental patterns of paraCSMA expression [5]. LbVGSC expression was lowest in the embryonic stage, which was most likely due to the fact that neuron tissue differentiation occurred at the beginning of the embryonic stage. In addition, LbVGSC mRNA transcripts were most abundant in the head and abdomen, in consistency with the fact that VGSC was mainly expressed in nerve cord and muscle, because the head and abdomen were the main parts containing most of these tissues [5]. As for DSC1, a recent study on two insect DSC1 orthologs (NlSC1 and BmSC1) showed that the relative expression levels in head (without antenna) and antenna were significantly higher than those in thorax (without leg), leg, and abdomen [40]. Our results showed that LbSC1 transcripts were observed in all three body parts of adult and were most abundant in the head, followed by the abdomen then the thorax, which was largely consistent with observations in NlSC1 [40].
It is well recognized that VGSCs underwent intensive alternative splicing to increase protein diversity. Internally-optional exon 23 has never been reported in LbVGSC, but was located next to the counterpart of exon 22ii in BmNav and shared the same splicing site, and partially overlaps, with the counterpart of exon h in para, implicating the relative conservation of this splicing site. Four mutually exclusive exons, 27A, 27B, 27C, and 27D, were first observed in IIIS3-IIIS4 in LbVGSC, where splicing events had been documented in VGSCs from species ranging from arthropods to mammals [18-19,21,25,27,47-48]. Interestingly, exon 27B differed from exon 27D by only a single Met>Leu substitution in IIIS4, hinting that this substitution might have a functional effect on the gating properties. In paraCSMA, exons G1/G2 were directly involved in modulating sodium current amplitude, voltage dependence of channel activation, and channel sensitivity to a pyrethroid [21]. It is likely that the role of those exons is also preserved in L. bostrychophila. Inclusion of exon 27C led to loss of IIIS4, a crucial element for generating functional channels, but no stop codon was generated. Exon 27C might play a similar role to the optional exon 3 in VmNav [47]. This further confirmed the conservation of this splicing site, from insects to humans, and suggested an ancient and indispensable role of the truncated or non-functional proteins [8]. More insights should be taken into the evolution of splicing in this site. Exons G1, G2 and G3 in paraCSMA exhibited distinct tissue-specific expression, but no stage-specific expression was observed [21]. Further research is required to determine the expression patterns of variants containing unique splicing exons in LbVGSC. In conclusion, we identified unique splicing patterns in LbVGSC, not highly conserved as previously reported, which hinted that more alternative splicing phenomena remained to be discovered.
Three exons, 11A, 11B, and 11C, have been identified in DSC1 at the position corresponding to exon 11 identified in LbSC1 [28]. No further variants of exon 11 were identified at this site in LbSC1. Exon 11 was only 15 bp long, encoding five amino acids, which was similar to exon 11B. Exon 11 was only detected in three clones from adults, so it might be adult-specific. Exons 13A and 13B have never been reported before. They were generated by intron retention, a rare splicing type in vertebrates and invertebrates. DSC1 had a constitutive exon 13 in this position, and 13A and 13B were located within exon 13. Similar splicing events in exon 13 in DSC1 were also possible and needed to be investigated further. Exon 13B contained a stop codon, and retention of exon 13B was expected to generate truncated proteins with only the first two domains, as production of truncated proteins had been reported in DSC1 [28]. This suggested the resulting two-domain proteins might be conserved in the evolution of voltage-gated ion channels and had potential biological significance. RT-PCR result showed that expression of variants containing exon 13B was highest in the embryonic stage, but low expression was also detected in adult and nymphal stages. Hence, we postulated variants containing exon 13B were the main forms in the embryonic stage and quantities of non-functional channels might exist in the embryonic stage. Furthermore, we found that the presence of exon 13A might promote the inclusion of exon 13B, which implicated complicated splicing mechanisms in DSC1 channels.
RNA editing has been reported in DSC1, and 17 potential RNA editing sites have been identified [28]. Similarly, a series of RNA editing sites were discovered in LbSC1. However, no functional expression was carried out to determine the consequences of each RNA editing site. Hence, to clarify the functional consequence of the co-occurring RNA editing sites in this study requires further work.
In conclusion, we provided the detailed molecular information for LbVGSC and LbSC1 in L. bostrychophila, performed a comparative analysis between them, and gained more insights into their splicing events. This study represents the first steps towards molecular characterization of VGSCs and DSC1 orthologs in Psocoptera, and also provides the first sequences of both channels. Our findings will contribute to the functional characterization of LbVGSC and LbSC1, and promote the cloning of more VGSCs and DSC1 orthologs in Psocoptera.
