10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(10):1089-1098. doi:10.7150/ijbs.7367 This issue Cite
Research Paper
Genistein Promotion of Osteogenic Differentiation through BMP2/SMAD5/RUNX2 Signaling
1. Division of Gastroenterology, The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong, 510080, China;
2. Institute of Clinical Pharmacology, Xiangya Medical College, Central South University, Changsha, Hunan, 410078, China;
3. Department of Medicine, the University of Tennessee Health Science Center, Memphis, TN, 38165, USA;
4. Department of Pharmacology, School of Pharmacy, Fudan University, Shanghai 201203, China.
* These authors contributed equally to this study
Received 2013-8-8; Accepted 2013-10-30; Published 2013-11-21
Abstract
To investigate the effects of Genistein on the osteogenic related gene expression profiles during osteoblastic differentiation of human bone marrow mesenchymal stem cell (hBMSC) cultures, the hBMSCs were cultured under osteogenic differentiation medium with the addition of Genistein (10-8∼10-5 M) for 12 days. The cell proliferation was measured by BrdU incorporation, while the osteoblastic differentiation in hBMSC cultures was assessed by cellular alkaline phosphatase (ALP) activity. The cell apoptosis was determined by caspase 3/7 activation. GEArray Q series human osteogenesis gene array was used to analyze large-scale gene expression in Genistein-treated hBMSC cultures compared to the control group. Quantitative real-time RT-PCR, small interfering RNA (siRNA), and western blot analysis were used to confirm the microarray data in five representative transcripts. Genistein (10-8∼10-6 M) dose- and time-dependently increased cell proliferation and cellular ALP activity, but had no significant effect on cell apoptosis in hBMSC cultures. The 96-gene array analysis indicated that 22 genes were upregulated more than 2-fold and 7 genes were downregulated at least 1.5-fold. The expressions of bone morphogenetic proteins (BMPs), small mothers against decapentaplegic homologs (SMADs), and Runt-related transcription factor 2 (RUNX2) were concomitantly increased under Genistein treatment while insulin-like growth factor 2 and inhibitory SMADs 6 and 7 expressions were significantly decreased. The results of the real-time RT-PCR had a correlation with the results of microarray analysis and were estrogen-receptor dependent. Specific gene siRNAs knock-down further confirmed the osteogenic effects of Genistein on BMP2, SMAD5 and RUNX2 protein expression. Genistein enhanced osteogenic differentiation in cultured hBMSCs mainly through the BMP-dependent SMADs and RUNX2 signaling.
Keywords: Genistein, bone marrow mesenchymal stem cell, cDNA microarray, osteogenic differentiation
Introduction
Osteoporosis occurs frequently in women following menopause, which is characterized by decreased bone density and increased risk of fracture 1. The incidence of osteoporosis in women over the age of 60 years is 60%∼70%, and the primary cause of postmenopausal osteoporosis is the deficiency of endogenous estrogen 2-4. In this regard, hormone replacement therapy (HRT) had a wide-spread use since 1960s 5, 6. However, the long-term utilization of HRT was limited due to its severe side effects, such as endometrial hyperplasia, hemorrhagic uterine, endometrial cancer, and breast cancer; moreover, HRT was observed to increase the incidence of colon carcinoma and ovarian cancer possibility 7-9. To replace the estrogen medications, many researchers have been seeking alternative natural medications with similar therapeutic effects but fewer side effects.
Genistein, rich in bean plants and soybean products, is categorized as plant estrogen (also called phytoestrogen). Genistein has a resemble structure with estrogen and therefore can bind to estrogen receptor (ER) 10-12. It has been reported by previous literature that soybean products and Genistein could prevent ovariectomized (OVX)-induced osteoporosis in rats 13, 14 and more importantly, the similar effect was also observed in postmenopausal women in their therapy 15. Furthermore, Genistein had milder side effect, especially to uterus and breast compared to estrogen treatment 15-18. Epidemiologic survey manifested that the incidence of osteoporosis and fracture of women in Asia was lower than women in western countries, which could be partially attributed to Asian diets containing more soybean products 19-21. In summary, Genistein is a potential substituent of estrogen in the therapy of osteoporosis. Although many in vitro studies have shown that Genistein promoted cell proliferation, osteogenic differentiation, and osteogenic gene expressions in mouse and human bone marrow mesenchymal stem cell cultures (mBMSC or hBMSC) 22-26, the mechanisms at the molecular level remain elusive. In addition, it is necessary to conduct more multifactorial evaluations based on the high-throughput screening of osteogenic-related genes to elucidate the molecular-level changes of cells treated by Genistein compared to those treated by vehicle control.
In the present study, we successfully verified a hypothesis that Genistein promotes cell proliferation and osteogenic differentiation, evidenced by increased cell growth and elevated cellular alkaline phosphatase (ALP) activity in the hBMSC cultures. We also identified that differentially-regulated genes were responsible for osteogenic differentiation by performing large-scale gene expression analyses in Genistein-induced hBMSC cultures with the use of GEArray Q series human osteogenesis gene array (Superarray Bioscience, Bethesda, MD, USA). Sequentially five critical transcripts closely related to osteogenic differentiation revealed by microarray analysis were confirmed by real-time RT-PCR analyses and specific gene siRNAs knock-down experiments. Our current study indicated that differentially-regulated genes linked with Genistein and their interactions contribute to the Genistein-induced osteogenic differentiation in the hBMSC cultures.
Materials and Methods
Reagents
Genistein, 17-estradiol (E2), ICI182780, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Alpha minimum essential medium (α-MEM), fetal bovine serum (FBS), trypsin-EDTA, and Trizol reagent were obtained from Invitrogen Corporation (Carlsbad, CA, USA). Rosiglitazone was purchased from Novo Nordisk (Denmark). Primary antibodies of CD44 and CD105 were obtained from Boster Co. (Shanghai, China). PE/FITC-conjugated antibodies of CD34 and CD45 were purchased from Becton-Dickinson (San Jose, CA, USA). GEArray Q series human osteogenesis gene array and SYBR Green qPCR reagents were obtained from SuperArray Bioscience Corporation (Frederick, MD, USA). Biotin-16-dUTP was purchased from Roche Applied Science (Indianapolis, IN, USA). BrdU Cell Proliferation Assay Kit (QIA58) was purchased from Calbiochem (Gibbstown, NJ, USA). RNase inhibitor, MMLV inverse transcriptase for cDNA synthesis, Caspase-3-GLO Assay, and Taq DNA polymerase were purchased from Promega Corporation (Madison, WI, USA).
