Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(3):245-256. doi:10.7150/ijbs.7401 This issue Cite
Research Paper
ZIP4, a Novel Determinant of Tumor Invasion in Hepatocellular Carcinoma, Contributes to Tumor Recurrence after Liver Transplantation
1. Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, First Affiliated, Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China;
2. Key Laboratory of Combined Multi-Organ Transplantation, Ministry of Public Health and Key Laboratory of Organ Transplantation of Zhejiang Province, Hangzhou, China;
3. Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Hangzhou, China.
* These two authors contribute equally to the work.
Received 2013-8-13; Accepted 2014-1-21; Published 2014-2-11
Abstract
Background and purpose: Recently, evidence that Zinc transporter ZRT/IRT-like protein 4 (ZIP4) is involved in invasiveness and apoptosis has emerged in pancreatic cancer and prostate cancer. Our aim was to assess the role of ZIP4 in invasiveness, migration and apoptosis of hepatocellular carcinoma (HCC). The prognostic value of ZIP4 in HCC after liver transplantation was evaluated.
Methods: The role of ZIP4 in HCC was investigated by overexpressing ZIP4 in BEL7402 and HepG2 cells and inhibiting ZIP4 in HuH-7 and HepG2 cells, using overexpression and shRNA plasmids in vitro studies. Immunohistochemical analysis was used to evaluate ZIP4 expression in HCC tissues from 60 patients undergoing liver transplantation, 36 cirrhotic tissue samples, and 6 normal tissue samples. Prognostic significance was assessed using the Kaplan-Meier method and the log-rank test.
Results: Specific suppression of ZIP4 reduced cell migration and invasiveness, whereas ZIP4 overexpression caused increases in cell migration and invasiveness. Furthermore, overexpression of ZIP4 resulted in increased expression of pro-metastatic genes (MMP-2, MMP-9) and decreased expression of pro-apoptotic genes (caspase-3, caspase-9, Bax). In contrast, suppression of ZIP4 resulted in an opposite effect. ZIP4 was more highly expressed in tumor tissues than non-tumor tissues (P < 0.0001). ZIP4 expression was significantly associated with tumor recurrence (P = 0.002), tumor node metastasis stage (P = 0.044), Child-Turcotte-Pugh score (P = 0.042), and tumor size (P = 0.022). Univariate analysis showed that ZIP4 expression was significantly associated with overall survival (P = 0.020) and tumor-free survival (P = 0.049). Multivariate analysis revealed that ZIP4 was an independent predictor of overall survival (P = 0.037) after liver transplantation.
Conclusions: ZIP4 could promote migration, invasiveness, and suppress apoptosis in hepatocellular carcinoma, and represent a novel predictor of poor prognosis and therapeutic target for patients with HCC who undergo liver transplantation.
Keywords: ZIP4, hepatocellular carcinoma, liver transplantation
Introduction
Hepatocellular carcinoma (HCC) is a major health problem and one of the most prevalent neoplasms worldwide, especially in China. An estimated 626,000 new cases occur around the world annually; about half of them occur in China [1]. HCC is commonly lethal because of its aggressive metastasis and most cases are diagnosed at an already advanced stage. Liver resection and liver transplantation (LT) remain the most effective treatments for HCC [2].
LT is the best treatment option for patients with HCC accompanied by cirrhosis and hepatitis [3]. Early results of LT for HCC in the late 1980s were disappointing [4-6], with high rates of tumor recurrence and poor (< 40%) 5-year survival. The main reason for these outcomes was the lack of stringent criteria for selecting patients with advanced tumors. In 1996, Mazzaferro et al. [7] introduced the Milan criteria (solitary tumor ≤ 5 cm or ≤ 3 tumors, each measuring < 3 cm), which have become the major criteria used to determine the indication for LT in patients with HCC worldwide [8]. However, the use of such restrictive inclusion criteria, which exclude a large pool of patients with HCC, has also been questioned [9]. Indeed, substantial evidence indicates good outcomes after LT among patients with HCC who do not satisfy the Milan criteria [10-13]. The establishment of optimal criteria for LT in HCC is necessary and important. Given developments in molecular biology, criteria based on molecular biomarkers for LT in HCC will be developed for the future. Thus, the identification of molecular biomarkers indicating LT for HCC is of great importance.
Zinc is an essential trace element and cells have developed a complex system to regulate the balance of zinc uptake, intracellular storage, and efflux [14, 15]. Two solute-linked carrier (SLC) gene families were identified in zinc transport: SLC30, which encodes zinc transporter (ZnT) proteins, and SLC39, which encodes Zrt-, Irt-like proteins (ZIP) [14, 16-18]. These proteins play opposite roles in cellular zinc homeostasis [19]. ZnT transporters promoting zinc efflux from cells or into intracellular vesicles cause decreased intracellular zinc levels. Simultaneously, ZIP transporters promoting extracellular zinc uptake and vesicular zinc release into the cytoplasm cause increased intracellular zinc levels. Recent studies demonstrated that ZIP proteins play important role in progression of solid tumors. ZIP6 (also known as LIV-1), a breast cancer-associated protein, which belongs to a new subfamily of ZIP transporters, has been found to be associated with estrogen-positive breast cancer and metastasis to lymph nodes [20]. ZIP6 and ZIP7 may play roles in the metastasis of estrogen receptor-positive breast cancer as well as in the proliferation, migration, and invasion of cervical cancer [21-23]. Similarly, ZIP10 is involved in the invasive behavior of breast cancer cells [24].