Supplementary Material
Figures S1-S2, and Tables S1-S5.
Acknowledgements
This work was supported by the National Natural Sciences Foundation of China (31000860), the Program for Innovative Research Team in University of China (IRT0976), the Specialized Research Fund for the Doctoral Program of Higher Education of China (20100182120022), and the Fundamental Research Funds for the Central Universities of China (XDJK2013C051, XDJK2013A005).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Catterall WA. From ionic currents to molecular mechanisms: The structure and function of voltage-gated sodium channels. Neuron. 2000;26:13-25
2. Salkoff L, Butler A, Wei A. et al. Genomic organization and deduced amino acid sequence of a putative sodium channel gene in Drosophila. Sci. 1987;237:744-749
3. Loughney K, Kreber R, Ganetzky B. Molecular analysis of the para locus, a sodium-channel gene in Drosophila. Cell. 1989;58:1143-1154
4. Littleton JT, Ganetzky B. Ion channels and synaptic viewpoint organization: analysis of the Drosophila genome. Neuron. 2000;26:35-43
5. Liu ZQ, Chung I, Dong K. Alternative splicing of the BSC1 gene generates tissue-specific isoforms in the German cockroach. Insect Biochem Mol Biol. 2001;31:703-713
6. Zhou W, Chung I, Liu Z, Goldin AL. et al. A voltage-gated calcium-selective channel encoded by a sodium channel-like gene. Neuron. 2004;42:101-112
7. Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2003;4:207
8. Dong K. Insect sodium channels and insecticide resistance. Invert Neurosci. 2007;7:17-30
9. Catterall WA, Cestele S, Yarov-Yarovoy V. et al. Voltage-gated ion channels and gating modifier toxins. Toxicon. 2007;49:124-141
10. Silver KS, Song WZ, Nomura Y. et al. Mechanism of action of sodium channel blocker insecticides (SCBIs) on insect sodium channels. Pestic Biochem Physiol. 2010;97:87-92
11. Wang SY, Wang GK. Voltage-gated sodium channels as primary targets of diverse lipid-soluble neurotoxins. Cell Signal. 2003;15:151-159
12. Soderlund DM. Pyrethroids, knockdown resistance and sodium channels. Pest Manag Sci. 2008;64:610-616
13. Rinkevich FD. et al. Diversity and convergence of sodium channel mutations involved in resistance to pyrethroids. Pestic Biochem Physiol. in press
14. Kulkarni NH, Yamamoto AH, Robinson KO. et al. The DSC1 channel, encoded by the smi60E locus, contributes to odor-guided behavior in Drosophila melanogaster. Genetics. 2002;161:1507-1516
15. Zhang T, Wang Z, Wang L. et al. Role of the DSC1 channel in regulating neuronal excitability in Drosophila melanogaster: extending nervous system stability under stress. PLoS Genet. 2013;9:e1003327
16. Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100-107
17. Bass BL. RNA editing. Oxford: Oxford University Press. 2001
18. Thackeray JR, Ganetzky B. Developmentally regulated alternative splicing generates a complex array of Drosophila para sodium channel isoforms. J Neurosci. 1994;14:2569-2578
19. Hanrahan CJ, Palladino MJ, Ganetzky B. et al. RNA editing of the Drosophila para Na+ channel transcript: Evolutionary conservation and developmental regulation. Genetics. 2000;155:1149-1160
20. Song WZ, Liu ZQ, Tan JG. et al. RNA editing generates tissue-specific sodium channels with distinct gating properties. Insect Biochem Mol Biol. 2004;279:32554-32561
21. Tan JG, Liu ZQ, Nomura Y. et al. Alternative splicing of an insect sodium channel gene generates pharmacologically distinct sodium channels. J Neurosci. 2002;22:5300-5309
22. Lee SH, Ingles PJ, Knipple DC. et al. Developmental regulation of alternative exon usage in the house fly Vssc1 sodium channel gene. Invert Neurosci. 2002;4:125-133
23. Olson ROD, Liu ZQ, Nomura Y. et al. Molecular and functional characterization of voltage-gated sodium channel variants from Drosohila melanogaster. Insect Biochem Mol Biol. 2008;38:604-610
24. Sonoda S, Igaki C, Tsumuki H. Alternatively spliced sodium channel transcripts expressed in field strains of the diamonback moth. Insect Biochem Mol Biol. 2008;38:883-890