Cell cultures
The hBMSCs were obtained from limb bones of a 5-month-old aborted fetus (Hunan Maternal and Child Health Hospital, Changsha, China), which was allowed by the parents and in accordance with the ethical standards of the Hunan Ethics Committee. Mononucleated cells were first isolated using Ficoll density gradient centrifugation method 27, followed by a step of seeding in α-MEM with 15% FBS (inactivated) loading and finally maintained in a humidified incubator filled with 5% CO2 and 95% air at 37°C. Three to five passages of hBMSCs were used in this study. Cell culture medium was prepared using the previous method reported by Abdallah et al. with minor modification27. To induce osteogenic differentiation, the cells were treated with osteogenic medium consisting of phenol red-free α-MEM, 10% FBS (dextran-coated charcoal stripped, DCS), dexamethasone (10-8 M), ascorbic acid (50 µg/ml), and β-glycerophosphate (10-2 M). To induce adipogenic differentiation, the cells were incubated with adipogenic medium consisting of DMEM, 10% FBS, dexamethasone (10-7 M), isobutyl methylxanthine (IBMX, 4.5 x10-4 M), insulin (2×10-6 M), and rosiglitazone (10-6 M) as previously reported 28. To evaluate the effect of Genistein on osteogenic differentiation, the hBMSCs were placed in 12-well-plate at a density of 5x104 cells/well and treated with either vehicle (0.01% DMSO), or E2 (10-8 M), or Genistein (10-8∼10-5 M), respectively. The medium was replaced every 3 days thereafter and the cellular samples for Alizarin Red S staining and ALP activity will be harvested at different time points as previously reported 28.
Alizarin red S staining
To test extracellular mineralization of hBMSC, the cells were seeded in 6-well-plate at a density of 1x105 cells/well and cultured in osteogenic differentiation medium for 28 days. At each pre-set time point, the cultured cells were fixed in 95% glutaraldehyde at 4 °C overnight, and subsequently stained with 40 mM Alizarin red-S for 15 minutes. After that, the cells were first rinsed with deionized water three times and then washed with 1x PBS for 15 minutes. The images were captured using Nikon Diaphot 300 inverted phase contrast microscope (Spectra Services, Ontario, NY USA).
Oil red-O staining
To examine adipogenic ability of hBMSC, the cells were seeded in 6-well-plate at a density of 1x105 cells/well and cultured in adipogenic differentiation medium for 28 days. At each pre-set time points, the cells were fixed in 10% formaldehyde at 4 °C for 1 hour, rinsed with 3% isopropanol, and stained with Oil red-O staining solution for 1 hour. Then the images were also captured using the same microscope mentioned above in Alizarin red S staining.
Flow cytometry and immunocytochemistry
For surface marker studies, primary hBMSCs were harvested and centrifuged at 3,000 rpm for 30 seconds; thereafter, the supernatants were discarded and the sediments were collected. Sequentially, for each sample, a cellular suspension was formed by reconstituting the sediment with pre-calculated amount of PBS and consequently its density was adjusted to 5×106 cells∙mL-1. Next, hBMSC cell suspensions were incubated with 20μl of FITC/PE-conjugated antibodies against CD34 (1:250) and CD45 (1:200) in dark for 30 minutes, and then washed twice with PBS containing 2% FBS and suspended in 500 μl of PBS and assayed by a FACScan flow cytometer linked with Cell-Quest 3.1 software (Becton-Dickinson). On the other hand, control groups were treated with isotype antibodies under identical conditions. For immunocytochemistry analysis, primary hBMSCs were cultured on coverslips and the immunostaining was conducted using SABC (Rabbit IgG)-POD kit. In brief, the coverslips were soaked in 4% paraformaldehyde for 30 minutes followed by incubation with 3% H2O2 for 5 minutes and 10% BSA for 1 hour. After that, the cells were first incubated with primary antibodies against CD105 (1:150) and CD44 (1:200) at 37 °C for 1 hour in a humidified box and then incubated with secondary antibodies. Finally, the cells were visualized with DAB staining according to the manufacturer's protocol. Control groups were treated without primary antibodies under identical conditions.
Cell proliferation and apoptosis assay
BrdU incorporation assay was used to determine dose-dependent effect of Genistein on cell proliferation following the manufacturer's instructions (QIA58, Calbiochem, Gibbstown, NJ, USA). Briefly, the cells were seeded into 96-well plates at a density of 1x104 cells/well in 200 µl medium and pre-treated with Genistein (10-8∼10-5 M) in the culture medium for 24 hours. The cells were incubated with 1:2000 BrdU fresh media at 37°C and 5% CO2 for 6 hour and fixed with 200µl of fixative/denaturing solution for 30 minutes at room temperature. Then 1:100 Anti-BrdU antibodies were added to each well and incubated for 1 hour at room temperature. An enzyme-linked immunosorbent assay (ELISA) was used to measure the values of optical densities (OD) at a wavelength of 450 nm. The relative rate of cell proliferation was calculated from the mean ratio of control group (OD450). For apoptosis, caspase 3/7 activity was measured using Apo-ONE® Homogeneous Caspase-3/7 Assay (Promega, Madison, WI) in Genistein (10-8∼10-5 M)-treated hBMSC cultures for 24 hours according to the manufacturer's instructions. Briefly, 20 µl of cell extract was added to each well of 96-well plates and mixed with 20 µl of Apo-ONE® Caspase-3/7 Reagent providing that the 1:1 ratio of Homogeneous Caspase-3/7 Reagent volume to sample volume is preserved. The reaction mixture was incubated at 37°C for 4 hours, and the caspase 3/7 activity was determined by analyzing the value of OD at 405 nm wavelength via ELISA. The relative rate of caspase 3/7 activity was calculated from the mean ratio of control group (OD405).
ALP activity
Cellular ALP activity was measured as previously described [27]. Enzyme activity was determined colorimetrically using p-NPP as the substrate at pH10.3 after incubation at 37ºC for 30 minutes. The optical density was read at 405 nm and total protein content was measured using a protein assay kit from Bio-Rad (Hercules, CA, USA). ALP activity was normalized to the protein content and expressed as nmol∙h-1∙µg protein-1.