As early as 2003, ZIP4 was reported to be associated with acrodermatitis enteropathica[25-31]. Recent studies found that ZIP4 was related to the occurrence and metastasis of pancreatic cancer and prostate carcinoma [14, 19, 20, 23]. In pancreatic cancer cells, ZIP4 was shown to promote the cell cycle, cell proliferation, and invasiveness. In vivo studies, silencing of ZIP4 increased the survival rate of nude mice with pancreatic cancer orthotopic xenografts significantly [32, 33]. Moreover, ZIP4 regulated the activity of the cyclic adenosine monophosphate response element-binding protein, an important zinc finger transcription factor, and increased the expression of interleukin-6 in pancreatic cancer, leading to signal transducer and activator of transcription 3 (STAT-3) activation [34]. In 2011, it was reported that overexpression of ZIP4 reduced the cell migration of prostate carcinoma cells significantly [35]. These findings suggest that ZIP4 plays an important, albeit not yet fully defined, role in the development of pancreatic and prostate cancer. In 2010, Glen Andrews' group reported a high expression of ZIP4 in 16/23 human HCC samples and a mouse model. They found that mZip4 (the murine equivalent of ZIP4) stimulated the cell cycle, reduced apoptosis, and induced cell migration in mouse Hepa cells [36].
Therefore, in the present study we evaluated whether ZIP4 could contribute to tumor invasiveness and reduced apoptosis in human hepatoma cells. We investigated the expression of ZIP4 in seven HCC cell lines and then explored how these changes in ZIP4 expression influenced MMPs, Caspases and Bcl-2 family. Finally, using immunohistochemical analysis in HCC samples, we firstly evaluated the prognostic value of ZIP4 in HCC after LT.
Materials and Methods
Cell culture
The HCC cell lines Hep-G2, PLC, SK-HEP-1, BEL7402, SMMC-7721, and HuH-7, and the normal liver cell lines HL-7702 and Chang liver, were obtained from the American Type Culture Collection. Hep-G2, PLC, SK-HEP-1, and Chang liver cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% foetal bovine serum (FBS); BEL7402, SMMC-7721, and HL-7702 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS.
Plasmid construction and transfection
ZIP4 shRNA with the following sequence was used: 5'-ACGTAGCACTCTGCGACATGGTCAGGATG-3'. The sequence was inserted into a pGPU6/GFP/Neo vector synthesised by GenePharma (Shanghai, China). A ZIP4 overexpression plasmid was generated by sub-cloning human ZIP4 cDNA (BC062625.1) into the mammalian expression plasmid EX-LV201 (provided by GeneCopoeia, Guangzhou, China). Hep-G2, BEL7402, and HuH-7 cells were transfected with 2 µg ZIP4 shRNA plasmid, 2 µg overexpression plasmid (Z1191), or 2 µg negative control plasmid (NC and NEG) per well using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA). The transfection efficiency was estimated using green fluorescence. Cells were harvested for total protein or mRNA extraction 72 h after transfection.
Real-time polymerase chain reaction
RNA was extracted using the TRIzol reagent (Invitrogen) and used for cDNA synthesis with a M-MLV Reverse Transcriptase (TaKaRa, Dalian, China). Quantitative real-time polymerase chain reaction (qRT-PCR) was conducted with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) using the ABI Prism 7500 Real-time PCR System (Applied Biosystems). The following primer sequences were used: ZIP4, 5'-ATGTCAGGAGCGGGTCTTGC-3' and 5'-GCTGCTGTGCTGCTGGAAC-3'; Bax, 5'-TGCAGAGGATGATCGCAGCTGTG-3'and 5'-CCAATGTCCAGCCCATCATGGTC-3'; Bcl-2, 5'-TTCTTTGAGTTCGGTGGGGTC-3' and 5'-TGCATATTTGTTTGGGGCAGG-3'; caspase-3, 5'-CAGTGGAGGCCGACTTCTTG-3'and 5'-TGGCACAAAGCGACTGGAT-3'; and caspase-9, 5'-TGTCCTACTCTACTTTCCCAGGTTTT-3' and 5'-GTGAGCCCACTGCTCAAAGAT-3'. The products were detected with SYBR Green (ABI, San Francisco, CA, USA). Melting curve analyses were performed to confirm specificity. The 2-ΔΔCt method was used for quantification [37]. Each sample was run in triplicate and β-actin was used as an internal control.
Cell migration and invasion assay
At 48 h after transfection, Hep-G2, HuH-7, and BEL-7402 cells were cultured in serum-free DMEM and RPMI 1640 medium for 24 h. The migration and invasion of cells were assessed using a Transwell chamber with 8-mm pore filters (Millipore, Billerica, USA). The filter used for invasion assays was coated with 100 mL Matrigel (BD Biosciences, San Jose, CA), diluted 1:10 in cold DMEM and RPMI 1640 to form a thin continuous film on the top of the filter. Cells were trypsinised and 5 × 104 cells in serum-free medium were added to the upper chamber. Then, 0.6 mL medium supplemented with 10% FBS was added to the lower chamber. After incubation for 48 h, the cells were washed with phosphate-buffered saline (PBS), fixed with methanol, rinsed in PBS, and stained with 0.2% crystal violet. The invaded cells were counted under a light microscope (at ×200 magnification). All experiments were performed in triplicate.
Cell cycle and apoptosis analysis
Cells were used 48 h after transfection. For cell cycle analysis, cells were fixed with ice-cold 75% ethyl alcohol at 4°C overnight and incubated with propidium iodide (BD Biosciences) at 4°C in the dark for 30-60 min. For apoptosis rate analysis, cells were incubated with Annexin V-fluorescein isothiocyanate (BD Biosciences) and propidium iodide for 5 min at 4°C in the dark. After staining, the cells were analysed using a Cytomics FC 500 flow cytometer (Beckman Coulter, Miami, FL, USA).
Immunoblot analysis
Proteins were extracted from HCC cell lines with radioimmunoprecipitation assay buffer (Beyotime, Shanghai, China), separated on an 8%-12% NUPAGE Bis-Tris Gel (Invitrogen) and transferred onto polyvinylidene difluoride membranes. Subsequent procedures were performed as described previously [38]. Primary antibodies against ZIP4 [rabbit monoclonal immunoglobulin G (IgG), dilution 1:2,000; Sigma-Aldrich, , USA], matrix metalloproteinase (MMP)-2 (rabbit IgG, dilution 1:1,000; Epitomics, , USA), and MMP-9 (rabbit IgG, dilution 1:1,000) were used. Horseradish peroxidase (HRP)-conjugated antibodies against rabbit IgG was used as secondary antibodies. Immunoreactive bands on the blots were visualised with an enhanced chemiluminescence reagent kit (Beit Haemek, Israel).