25. Shao YM, Dong K, Tang ZH. et al. Molecular characterization of a sodium channel gene from the silkworm Bombyx mori. Insect Biochem Mol Biol. 2009;39:145-151
26. Lin H, Li T, Zhang L. et al. Multiple sodium channel variants in the mosquito Culex quinquefasciatus. Int J Biol Sci. 2012;8:1291-1309
27. Li F, Zhu YL, Xie ZJ. et al. Molecular characterization of a sodium channel gene in the rice leaffolder, Cnaphalocrocis medinalis (Guenée). Pesticide Biochemistry and Physiology. 2013;105:111-117
28. Zhang TX, Liu ZQ, Song WZ. et al. Molecular characterization and functional expression of the DSC1 channel. Insect Biochem Mol Biol. 2011;41:451-458
29. Ahmedani MS, Shagufta N, Aslam M. et al. Psocid: A new risk for global food security and safety. Appl Entomol Zool. 2010;45:89-100
30. Turner BD. Liposcelis bostrychophila (Psocoptera: Liposcelididae), a stored food pest in the UK. Int J Pest Manag. 1994;40:179-190
31. Pascual-Villalobos MJ, Baz A, Del Estal P. Occurrence of psocids and natural predators on organic rice in Calasparra (Murcia, Spain). J Stored Product Res. 2005;41:231-235
32. Wang JJ, Tasi JH, Zhao ZM. et al. Development and reproduction of the psocid Liposcelis bostrychophila (Psocoptera: Liposcelididae) as a function of temperature. Ann Entomol Soc Am. 2000;93:261-270
33. Jiang XZ, Wei DD, Shen GM. et al. Cloning and bioinformatics analysis of voltage-gated sodium channel gene cDNA in Bactrocera dorsalis (Hendel). Sci Agric Sin. 2012;45:3996-4003
34. Sievers F, Wilm A, Dineen D. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2007;7:539
35. Huang HD, Lee TY, Tseng SW. et al. KinasePhos: a web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res. 2005;33:W226-W229
36. Gupta R, Jung E, Brunak S. Prediction of N-glycosylation sites in human proteins. 2004; in preparation.
37. Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564-577
38. Tamura K, Peterson D, Peterson N. et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731-2739
39. Jiang HB, Liu YH, Tang PA. et al. Validation of endogenous reference genes for insecticide-induced and developmental expression profiling of Liposcelis bostrychophila (Psocoptera: Liposcelididae). Mol Biol Rep. 2010;37:1019-1029
40. Cui YJ, Yu LL, Xu HJ. et al. Molecular characterization of DSC1 orthologs in invertebrate species. Insect Biochem Mol Biol. 2012;42:353-359
41. Jeziorski MC, Greenberg RM, Anderson PA. The molecular biology of invertebrate voltage-gated Ca2+ channels. J Exp Biol. 2000;203:841-856
42. Heinemann SH, Terlau H Stuhmer W. et al. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 1992;356:441-443
43. Denac H, Mevissen M, Scholtysik G. Structure, function and pharmacology of voltage-gated sodium channels. Naunyn-Schmiedebergs Arch Pharmacol. 2000;362:453-479
44. O'Dowd DK, Gee JR, Smith MA. Sodium current density correlates with expression of specific alternatively spliced sodium channel mRNAs in single neurons. J Neurosci. 1995;15:4005-4012
45. Waechter C, Schmidt J, Catterall W. Glycosylation is required for maintenance of functional sodium channels in neuroblastoma cells. J Biol Chem. 1983;258:5117-5123
46. Barzilai MG, Reitzel AM, Kraus JE. et al. Convergent Evolution of Sodium Ion Selectivity in Metazoan Neuronal Signaling. Cell Rep. 2012;2:242-248
47. Wang RW, Huang ZY, Dong K. Molecular characterization of an arachnid sodium channel gene from the varroa mite (Varroa destructor). Insect Biochem Mol Biol. 2003;33:733-739
48. Plummer NW, McBurney MW, Meisler MH. Alternative splicing of the sodium channel SCN8A predicts a truncated two-domain protein in fetal brain and non-neuronal cells. J Biol Chem. 1997;272:24008-24015
Author contact
![]() Corresponding author: Dr. Jin-Jun Wang, College of Plant Protection, Southwest University, Chongqing 400716, China. Tel: +86-23-68250255; Fax: +86-23-68251269; Email: jjwang7008com; wangjinjunedu.cn.
Corresponding author: Dr. Jin-Jun Wang, College of Plant Protection, Southwest University, Chongqing 400716, China. Tel: +86-23-68250255; Fax: +86-23-68251269; Email: jjwang7008com; wangjinjunedu.cn.

 Global reach, higher impact
Global reach, higher impact