Expression profiling and data analysis
The hBMSCs were seeded into a 25 cm2 flask and treated with either 1 µmol·L-1 Genistein or vehicle (DMSO) control for 12 days in the absence or presence of a complete estrogen receptor (ER) antagonist, 0.1 µmol·L-1 ICI182780 (ICI). Total RNA was isolated from the Genistein-treated and control hBMSC cultures without or with ICI using Trizol reagent and then treated with DNase I (Invitrogen Corporation, Carlsbad, CA, USA) to avoid any potential DNA contamination. The quantity and purity of the RNA were measured by a spectrophotometer. Genistein-treated and control RNA (5.0 µg total) samples were used as a template to generate biotin-16-dUTP-labeled cDNA probes, according to the manufacturer's instructions. Expression profiling was performed with the use of the GEArray Q series human osteogenesis gene array containing 96 bone-formation-correlated genes. The cDNA probes were denatured and hybridized at 60°C with the osteogenesis SuperArray membrane. This membrane was then washed and exposed with the use of a chemiluminescent substrate to obtain an X-ray film. To analyze the SuperArray membrane, we scanned the X-ray film and imported it into Adobe Photoshop (Adobe Systems Inc, San Jose, California, USA) as a TIFF file. The image file was inverted; consequentially spots were digitized using ScanAlyze software (Version 2.51; http://rana.lbl.gov/EisenSoftware.htm), and normalized by subtracting the background as the average intensity value of 3 spots containing plasmid DNA (PUC18). The average of 2 GAPDH spots were used as positive controls and set as baseline values to which the signal intensity of other spots was compared. Based on this normalized data, the signal intensity from the membranes was ranked using the GEArray analyzer program. To assess robustness of the methods, the cDNA probes from original total RNA were respectively synthesized in the cells from the same batch but 3 parallel groups, and hybridization experiments were conducted using membranes stripped by boiled 0.5% SDS solution in triplicate. In the present study, a significant change in the gene expression level was defined to be not less than a 2-fold in increments or not more than one-third of magnitude reduction in signal density. All differentially expressed spots on filters were singled out and confirmed visually, and representative images were chosen for demonstration.
Quantitative real-time RT-PCR
To confirm the gene expression data of DNA microarrays, we selected 5 important transcripts for real-time RT-PCR analysis. In brief, total RNA, which was the identical RNAs used as targets for array hybridization, were treated by DNase I to remove genomic DNA contamination and then reverse-transcribed into cDNA using the MMLV reverse transcriptase (Promega Corporation, Madison, WI, USA). Human β-ACTIN was used as the normalization control in the quantitative analysis. The sequences for the primers were as follows: Bone morphogenetic protein 2 (BMP2) forward: 5'-GAG GTC CTG AGC GAG TTC GA-3', BMP2 reverse: 5'-ACC TGA GTG CCT GCG ATA CA-3'; small mothers against decapentaplegic homologs 5 (SMAD5) forward: 5'-TCC ATC GTC TAC TAC GAA CTC AA-3', SMAD5 reverse: 5'-TGT GAC GCC TGT CGG TGA TA-3'; RUNX2/CBFA1 forward: 5'-GGC TGT GGA GTT TGG TGT CTA-3', RUNX2/CBFA1 reverse: 5'-TCT GCT AAA TTC TGC TTG GGT-3'; Alkaline phosphatase (ALP) forward: 5'-ACC TCC TCG GAA GAC ACT CTG-3', ALP reverse: 5'-ACT GCG CCT GGT AGT TGT TG-3'; OSTEOCALCIN (OSC) forward: 5'-AAG GTG GTG A AT AGA CTC CG-3', OSC reverse: 5'-AAA CGG TGG TGC CAT AGA TG-3'. Quantitative real-time PCR was performed in an ABI Prism 7100 system (Applied Biosystems Foster City, CA, USA) using approximate 200 ng of cDNA and 1X SYBR Green PCR Master Mix together with the 300 nM of primer pairs according to the standard protocol. The PCR conditions were: 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s for 40 cycles. The relative mRNAs abundance was expressed as the fold changes relative to housekeeping gene β-ACTIN and normalized to the control group.
Small interfering RNA (siRNA) and western blot analysis
All hBMSC samples were cultured in osteogenic medium with either Genistein (1 µmol·L-1) or DMSO for 10 days. The control siRNA (siControl) and siRNA duplexes specific for SMAD5 (siSMAD5) and BMP-2 (siBMP2) were transfected into the Genistein-treated hBMSCs using Lipofectamine 2000 (Invitrogen Life Technology) according to the manufacturer's instructions. siSMAD5 (sc-38378) and siBMP-2 (sc-39738) were purchased from Santa Cruz Biotechnology. Total proteins were extracted from the siRNA-treated cells after 48 hours using Tissue or Cell Total Protein Extraction Kit (KeyGEN Biotech, Nanjing, China) according to the manufacturer's instructions. Equal quantities of proteins were added to NuPAGETM 6-12% Bis-Tris Gel (Invitrogen, Carlsbad, CA) and analyzed with standard Western blot protocols (HRP-conjugated secondary antibodies from Santa Cruz Biotechnology and ECL from Amersham Biosciences (Buckinghamshire, UK). Antibodies against β-ACTIN (ab14128), GAPDH (ab37168), BMP2 (ab82511), SMAD5 (ab88559), RUNX2 (ab76956), ALP (ab95462), and Osteocalcin (OSC) (ab13418) were purchased from Abcam (Cambridge, UK). The relative densities of proteins were quantified with FluorChem 8900 and normalized to β-ACTIN control.
Statistical analysis
We evaluated differences between two groups by unpaired t-test and among multiple groups by one-way analysis of variance. All values are expressed as means ± S.D. All data was calculated and analyzed statistically with the aid of GraphPad Prism5 (GraphPad Software, Inc., La Jolla, CA).
Results
Characterization of isolated human primary BMSCs
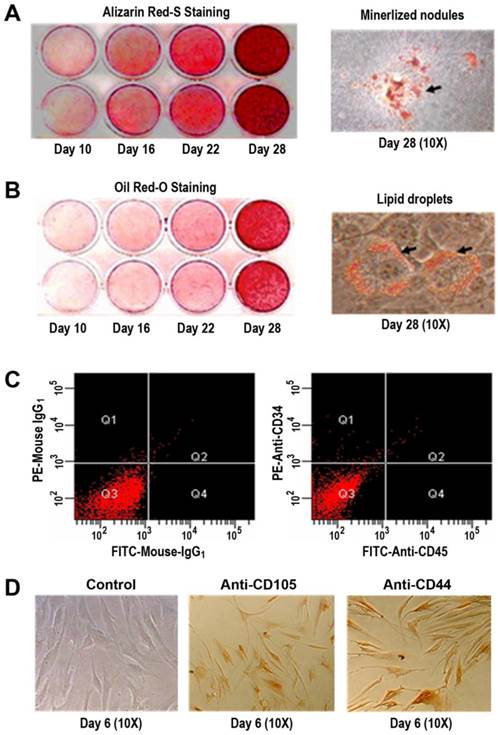
Primary hBMSCs were induced in osteogenic and adipogenic medium for 10, 16, 22, and 28 days, respectively. Alizarin Red-S staining indicated that the formation of mineralized nodules was positively dependent on time in osteogenic cultures (Figure 1A), which was an evidence for a potential osteogenesis of the primary hBMSCs. With the use of Oil Red O staining, it was observed that the lipid droplets accumulated as a function of time (Figure 1B), suggesting an adipogenic differentiation of the primary hBMSCs. Using flow cytometry and immunocytochemistry, it was found that the primary hBMSCs were negative for CD34 (a marker protein of hematopoietic lineages) and CD45 (the common leukocyte antigen) shown in Figure 1C. Conversely, the primary hBMSCs exhibited positive staining for mesenchymal stem cell hall markers CD105 and CD44 (Figure 1D).