Confocal immunofluorescence analysis
Cells (5×103) were incubated in a cell culture dish (NEST Biotech, Hong Kong) for 24 h, fixed with paraformaldehyde for 30 min, and permeabilized with 0.1% Triton X-100 for 5 min at room temperature, and then incubated with the primary polyclonal rabbit anti-human ZIP4 antibody (Sigma-Aldrich; dilution 1:200) in the dark for 24 h at 4°C. Next, cells were incubated with Alexa Fluor 488 donkey anti-rabbit IgG (1:500, Invitrogen, USA) for 1 h at room temperature. Fluorescence images were captured using a confocal microscope.
Patients and clinical specimens
The tumour specimens used in immunohistochemical analysis were obtained from 60 patients with HCC who underwent LT, 36 patients with liver cirrhosis, and six LT donors at the First Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, between 2006 and 2009. Written informed consent was obtained from all donors and recipients. No donor liver was harvested from an executed prisoner. The characteristics of patients with HCC are summarised in Table 1. Two independent pathologists diagnosed HCC based on the World Health Organization criteria [39]. No patient received chemotherapy or radiotherapy before surgery. Tumour specimens, adjacent non-tumorous tissues, and liver cirrhosis tissues were collected, fixed in 10% formaldehyde solution, and embedded in paraffin. Tumour-node-metastasis (TNM) staging was performed according to the Sixth edition of the Classification of the International Union against Cancer [40]. All data were analysed anonymously.
Ethics statement
The study protocol was approved by the Institutional Review Board of the Key Lab of Combined Multi-Organ Transplantation, Ministry of Public Health. Informed written consent was obtained from patients in accordance with the Declaration of Helsinki.
Immunohistochemical staining
All samples embedded in paraffin blocks were cut into 4-μm-thick sections, which were mounted on glass slides. Each slide was dewaxed in xylene, rehydrated in a graded series of alcohol solutions, and boiled in 10 mmol/L citrate buffer (pH 6.0) for antigen retrieval. After inhibition of endogenous peroxidase activity by incubation for 10 min with methanol containing 0.3% H2O2, the sections were blocked with 2% bovine serum albumin for 30 min and incubated overnight at 4°C with a primary polyclonal rabbit anti-human ZIP4 antibody (Sigma-Aldrich; dilution 1:200). After three washes with PBS, the slides were incubated with HRP-conjugated goat anti-rabbit IgG for 30 min, reacted with diaminobenzidine, and counterstained with haematoxylin. As a negative control, the primary antibody was omitted and substituted.
Immunohistochemical evaluation
To estimate the expression of ZIP4, 10 fields with a total of at least 1,000 tumour cells were evaluated by high-power microscopy. Two independent technicians evaluated the immunostaining in a blinded manner, and re-evaluated immunostaining together to reach consensus in the case of a discrepancy. ZIP4 expression was calculated as the percentage of positive cells.
Statistical analysis
All statistical data were analysed using SPSS 10.0 (SPSS Inc. Chicago, IL, USA). Differences between groups were analysed by one-way analysis of variance, the Mann-Whitney U-test, or the chi-squared test. Cumulative survival was compared by the Kaplan-Meier test using the log-rank method. A multivariate Cox proportional-hazards regression model was used to identify independent prognostic factors. Statistical significance was set at P < 0.05.
Results
Location and expression of ZIP4 in HCC
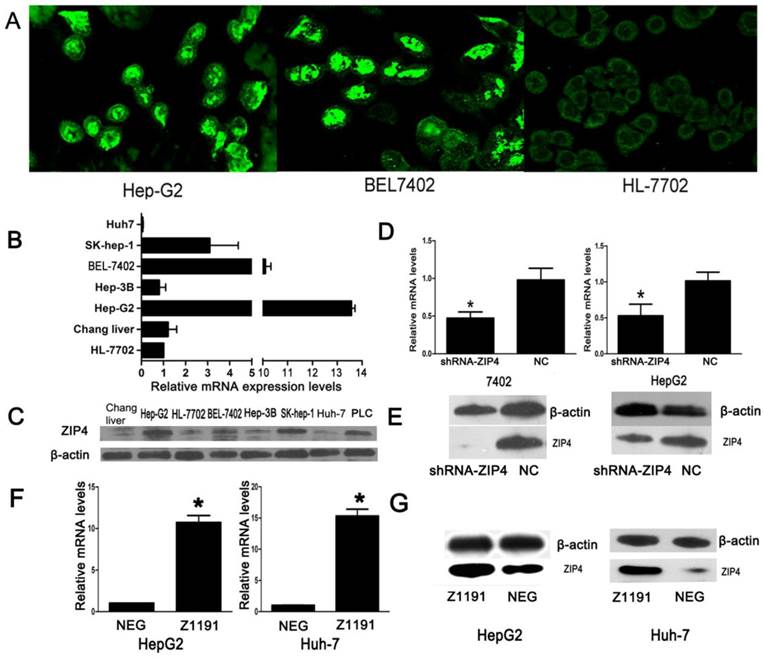
Using a laser-scanning confocal microscope, ZIP4 was detected in cell membranes and nuclei in cancer cell lines (HepG2 and BEL7402), but only at cell membranes in immortalized liver cells (HL-7702; Fig. 1A). Expression of ZIP4 was higher in HCC cell lines (HepG2, SK-HEP-1, and BEL-7402) than in immortalized liver cells (HL-7702 and Chang liver; Fig. 1B, 1C), and was highest in HepG2 cells. These results showed that ZIP4 was more highly expressed in HCC cells than in normal liver cells.