Multiple differentiation potential and cell surface markers of the primary hBMSCs. (A) Alizarin Red-S staining of mineralized nodules under osteogenic condition for 10, 16, 22, and 28 days (left panel) and the images at 10x magnification of mineralized nodules at day 28 (right panel). (B) Oil red-O staining of lipid droplets under adipogenic condition for 10, 16, 22, and 28 days (left panel) and the images at 10x magnification of lipid droplets at day 28 (right panel). (C) CD34 and CD45 expressions on the surface of the primary hBMSCs by flow cytometry. Negative controls with isotype antibodies of mouse IgG1 (left panel); CD34 and CD45 expressions with PE/FITC-conjugated antibodies (right panel). (D) CD105 and CD44 expressions on the surface of the primary hBMSCs by immunocytochemistry. Negative control images by omitting the primary antibody in staining procedures at day 6 (left, 10x); Anti-CD105 staining images under identical conditions at day 6 (middle, 10x); Anti-CD44 staining images under identical conditions at day 6 (right, 10x).
Genistein enhanced cell proliferation and osteoblastic differentiation but had no effects on cell apoptosis in cultured hBMSCs
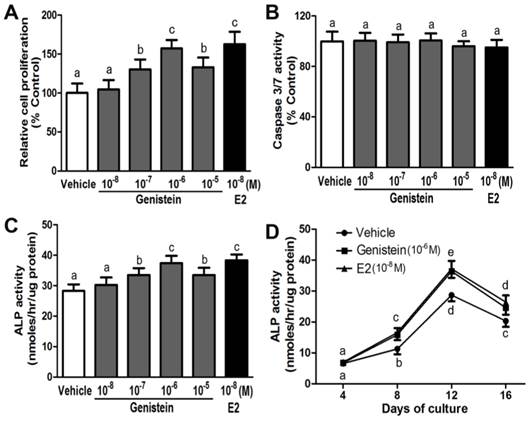
Genistein (10-8~10-5 M) stimulated cell proliferation in a manner dependent on dose and time in the hBMSC cultures (Figure 2A). The stimulations were statistically significant at 10-7 M, and Genistein (10-6 M) was found to give the maximal response when compared with control (Figure 2A). This concentration was then used for the proceeding experiments. In contrast, Caspase 3/7 activities were not elevated by genistein (10-8~10-5 M) treatment in cultured hBMSCs, suggesting the Genistein over this dose range could not cause significant effects on cell apoptosis (Figure 2B). However, Genistein (10-8~10-5 M) dose- and time-dependently induced osteoblastic differentiation within 16 days in culture (Figure 2C and 2D). It is noticeable that, during the period of this treatment, Genistein (10-6 M) led to the maximal effect on ALP activity at day 12. Additionally, E2 (10-8 M), similar to Genistein (10-6 M), was successfully utilized in this experiment to serve as a positive control since it also increased the cell proliferation and ALP activity respectively (Figure 2A, 2C, and 2D).
Gene expression profiling by microarray analysis
The GEArray analyzer software was applied to perform a global expression analysis of genes associated with osteogenic differentiation in the Genistein-treated hBMSC cultures. It was observed that, in the presence of Genistein, there were evident changes in the expression of 29 genes in the hBMSC cultures (Table 1). The results were expressed as a ratio of relative expression in GEN (+)/GEN (-). Among the 29 genes, cumulatively 22 genes were identified as Genistein-promoted genes since these genes had an at least two-fold upregulation in Genistein-treated cells as compared to those cells in the control group. Whereas, the other 7 genes were identified as Genistein-downregulated genes, which showed a more than 1.5-fold decrease of expression in Genistein-treated cells compared to the cells in the control group. Moreover, it was noticed that the level of several growth factors (e.g., fibroblast growth factors, fibroblast growth factor receptors, and colony stimulating factors) increased significantly as Genistein was treated to the cells. As we expected, this phenomenon was consistent with the Genistein-stimulated cell proliferation. In agreement with Genistein-induced osteogenesis, the genes associated with BMP/SMAD signaling pathway had the strongest enhancement amongst genes in the Genistein-treated hBMSC cultures; simultaneously, a majority most types of collagen genes representative of extracellular matrixes were also increased. As inhibitors of BMP signaling, SMADs 6 and 7 were remarkably decreased. RUNX2/CBFA1 and its down-stream gene OSC were significantly up-regulated in the Genistein-treated hBMSC cultures, indicating that Genistein played a role in facilitating hBMSC towards osteoblastic differentiation and maturation.
Effects of Genistein on cell proliferation, apoptosis, and osteoblastic differentiation in the cultured hBMSCs. (A) Dose-dependent stimulation of cell growth in response to different concentration of Genistein (10-8~10-5 M) through BrdU incorporation assay; (B) No significant effects of Genistein (10-8~10-5 M) on cell apoptosis by measuring activation of caspase 3/7 activity; (C) Dose-dependent enhancement of ALP activity in response to different concentration of Genistein (10-8~10-5 M) at day 12; (D) Time-dependent acceleration of ALP activity in response to Genistein (10-6 M) treatment during 16 days of culture. Data are mean ± S.D. from triple independent experiments. E2 (10-8 M) served as a positive control. Values sharing the same superscript are not significantly different at P<0.05.
Most differentially-expressed genes in GEN-treated hBMSCs as compared with control samples. Expression changes are expressed as a ratio of relative expression in GEN (+)/GEN (-) in the form of mean±SD. Genes are ordered by the ratio.
| GenBank | Description | Gene Name | Ratio(GEN+/GEN-) |
|---|---|---|---|
| Up-regulated genes(>2 fold) | |||
| NM_005247 | Fibroblast growth factor 3 | FGF3 | 12.028±4.223 |
| NM_001718 | Bone morphogenetic protein 6 | BMP6 | 6.234±0.306 |
| NM_001200 | Bone morphogenetic protein 2 | BMP2 | 5.954±0.506 |
| NM_001719 | Bone morphogenetic protein 7 | BMP7 | 4.913±1.545 |
| NM_001855 | Collagen, type XV, alpha 1 | COL15A1 | 4.720±0.436 |
| NM_005900 | MAD, mothers against decapentaplegic homolog 1 | SMAD1 | 4.427±1.273 |
| NM_000759 | Colony stimulating factor 3 | CSF-3 | 4.366±2.294 |
| NM_001844 | Collagen, type II, alpha 1 | COL2A1 | 3.827±0.358 |
| NM_000711 | Bone gamma-carboxyglutamate (gla) protein | OSTEOCALCIN | 3.782±0.692 |
| NM_000758 | Colony stimulating factor 2 | CSF-2 | 3.390±0.439 |
| NM_001202 | Bone morphogenetic protein 4 | BMP4 | 3.352±1.234 |
| NM_004348 | Runt-related transcription factor 2 | RUNX2/CBFA1 | 3.301±0.202 |
| NM_000604 | Fibroblast growth factor receptor 1 | FGFR1 | 3.152±0.285 |
| NM_001856 | Collagen, type XVI, alpha 1 | COL16A1 | 3.081±0.911 |
| NM_000088 | Collagen, type I, alpha 1 | COL1A1 | 2.711±0.522 |
| XM_044622 | Collagen, type XIV, alpha 1 | COL14A1 | 2.675±0.485 |
| NM_001201 | Bone morphogenetic protein 3 | BMP3 | 2.595±0.223 |
| NM_005903 | MAD, mothers against decapentaplegic homolog 5 | SMAD5 | 2.435±0.429 |
| NM_021073 | Bone morphogenetic protein 5 | BMP5 | 2.434±0.380 |
| NM_006128 | Bone morphogenetic protein 1 | BMP1 | 2.293±0.213 |
| NM_000800 | Fibroblast growth factor 1 | FGF1 | 2.169±0.160 |
| NM_000478 | Alkaline phosphatase | ALP | 2.168±0.136 |
| Down-regulate genes(<1/3 fold) | |||
| NM_002026 | Fibronectin-1 | FN1 | 0.607±0.076 |
| NM_002449 | Msh homeo box homolog 2 | MSX2 | 0.596±0.081 |
| NM_000093 | Collagen, type V, alpha 1 | COL5A1 | 0.569±0.050 |
| NM_005904 | MAD, mothers against decapentaplegic homolog 7 | SMAD7 | 0.490±0.122 |
| NM_005585 | MAD, mothers against decapentaplegic homolog 6 | SMAD6 | 0.432±0.096 |
| NM_001711 | Biglycan | BGN | 0.261±0.070 |
| NM_000612 | Insulin-like growth factor 2 (somatomedin A) | IGF-II | 0.225±0.053 |
Validation of microarray data by real-time RT-PCR, siRNAs knock-down, and western blot analysis
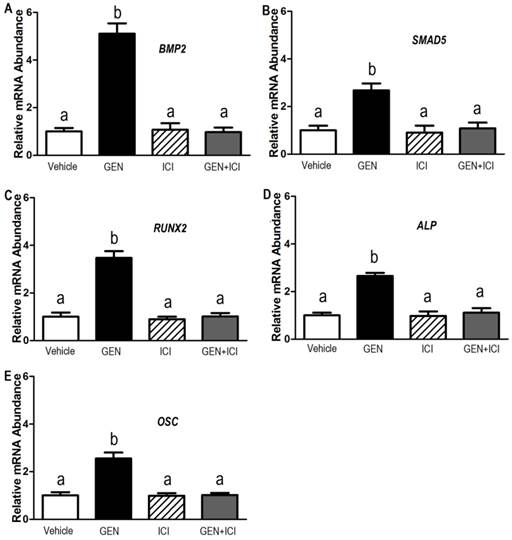
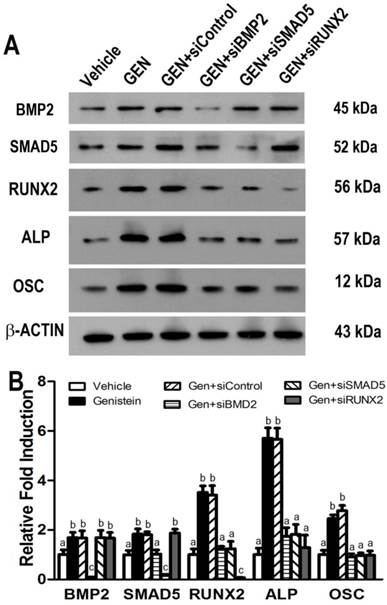
To verify the molecular signature of the Genistein-induced osteogenic differentiation in the hBMSC cultures revealed by the microarray results, we selected five representative genes to do real-time RT-PCR analysis. The data illustrated that these five genes including BMP2, SMAD5, RUNX2, ALP, and OSC had significant different expressions between Genistein-treated cells and vehicle-treated control cells (Figure 3A-3E), corroborating the results obtained in the microarray (Table 1). Estrogen receptor (ER) antagonist ICI182780 completely abolished the GEN-induced osteogenic gene transcription in cultured hBMSCs. However, ICI182780 alone had no effect on these genes expression. In addition, the levels of BMP2, SMAD5, RUNX2, ALP, and OSC proteins were significantly higher in cells treated by Genistein than those treated by vehicle control (Figure 4A and 4B), consistent with microarray analysis and real-time RT-PCR results. siRNAs directed against either BMP2, or SMAD5, or RUNX2 markedly attenuated Genistein-induced increments of RUNX2, ALP, and OSC protein expression, respectively, when compared with control siRNA (Figure 4A and 4B), suggesting that BMP2-dependent SMAD5/RUNX2 signaling mediated Genistein-induced osteoblastic differentiation and related gene expressions in the cultured hBMSCs.