We performed knockdown and overexpression experiments with shRNA-ZIP4 and Z1191 plasmids, respectively. ZIP4 expression was relatively lower in HuH-7 cells and higher in BEL-7402 cells than HL-7702 cells, respectively. Thus, we overexpressed ZIP4 in HuH-7 cells and knocked it down in BEL-7402 cells. The efficiencies of knockdown and overexpression were confirmed by qRT-PCR and immunoblotting (Fig. 1D‑G).
Effect of ZIP4 on invasiveness and migration in HCC
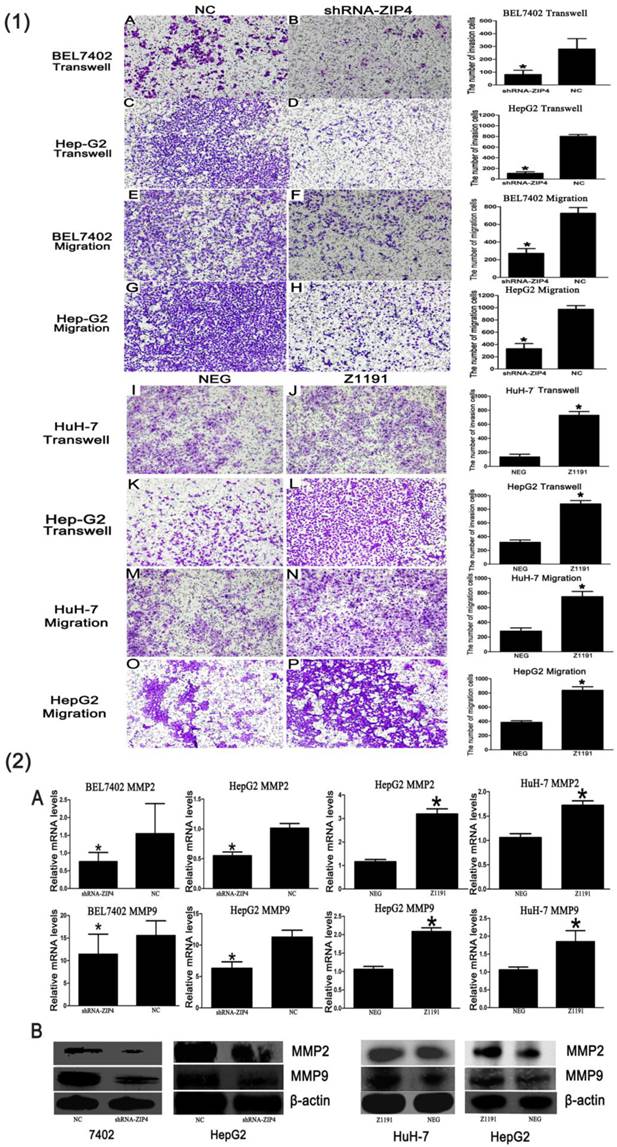
In an invasion assay, we found significantly fewer migrated/invaded cells in the ZIP4-silenced group than in the negative control, but many more in the ZIP4 overexpression group than in the negative control (P < 0.05; Fig. 2(1)). It demonstrates that ZIP4 can enhance HCC migration and invasion.
Because MMPs are involved in the digestion of extracellular matrices, penetration of the stroma, and cancer cell motility, we examined the expression of MMP family members in different groups to explore the potential mechanisms of ZIP4-promoted invasiveness. We found that expression of MMP-2 and MMP-9 was markedly reduced in the ZIP4-silenced group and increased in the ZIP4 overexpression group, compared with their respective negative controls (P < 0.05; Fig. 2(2)). Negative results are not shown. These data indicate that ZIP4 was involved in regulating the expression of MMP2 and MMP9.
Effect of ZIP4 on Cell cycle in HCC
As shown in Figure 3A, silencing of ZIP4 induced the accumulation of HepG2 cells in the G1 phase and reduced the proportions of HepG2 cells in the S and G2 phases, whereas overexpression of ZIP4 decreased the proportion of HepG2 cells in the G1 phase and increased the proportions of HepG2 cells in the S and G2 phases. These demonstrate that ZIP4 accelerates HepG2 cells growth.
Effect of ZIP4 on apoptosis in HCC
Decreased or increased ZIP4 expression led to an elevated or reduced rate of HCC cell apoptosis (P < 0.05; Fig. 3B). Results show that ZIP4 inhibited HCC apoptosis.
The Bcl-2 and caspase pathways are considered classical apoptotic pathways. Thus, we next examined members of the Bcl-2 and caspase families. We found that ZIP4 silencing resulted in increased expression of pro-apoptotic genes (caspase-3, caspase-9, Bax) and decreased expression of Bcl-2, whereas ZIP4 overexpression had the opposite effect (P < 0.05; Fig. 3C‑F). Negative results are not shown. It turned out that ZIP4 was involved in regulating the expression of caspase-3, caspase-9, Bax, and Bcl-2.
(A) Laser-scanning confocal microscope was used to detect the distribution of ZIP4 in HepG2, BEL7402, and HL-7702 cells (magnification: ×1260). Cells were incubated in a cell culture dish, fixed with paraformaldehyde for 30 min, and permeabilized with 0.1% Triton X-100 for 5 min at room temperature and incubated with primary polyclonal rabbit anti-human ZIP4 antibody. Cells were then washed and incubated with Alexa Fluor 488 donkey anti-rabbit IgG. The green signal represents staining for ZIP4 protein. ZIP4 located in cell membranes of HL-7702 cells, but not in nuclei. ZIP4 accumulated in the nuclei in HepG2 and BEL7402 cells. (B) The mRNA levels of ZIP4 in seven liver cell lines. We used the TRIzol reagent (Invitrogen) to extract total RNA and performed cDNA synthesis using M-MLV reverse transcriptase (TaKaRa, Dalian, China). ß-actin was the endogenous control. All the samples were normalized to human ß-actin according to the 2-ΔΔCT method. ZIP4 mRNA levels in immortalized liver cells (HL-7702) was the negative control. The x-axis represents multiples of mRNA levels in HL-7702 cells. (C) ZIP4 protein levels in eight liver cell lines. (D) The mRNA levels of ZIP4 in BEL7402 and HepG2 cells with ZIP4 silencing (shRNA-ZIP4) and negative control (NC) cells. β-actin was used as an internal control. (E) ZIP4 protein levels in BEL7402 and HepG2 cells with ZIP4 silencing (shRNA-ZIP4) and negative control (NC) cells. (F) ZIP4 mRNA levels in HuH-7 and HepG2 cells with ZIP4 overexpression (Z1191) and negative control (NEG). β-actin was used as an internal control. (G) ZIP4 protein levels in HuH-7 and HepG2 cells with ZIP4 overexpression (Z1191) and a negative control (NEG). Data are from one of three repeated independent experiments. *P < 0.05.