Discussion
It has been reported that both human and mouse bone marrow contains pluripotent progenitor cells or mesenchymal stem cells that can give rise to several lineages such as osteoblasts, adipocytes, chondrocytes, and myoblasts 29-32. In the current study, we established that our isolated primary hBMSCs possess the phenotypic and functional characteristics of mesenchymal stem cells, evidenced by osteogenic and adipogenic differentiation potentials and positive immunostaining with mesenchymal stem cell hall markers. We also found that Genistein (10-8~10-5 M) dose- and time-dependently promoted cell proliferation and osteoblastic differentiation in cultured primary hBMSCs, consistent with our previous observation that Genistein increased cell growth and enhanced ALP activity in cultured primary mBMSCs 22, 23, 26. However, Genistein had no significant effect on cell apoptosis in the hBMSC cultures. More importantly, using microarray analysis, we identified that the BMP-dependent SMAD signaling and BMP-induced collagen synthesis are two major pathways in Genistein-induced osteogenic transcriptional networks. Furthermore, a series of tests including quantitative real-time RT-PCR, specific gene siRNAs knock down, and western blot analysis were successfully conducted to confirm that BMP2-dependent SMAD5/RUNX2 signaling mediated Genistein-induced osteoblastic differentiation and maturation. These findings provide novel molecular insights for Genistein-induced osteogenesis in the hBMSC cultures.
Bone morphogenetic proteins (BMPs) are members of the TGF-β superfamily that tends to be key regulators involved in bone formation and remodeling 33-35. BMPs also play an important role in the differentiation process of hBMSCs to osteoblast-like cells 36, 37. In the present study, it was observed that, after 12 days, aside from BMP8, a series of BMP (1 to 7) transcripts were upregulated in the Genistein-treated cells. It was reported that the expression of BMP2 and BMP6 in promoting osteoblast differentiation and mineralization was significantly increased via ERalpha-dependent mechanism after in vitro and/or in vivo administration of E2 and Genistein 38-41, consistent with our observations in this study. It is generally acceptable that BMP receptor signaling through SMADs 1, 5, and 8 proteins regulates gene transcription during osteoblastic differentiation and bone formation 42-45, whereas SMADs 6 and 7 are inhibitors of BMP signaling 46, 47. Consistent with these previous observations, our current microarray data showed that Genistein-treated hBMSCs had a significant increase in BMP signaling along with SMADs 1 and 5 expressions and concomitantly a decrease in the expression of SMADs 6 and 7. Furthermore, results from quantitative real-time RT-PCR as well as BMP2 and SMAD5 gene siRNAs knock-down confirmed that BMP2-dependent SMAD5 signaling was the downstream pathway for Genistein-induced osteogenic effects. These findings proved that BMP/SMAD signaling was a considerable factor on enhancing hBMSC differentiation into osteoblast lineage under Genistein induction.
Effects of Genistein on BMP2/SMAD5/RUNX2 transcription and downstream gene expression in cultured hBMSCs. Real-time RT-PCR analyses revealed that five representative genes (A) BMP2, (B) SMAD5, (C), RUNX2, (D), ALP, and (E) OSTEOCALCIN (OSC) were differentially expressed in Genistein (GEN, 10-6 M)-treated and vehicle control hBMSC cultures in the absence or presence of a complete estrogen receptor (ER) antagonist, ICI182780 (ICI, 10-7 M). Consistent with the results of microarray analysis, normalized transcript levels of BMP2, SMAD5, RUNX2, ALP, and OSC are shown to be upregulated in GEN-treated cells, and these five genes have significant difference between GEN-treated and the vehicle control group. In addition, ER antagonist ICI completely abolished the GEN-induced osteogenic gene transcription in cultured hBMSCs. Data are mean ± S.D. from triple independent experiments. Values sharing the same superscript are not significantly different at P<0.05.
Effects of gene-specific siRNAs knock-down on Genistein-induced BMP2/SMAD5/RUNX2 signaling in cultured hBMSCs. (A) Representative western blot for BMP2, SMAD5, RUNX2, ALP, and OSTEOCALCIN (OSC) protein expressions in Genistein-treated and untreated hBMSC cultures after transfected with different gene-specific siRNAs. (B) Band densitometry quantified by fluorChem 8900 and normalized to β-ACTIN. Consistent with microarray analysis and real-time RT-PCR results, siRNAs directed against BMP2, or SMAD5, or RUNX2 significantly attenuated Genistein-induced increments of BMP2, SMAD5, RUNX2, ALP, and OSC protein expression, respectively, when compared with control siRNA treatment. Data are mean ± S.D. from triple independent experiments. Values sharing the same superscript are not significantly different at P<0.05.
Extracellular matrix synthesis including collagen and non-collagenous proteins is a major marker of the osteogenic lineage differentiation of hBMSC cultures 48, 49. In this experiment, the majority of collagen transcripts such as COL1A1, COL14A1, COL15A1, COL16A1, and COL2A1 showed significant increases after Genistein induction. These results confirmed that we successfully promoted osteogenic differentiation of hBMSC cultures through BMP-induced collagen synthesis using Genistein on a persuasive basis. In addition, the expressions of RUNX2/CBFA1, a key osteogenic transcriptional factor and its downstream gene OSC, a non-collagenous protein were markedly upregulated 23, 50. In this regard, results from microarray, quantitative real-time RT-PCR, and BMP2, SMAD5, and RUNX2 gene siRNAs knock-down confirmed that RUNX2 was the downstream target for Genistein-induced BMP2/SMAD5 osteogenic signaling. It has been reported that BMPs need extracellular collagen matrix to enhance osteoblastic differentiation and osteogenic gene expression 51, 52, and RUNX2/CBFA1 requires BMP/SMADs signaling to induce osteoblast-specific gene expression 53, 54. Based on our current findings and literature reports, we could draw the conclusion that Genistein-induced BMP/SMADs/RUNX2 transcriptions may form a synergistic and antagonistic network to drive hBMSC towards osteogenic lineage.
Besides BMPs, fibroblast growth factor (FGFs) and colony stimulating factors (CSFs), also increased in the Genistein-induced hBMSC cultures, had synergistic effects with BMP signaling, proven by a synchronicity of expression levels in development and osteogenic differentiation in vitro and in vivo 55, 56. In addition, the expressions of FGF receptor variants, including FGFR1, FGFR2, and FGFR3 were also promoted in the Genistein-induced hBMSC cultures 57. Genistein simultaneously enhanced both cell growth FGF signaling and cell differentiation BMP signaling, consistent with the observation of increased cell proliferation and osteoblastic differentiation in the Genistein-induced hBMSC cultures in vitro.
In conclusion, Genistein promoted cell proliferation and osteoblastic differentiation in the cultured hBMSCs through ER-dependent mechanism. Using pathway-specific microarray analyses, we have identified that BMP-dependent SMADs and RUNX2 signaling plays an essential role in Genistein-induced osteoblastic differentiation of hBMSC cultures. Our result was a meaningful evidence for the prospective use of Genistein in preventing and treating osteoporosis disease.
Abbreviations
hBMSC, human bone marrow mesenchymal stem cell; ALP, alkaline phosphatase; BMPs, bone morphogenetic proteins; SMADs, small mothers against decapentaplegic homologs; HRT, hormone replacement therapy.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grants No. 30171085), and the grant R21-AR056794 from the National Institutes of Health, USA. We would also like to thank Hao Lu and Tianyun Wang for English editing.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Liberman UA, Weiss SR, Bröll J. et al. Effect of oral alendronate on bone mineral density and the incidence of fractures in postmenopausal osteoporosis. The Alendronate Phase III Osteoporosis Treatment Study Group. N Engl J Med. 1995;333(22):1437-43
2. Kanis JA, Melton LJ, Christiansen C, Johnston CC, Khaltaev N. The diagnosis of osteoporosis. J Bone Miner Res. 1994;9(8):1137-41
3. Han ZH, Palnitkar S, Rao DS, Nelson D, Parfitt AM. Effects of ethnicity and age or menopause on the remodeling and turnover of iliac bone: implications for mechanisms of bone loss. J Bone Miner Res. 1997;12(4):498-508
4. Järvinen TL, Kannus P, Sievänen H. Estrogen and bone--a reproductive and locomotive perspective. J Bone Miner Res. 2003;18(11):1921-31
5. Michaëlsson K, Baron JA, Farahmand BY. et al. Hormone replacement therapy and risk of hip fracture: population based case-control study. The Swedish Hip Fracture Study Group. BMJ. 1998;316(7148):1858-63
6. Blank RD, Bockman RS. A review of clinical trials of therapies for osteoporosis using fracture as an end point. J Clin Densitom. 1999;2(4):435-52
7. Recker RR. Clinical review 41: Current therapy for osteoporosis. J Clin Endocrinol Metab. 1993;76(1):14-6
8. Vihtamäki T, Savilahti R, Tuimala R. Why do postmenopausal women discontinue hormone replacement therapy? Maturitas. 1999;33(2):99-105
9. Toth S, Blanchard E, Herman L. et al. Weighing the risks and benefits of hormone therapy. JAAPA. 2003;16(6):45-7
10. Rickard DJ, Monroe DG, Ruesink TJ. et al. Phytoestrogen genistein acts as an estrogen agonist on human osteoblastic cells through estrogen receptors alpha and beta. J Cell Biochem. 2003;89(3):633-46
11. Wang TT, Sathyamoorthy N, Phang JM. Molecular effects of genistein on estrogen receptor mediated pathways. Carcinogenesis. 1996;17(2):271-5
12. Wang H-J, Murphy PA. Isoflavone Composition of American and Japanese Soybeans in Iowa: Effects of Variety, Crop Year, and Location. Journal of Agricultural and Food Chemistry. 1994;42(8):1674-77
13. Arjmandi BH, Alekel L, Hollis BW. et al. Dietary soybean protein prevents bone loss in an ovariectomized rat model of osteoporosis. Journal of Nutrition. 1996;126(1):161-67
14. Uesugi T, Toda T, Tsuji K, Ishida H. Comparative study on reduction of bone loss and lipid metabolism abnormality in ovariectomized rats by soy isoflavones, daidzin, genistin, and glycitin. Biol Pharm Bull. 2001;24(4):368-72
15. Morabito N, Crisafulli A, Vergara C. et al. Effects of genistein and hormone-replacement therapy on bone loss in early postmenopausal women: a randomized double-blind placebo-controlled study. J Bone Miner Res. 2002;17(10):1904-12
16. D'Anna R, Cannata ML, Marini H. et al. Effects of the phytoestrogen genistein on hot flushes, endometrium, and vaginal epithelium in postmenopausal women: a 2-year randomized, double-blind, placebo-controlled study. Menopause. 2009;16(2):301-6
17. Atteritano M, Marini H, Minutoli L. et al. Effects of the phytoestrogen genistein on some predictors of cardiovascular risk in osteopenic, postmenopausal women: a two-year randomized, double-blind, placebo-controlled study. J Clin Endocrinol Metab. 2007;92(8):3068-75
18. D'Anna R, Cannata ML, Atteritano M. et al. Effects of the phytoestrogen genistein on hot flushes, endometrium, and vaginal epithelium in postmenopausal women: a 1-year randomized, double-blind, placebo-controlled study. Menopause. 2007;14(4):648-55
19. Ross PD, Norimatsu H, Davis JW. et al. A comparison of hip fracture incidence among native Japanese, Japanese Americans, and American Caucasians. Am J Epidemiol. 1991;133(8):801-9
20. Fujita T, Fukase M. Comparison of osteoporosis and calcium intake between Japan and the United States. Proc Soc Exp Biol Med. 1992;200(2):149-52
21. Kin K, Lee JH, Kushida K. et al. Bone density and body composition on the Pacific rim: a comparison between Japan-born and U.S.-born Japanese-American women. J Bone Miner Res. 1993;8(7):861-9
22. Liao QC, Xiao ZS, Qin YF, Zhou HH. Genistein stimulates osteoblastic differentiation via p38 MAPK-Cbfa1 pathway in bone marrow culture. Acta Pharmacol Sin. 2007;28(10):1597-602
23. Pan W, Quarles LD, Song LH. et al. Genistein stimulates the osteoblastic differentiation via NO/cGMP in bone marrow culture. J Cell Biochem. 2005;94(2):307-16
24. Chen X, Garner SC, Quarles LD, Anderson JJ. Effects of genistein on expression of bone markers during MC3T3-E1 osteoblastic cell differentiation. J Nutr Biochem. 2003;14(6):342-9
25. Heim M, Frank O, Kampmann G. et al. The phytoestrogen genistein enhances osteogenesis and represses adipogenic differentiation of human primary bone marrow stromal cells. Endocrinology. 2004;145(2):848-59
26. Okumura N, Yoshikawa T, Iida J, Nonomura A, Takakura Y. Bone formation-promoting effect of genistein on marrow mesenchymal cell culture. Biomed Mater Eng. 2006;16(1):23-32
27. Dai Z, Li Y, Quarles LD. et al. Resveratrol enhances proliferation and osteoblastic differentiation in human mesenchymal stem cells via ER-dependent ERK1/2 activation. Phytomedicine. 2007;14(12):806-14
28. Abdallah BM, Haack-Sorensen M, Burns JS. et al. Maintenance of differentiation potential of human bone marrow mesenchymal stem cells immortalized by human telomerase reverse transcriptase gene despite [corrected] extensive proliferation. Biochem Biophys Res Commun. 2005;326(3):527-38