(1-B), (1-D), (1-F), and (1-H) Cells were transfected with ZIP4-shRNA plasmid (shRNA-ZIP4). (1-J), (1-L), (1-N), and (1-P) Cells were transfected with ZIP4 overexpression plasmid (Z1191). (1-A), (1-C), (1-E), (1-G), (1-I), (1-K), (1-M), and (1-O) Cells transfected with a negative control plasmid (NC or NEG; magnification: ×200). Right: statistical analysis of the numbers of invaded/migrated HepG2, HuH-7, and BEL7402 cells. Student's t-test was used for statistical comparisons. Data are from one of three repeated independent experiments. *P < 0.05. (2-A) mRNA of MMP-2 and MMP-9 in BEL7402 and HepG2 cells with ZIP4 silencing (shRNA-ZIP4) and HuH-7 and HepG2 cells with ZIP4 overexpression (Z1191) was detected by quantitative real-time polymerase chain reaction (qRT-PCR). (2-B) Protein levels of MMP-2 and MMP-9 in BEL7402 and HepG2 cells with ZIP4 silencing (shRNA-ZIP4) and HuH-7 and Hep-G2 cells with ZIP4 overexpression (Z1191) were detected by immunoblotting.
(A) Cell cycle distribution of HepG2 cells transfected with shRNA (shRNA-ZIP4), overexpression (Z1191), or negative control plasmids (NC or NEG). 2N, diploid DNA content; 4N, tetraploid DNA content. (B) Percentages of apoptotic cells in BEL7402 and HepG2 cells transfected with shRNA (shRNA-ZIP4) and HuH-7 and Hep-G2 cells transfected with an overexpression plasmid (Z1191). Student's t-test was used for statistical comparisons. *P < 0.05. (C), (D), (E), and (F) mRNA of caspase-3, caspase-9, Bax, and Bcl-2 in HepG2 cells with ZIP4 silencing (shRNA-ZIP4) or ZIP4 overexpression (Z1191) were detected by qRT-PCR. NC and NEG denote cells transfected with a negative control plasmid. Data shown are representative of at least three separate experiments. *P < 0.05.

Relationship between ZIP4 and clinicopathological features
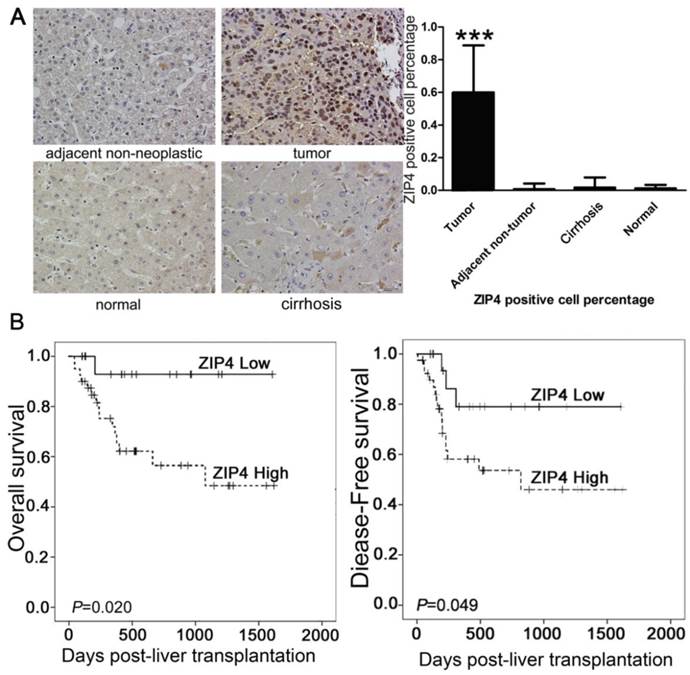
ZIP4-positive cells showed stained nuclei in immunohistochemical staining (Fig. 4A). The density of ZIP4-positive cells was significantly higher in tumor tissues than in peritumoral tissues, cirrhotic liver tissue, and normal liver tissue (P < 0.0001; Fig. 4A).
The relationship between ZIP4 expression and clinicopathological parameters is described in Supplemental Table 1. Using the median (50%) density of ZIP4-positive cells as the cut-off value [41], patients were divided into high (n = 40) and low expression (n = 20) groups. High expression of ZIP4 correlated significantly with high Child-Turcotte-Pugh (CTP) score (P = 0.042), late TNM stage (P = 0.044), tumor recurrence (P = 0.002), large tumor size (P = 0.022), and high alanine aminotransferase level (P = 0.043) (Table 1). ZIP4 expression did not correlate significantly with gender, age, cirrhosis, α‑fetoprotein (AFP) level, or hepatitis B surface antigen (Supplemental Table 1).
Relationship between ZIP4 and survival
In univariate analyses, tumor size, vascular invasion, cancer embolus, AFP level, history of hepatitis, distant metastasis, bile duct invasion, prothrombin time, total bilirubin level, and TNM stage were prognostic factors for overall survival and/or disease-free survival (Table 2). The density of ZIP4-positive cells was correlated significantly with overall survival (P = 0.020) and tumor -free survival (P = 0.049). Overall survival and tumor-free survival rates were poorer in patients with higher ZIP4 expression than in those with lower ZIP4 expression (Fig. 4B).