29. Majumdar MK, Thiede MA, Mosca JD, Moorman M, Gerson SL. Phenotypic and functional comparison of cultures of marrow-derived mesenchymal stem cells (MSCs) and stromal cells. J Cell Physiol. 1998;176(1):57-66
30. Pereira RF, O'Hara MD, Laptev AV. et al. Marrow stromal cells as a source of progenitor cells for nonhematopoietic tissues in transgenic mice with a phenotype of osteogenesis imperfecta. Proc Natl Acad Sci U S A. 1998;95(3):1142-7
31. Pittenger MF, Mackay AM, Beck SC. et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143-7
32. Bianco P, Gehron Robey P. Marrow stromal stem cells. J Clin Invest. 2000;105(12):1663-8
33. Yu YY, Lieu S, Lu C, Colnot C. Bone morphogenetic protein 2 stimulates endochondral ossification by regulating periosteal cell fate during bone repair. Bone. 2010;47(1):65-73
34. Ghodadra N, Singh K. Recombinant human bone morphogenetic protein-2 in the treatment of bone fractures. Biologics. 2008;2(3):345-54
35. Lissenberg-Thunnissen SN, de Gorter DJ, Sier CF, Schipper IB. Use and efficacy of bone morphogenetic proteins in fracture healing. Int Orthop. 2011;35(9):1271-80
36. Carpenter RS, Goodrich LR, Frisbie DD. et al. Osteoblastic differentiation of human and equine adult bone marrow-derived mesenchymal stem cells when BMP-2 or BMP-7 homodimer genetic modification is compared to BMP-2/7 heterodimer genetic modification in the presence and absence of dexamethasone. J Orthop Res. 2010;28(10):1330-7
37. Seib FP, Franke M, Jing D, Werner C, Bornhauser M. Endogenous bone morphogenetic proteins in human bone marrow-derived multipotent mesenchymal stromal cells. Eur J Cell Biol. 2009;88(5):257-71
38. Zhou S, Turgeman G, Harris SE. et al. Estrogens activate bone morphogenetic protein-2 gene transcription in mouse mesenchymal stem cells. Mol Endocrinol. 2003;17(1):56-66
39. Hertrampf T, Gruca MJ, Seibel J. et al. The bone-protective effect of the phytoestrogen genistein is mediated via ER alpha-dependent mechanisms and strongly enhanced by physical activity. Bone. 2007;40(6):1529-35
40. Ma HP, Ming LG, Ge BF. et al. Icariin is more potent than genistein in promoting osteoblast differentiation and mineralization in vitro. J Cell Biochem. 2011;112(3):916-23
41. Plant A, Tobias JH. Increased bone morphogenetic protein-6 expression in mouse long bones after estrogen administration. J Bone Miner Res. 2002;17(5):782-90
42. Cao X, Chen D. The BMP signaling and in vivo bone formation. Gene. 2005;357(1):1-8
43. Miyazono K. Signal transduction by bone morphogenetic protein receptors: functional roles of Smad proteins. Bone. 1999;25(1):91-3
44. Retting KN, Song B, Yoon BS, Lyons KM. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development. 2009;136(7):1093-104
45. Rath B, Nam J, Deschner J. et al. Biomechanical forces exert anabolic effects on osteoblasts by activation of SMAD 1/5/8 through type 1 BMP receptor. Biorheology. 2011;48(1):37-48
46. Ishida W, Hamamoto T, Kusanagi K. et al. Smad6 is a Smad1/5-induced smad inhibitor. Characterization of bone morphogenetic protein-responsive element in the mouse Smad6 promoter. J Biol Chem. 2000;275(9):6075-9
47. Fujii M, Takeda K, Imamura T. et al. Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol Biol Cell. 1999;10(11):3801-13
48. Fried A, Benayahu D, Wientroub S. Marrow stroma-derived osteogenic clonal cell lines: putative stages in osteoblastic differentiation. J Cell Physiol. 1993;155(3):472-82
49. Shindo K, Kawashima N, Sakamoto K. et al. Osteogenic differentiation of the mesenchymal progenitor cells, Kusa is suppressed by Notch signaling. Exp Cell Res. 2003;290(2):370-80
50. Komori T, Yagi H, Nomura S. et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755-64
51. Xiao G, Gopalakrishnan R, Jiang D. et al. Bone morphogenetic proteins, extracellular matrix, and mitogen-activated protein kinase signaling pathways are required for osteoblast-specific gene expression and differentiation in MC3T3-E1 cells. J Bone Miner Res. 2002;17(1):101-10
52. Suzawa M, Takeuchi Y, Fukumoto S. et al. Extracellular matrix-associated bone morphogenetic proteins are essential for differentiation of murine osteoblastic cells in vitro. Endocrinology. 1999;140(5):2125-33
53. Lee KS, Kim HJ, Li QL. et al. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20(23):8783-92
54. Bae JS, Gutierrez S, Narla R. et al. Reconstitution of Runx2/Cbfa1-null cells identifies a requirement for BMP2 signaling through a Runx2 functional domain during osteoblast differentiation. J Cell Biochem. 2007;100(2):434-49
55. Fakhry A, Ratisoontorn C, Vedhachalam C. et al. Effects of FGF-2/-9 in calvarial bone cell cultures: differentiation stage-dependent mitogenic effect, inverse regulation of BMP-2 and noggin, and enhancement of osteogenic potential. Bone. 2005;36(2):254-66
56. Aberg T, Wozney J, Thesleff I. Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Dev Dyn. 1997;210(4):383-96
57. Morsczeck C, Gotz W, Schierholz J. et al. Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol. 2005;24(2):155-65
Author contact
![]() Corresponding author: Zhousheng Xiao, Ph.D., the University of Tennessee Health Science Center, Memphis, TN, 38163, USA. Tel. 901-448-1489; Fax: 901-448-1188; E-mail: zxiao2edu or Minhu Chen, M.D., the First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong, 510080, China. Tel.86-020-87755766; E-mail: chenminhu163.com.
Corresponding author: Zhousheng Xiao, Ph.D., the University of Tennessee Health Science Center, Memphis, TN, 38163, USA. Tel. 901-448-1489; Fax: 901-448-1188; E-mail: zxiao2edu or Minhu Chen, M.D., the First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong, 510080, China. Tel.86-020-87755766; E-mail: chenminhu163.com.

 Global reach, higher impact
Global reach, higher impact