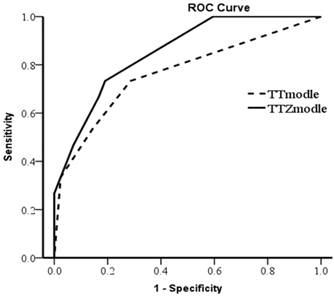
In a multivariate analysis, the expression of ZIP4 was an independent prognostic factor for overall survival (hazard ratio = 4.343, P = 0.037), but not tumor-free survival (P = 0.075; Table 2). Multivariate analysis (without ZIP4) showed that tumor size (P = 0.002) and total bilirubin level (P = 0.011) were two independent prognostic factors for overall survival. To verify the effectiveness of ZIP4, we constructed TT Model (based on tumor size and total bilirubin level) and TTZ Model (based on tumor size, total bilirubin level and ZIP4). The equation of TT Model was TT model=- 1.701×Tumor size+ (-1.362×TBIL level) (tumor size, 1 for tumor size≥8cm, 2 for tumor size<8cm; TBIL level, 1= Normal; 2= Abnormal). The equation of TTZ Model was TTZ model=(- 1.622)×tumor size+ (-1.249×TBIL level) +1.723×ZIP4 (tumor size, 1 for tumor size≥8cm, 2 for tumor size<8cm; TBIL level, 1= Normal, 2= Abnormal; ZIP4, Low=1, High=2). The receiver operating characteristic curve (ROC) was used to assess which model was better (Fig 5). Area of the TTZ Model under the curve is 0.852 and the TT Model is 0.765. The ROC indicated the prediction was improved when combined with ZIP4. These data suggest ZIP4 was a very effective prognostic factor for overall survival. These results show that the expression of ZIP4 can serve as an effective predictor for survival in HCC patients after LT.
Correlation between ZIP4 expression and clinicopathologic characteristics.
| Characteristics | ZIP4 positive cell density of cells (%) | ||
|---|---|---|---|
| Low (n=20) (33.3%) | High (n=40) (66.7%) | P | |
| Tumor size(cm) | 0.022 | ||
| <5 | 11(55%) | 10(25%) | |
| ≥5 | 9(45%) | 30(75%) | |
| Tumor number | 0.041 | ||
| Single | 13(65%) | 14(35%) | |
| Multiple | 7(35%) | 24(65%) | |
| Tumor recurrence | 0.002 | ||
| Yes | 3(15%) | 23(57.5%) | |
| No | 17(85%) | 17(42.5%) | |
| Ascites | 0.022 | ||
| Yes | 5(25%) | 1(2.5%) | |
| no | 15(75%) | 39(97.5%) | |
| ALT level | 0.043 | ||
| Normal | 9(45%) | 8(20%) | |
| Abnormal | 11(55%) | 32(80%) | |
| TNM stage | 0.044 | ||
| I+II | 13(65%) | 15(37.5%) | |
| III+IV | 7(35%) | 25(62.5%) | |
| CTP | 0.042 | ||
| A | 12(60%) | 13(32.5%) | |
| B/C | 8(40%) | 27(67.5%) | |
Abbreviation: ALT: alanine aminotransferase; CTP Child-Turcotte-Pugh scores. AFP: alpha fetoprotein;γGT: γ- glutamyl transferase; CEA: carcinoembryonic antigen; CHE: cholinesterase; TBIL: total bilirubin; ALB: Serum albumin; PT: prothrombintime; INR: International Normalized Ratio; ALT: alanine aminotransferase; DBIL: bilirubin direct. Chi-square tests for all other analyses.
Univariate and Multivariate analysis of factors associated with survival and recurrence (n=60).
| Variables | Overall survival | Disease-free survival | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate* | Univariate | Multivariate* | |||||
| HR | P | HR | P | HR | P | HR | P | |
| Age, <50/≥50 years | 0.725 | 0.522 | 0.598 | 0.258 | ||||
| Tumor size, <8/≥8cm | 0.161 | 0.001 | NA | 0.179 | 0.349 | 0.031 | NA | 0.545 |
| Tumor number, Single/Multiple | 2.192 | 0.122 | 1.607 | 0.325 | ||||
| Distant metastasis, Yes/no | 2.072 | 0.327 | 4.948 | 0.005 | NA | 0.209 | ||
| Tumor differentiation, I/II-III | 0.610 | 0.334 | 0.732 | 0.502 | ||||
| Liver cirrhosis, Yes/No | 0.483 | 0.199 | 1.239 | 0.774 | ||||
| History of hepatitis, Yes/No | 0.205 | 0.001 | 0.156 | 0.029 | 0.472 | 0.224 | ||
| HBsAg, Negative/Positive | 0.761 | 0.682 | 2.757 | 0.214 | ||||
| AFP level, ≤400/>400 ng/mL | 1.050 | 0.928 | 2.799 | 0.022 | NA | 0.277 | ||
| AFP level, ≤20/>20 ng/mL | 2.068 | 0.249 | 2.322 | 0.169 | ||||
| γGT level, Normal/Abnormal | 1.863 | 0.325 | 1.537 | 0.443 | ||||
| CEA level, Normal/Abnormal | 1.751 | 0.378 | 0.401 | 0.357 | ||||
| CHE level, Normal/Abnormal | 0.484 | 0.143 | 0.751 | 0.532 | ||||
| Ascites , Yes/no | 0.044 | 0.251 | 0.043 | 0.184 | ||||
| TBIL level, Normal/Abnormal | 0.261 | 0.004 | NA | 0.410 | 0.914 | 0.863 | ||
| ALB level, Normal/Abnormal | 0.378 | 0.061 | 0.866 | 0.753 | ||||
| PT, Normal/Abnormal | 0.369 | 0.072 | 0.329 | 0.025 | 4.192 | 0.041 | ||
| INR level, Normal/Abnormal | 0.774 | 0.624 | 1.300 | 0.587 | ||||
| Serum creatinine | 0.669 | 0.450 | 1.252 | 0.633 | ||||
| ALT level, Normal/Abnormal | 1.289 | 0.647 | 1.151 | 0.778 | ||||
| DBIL level, Normal/Abnormal | 0.458 | 0.110 | 0.946 | 0.910 | ||||
| ZIP4 positive rate of cells, Low/High | 7.613 | 0.020 | 10.071 | 0.037 | 3.216 | 0.049 | NA | 0.075 |
| CTP, A/B+C | 1.506 | 0.445 | 1.005 | 0.991 | ||||
| TNM stage, I+II/III+IV | 3.820 | 0.005 | NA | 0.330 | 3.396 | 0.005 | NA | 0.301 |
Univariate and multivariate analysis: Cox proportional hazards regression model. Abbreviation: HR, Hazard Ratio; NA, not applicable; AFP: alpha fetoprotein;γGT: γ- glutamyl transferase; CEA: carcinoembryonic antigen; CHE: cholinesterase; TBIL: total bilirubin; ALB: Serum albumin; PT: prothrombintime; INR: International Normalized Ratio; ALT: alanine aminotransferase; DBIL: bilirubin direct. *Multivariate variables were adopted by univariate analysis (P<0.05).
(A) Immunohistochemical staining of ZIP4 (magnification: ×200). Numerous tumor cells were ZIP4-positive. In peritumoral, cirrhotic, and normal tissues, almost no cells were ZIP4-positive. ***P < 0.0001. (B) Patients with high ZIP4-positive cell densities (dashed lines) had significantly poorer survival compared with individuals with low ZIP4-positive cell densities (solid lines).
Multivariate analysis of factors associated with survival without ZIP4.
| *Variables | P | OR (95% CI) |
|---|---|---|
| Tumor size, <8/≥8cm | 0.002 | 0.182(0.061-0.545) |
| TBIL level, Normal/Abnormal | 0.001 | 0.256(0.090-0.730) |
Abbreviations: TBIL, total bilirubin; CI, confidence interval; OR, odds ratio.
The prediction of TT Model and TTZ Model were assessed by receiver operating characteristic curve (ROC). The area under the curve of TTZ Model was 0.852 and TT Model was 0.765.
Discussion
Carcinogenesis is a complex and multifactorial process, resulting from many environmental effects. It involves the combined actions of multiple oncogenes and tumor suppressor genes. HCC is commonly lethal due to its aggressive metastasis, and migration and invasion are important features of cancer cells generally [42]. However, the molecular mechanism underlying HCC metastasis remains unclear. Therefore, the identification of novel molecular markers will provide new opportunities for the prevention of HCC recurrence and metastasis. In previous studies, ZIP4 was reported it could promote the cell invasiveness in the human pancreatic cancer and induced cell migration in mouse Hepa cells. In this study, we found that ZIP4 contributed to invasiveness and migration of hepatoma cells. Furthermore, we discovered ZIP4 could influence the expression of MMP2 and MMP9, which are directly linked to angiogenesis and the degradation of basement membrane collagen, and their expression and activity correlate with metastatic ability and the prognosis of cancer [43]. Therefore, ZIP4 increasing the gene expression of MMP2 and MMP9 lead to promote invasiveness and migration of hepatoma cells. Recent studies indicate that MMP consist of a family of zinc-containing enzymes, and their activities can be activated by zinc. ZIP4, zinc transporter, might regulate the expression of MMP2 and MMP9 by influence on the concentration of Zinc.
Apoptosis is intimately connected with the elimination of potentially malignant cells, hyperplasia and tumor progression [44]. Apoptosis of tumor cells was related to the tumor growth and the apoptosis was also an important feature of cancer cells. In the process of the cell culture, we observed cells growth status and found more dead cells in the ZIP4 suppressive group than the negative control. The previous studies showed that mZip4 (the murine equivalent of ZIP4) reduced apoptosis in mouse Hepa cells. Thus, we speculated ZIP4 might reduce apoptosis of human hepatoma cells. As we surmised, ZIP4 could reduce apoptosis of heptama cells in this study. Bcl-2 and Caspases pathways have been considered as classical apoptotic pathways and these two pathways mediated most of the cell apoptosis. Therefore, we firstly assessed the effect of ZIP4 on these two pathways. The results showed exactly what we expected: ZIP4 upregulated apoptosis-related proteins caspase-3, caspases-9, and Bax, whereas expression of Bcl-2 was decreased. From this, ZIP4 decreasing gene expression of Caspase-3, Caspase-9 and Bax and increasing the gene expression of Bcl-2 result in inhibition of apoptosis in hepatoma cells.
These results consistently indicate that knocking down ZIP4 by shRNA, might be an effective method to control HCC growth, and represent a novel and potent treatment method for ZIP4-overexpressing HCC. It may ultimately have potential use in the clinical treatment of HCC. However, individuals may respond to ZIP4 shRNA therapy differently. It may be necessary to use multiple cycles of shRNA treatment due to the short lifespan of shRNA. Thus, comprehensive treatment with ZIP4 shRNA and chemo/radiation therapy may increase the efficacy of each individual therapy. These results show that ZIP4 represents a novel and potent target in ZIP4-overexpressing HCC.
HCC recurrence, a major lethal problem of LT for HCC, is well known to be a close correlation with TNM stage, tumor size and tumor metastasis. Highly metastatic tumour, such as HCC with large tumor size, was the key risk factor of tumor recurrence. Since, in vitro study, ZIP4 induced invasiveness and migration and inhibited apoptosis of hepatoma cells, which contributed to tumor size and tumor metastasis. We speculated that ZIP4 expression is correlated with clinicopathological parameters of LT and could be an important factor related to clinical outcome of HCC patients after LT. As we expected, ZIP4 was associated with tumor recurrence, tumor size, TNM stage, and CTP score. Further, our results indicated that patients with high expression of ZIP4 have a higher recurrence rate and shorter survival time. Thereafter, through assessing TT Model and TTZ Model, we found ZIP4 was a very effective prognostic factor for overall survival. Thus, ZIP4 expression was an independent and effective prognostic factor for overall survival after LT for HCC.
The selection criterion for LT in HCC is very important in determining outcome of transplantation in the recipient. All of those conventional criteria (Milan criteria et al) are pathological TNM criteria, based on the morphological characteristics of HCC [9, 45] and do not appear to be sufficiently prognostic with regard to survival and risk of HCC recurrence after LT [46]. As molecular biomarker of tumor development, biomarkers of tumors can provide a more standardized approach at the molecular level for tumor classification [47]. Therefore, we've been committed to finding biomarkers can be used in criteria for selecting LT candidates among patients with HCC. Our results indicate that ZIP4 may be one of tumor biomarkers probably used in LT criteria.
The detection of a membrane protein in the nucleus is difficult to explain. Several studies have found that the locations of some members of the ZIP family, such as ZIP1, ZIP3, and ZIP7, may move from intracellular organelles to cell surfaces when zinc is limited [48-50]. This process may partly explain the presence of ZIP4 in nuclei. Unlike in immortalized liver cells, ZIP4 was located not only at membranes, but also in nuclei, in cancer cells. Thus, cancer may enable the presence of ZIP4 in cell nuclei. We speculated that ZIP4 may take part in some as yet unknown functional change during the transformation of normal cells into tumor cells, and that its transfer to the nucleus may be an important step. Whether ZIP4 can function as a transcription factor or a kinase in the nucleus is unknown and needs to be studied further.
This report is the first to show the potential of ZIP4 as a predictive biomarker for HCC recurrence or patient survival in patients with HCC undergoing LT. Limitations of this study include the small number of patients and its retrospective design. It is unclear that whether ZIP4 directly or indirectly influence the expression of MMP2 and MMP9 and the specific mechanisms that ZIP4 regulating apoptosis-related proteins need to be further study. Well-designed studies with larger samples are needed to further evaluate the role of ZIP4 in predicting HCC recurrence after transplantation.
In summary, ZIP4 increasing the expression of MMP-2 and MMP-9 result in increased invasiveness, and reducing expression of the apoptosis-related genes caspase-3, capase-9, and Bax result in reduced apoptosis. More importantly, expression of ZIP4 was associated with outcome after LT in patients with HCC. ZIP4 is a novel treatment target and a suitable prognostic marker for LT in HCC.
Supplementary Material
Supplementary Table 1Abbreviations
AFP, alpha fetoprotein; CTP, Child-Turcotte-Pugh; DMEM, Dulbecco's modified Eagle's medium; FBS, foetal bovine serum; HCC, hepatocellular carcinoma; HRP, horseradish peroxidase; IgG, immunoglobulin G; LT, liver transplantation; MMP, matrix metalloproteinase; PBS, phosphate-buffered saline; qRT-PCR, quantitative real-time polymerase chain reaction; RPMI, Roswell Park Memorial Institute; TNM, tumour-node-metastasis; ZIP4, ZRT/IRT-like protein 4.
Acknowledgements
We thank all patients enrolled in this study for their kind understanding and support. This study was supported by grants from the National High Technology Research and Development Program 863 (no. 2012AA020204), the National Science and Technology Major Project (no. 2012ZX10002010-001-005) and Zhejiang Provincial Program for the Cultivation of High-Level Innovative health Talents. The funders had no role in study design, data collection and analysis, the decision to publish, or the preparation of the manuscript.
Competing interests
The authors declare that they have no competing interests.
References
1. Xu ZY, Ding SM, Zhou L. et al. FOXC1 contributes to microvascular invasion in primary hepatocellular carcinoma via regulating epithelial-mesenchymal transition. Int J Biol Sci. 2012;8:1130-1141
2. Shim JH, Lee HC, Han S. et al. Hepatocyte nuclear factor 1beta is a novel prognostic marker independent of the Milan criteria in transplantable hepatocellular carcinoma: a retrospective analysis based on tissue microarrays. Liver Transpl. 2013;19:336-345
3. Ng KK, Lo CM, Chan SC. et al. Liver transplantation for hepatocellular carcinoma: the Hong Kong experience. J Hepatobiliary Pancreat Sci. 2010;17:548-554
4. Ringe B, Pichlmayr R, Wittekind C. et al. Surgical treatment of hepatocellular carcinoma: experience with liver resection and transplantation in 198 patients. World J Surg. 1991;15:270-285
5. O'Grady JG, Polson RJ, Rolles K. et al. Liver transplantation for malignant disease. Results in 93 consecutive patients. Ann Surg. 1988;207:373-379
6. Iwatsuki S, Gordon RD, Shaw BW Jr. et al. Role of liver transplantation in cancer therapy. Ann Surg. 1985;202:401-407
7. Mazzaferro V, Regalia E, Doci R. et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med. 1996;334:693-699
8. Volk ML, Vijan S, Marrero JA. A novel model measuring the harm of transplanting hepatocellular carcinoma exceeding Milan criteria. Am J Transplant. 2008;8:839-846
9. Lei J, Yan L. Outcome comparisons among the Hangzhou, Chengdu, and UCSF criteria for hepatocellular carcinoma liver transplantation after successful downstaging therapies. J Gastrointest Surg. 2013;17:1116-1122
10. Malago M, Sotiropoulos GC, Nadalin S. et al. Living donor liver transplantation for hepatocellular carcinoma: a single-center preliminary report. Liver Transpl. 2006;12:934-940
11. Gondolesi GE, Roayaie S, Munoz L. et al. Adult living donor liver transplantation for patients with hepatocellular carcinoma: extending UNOS priority criteria. Ann Surg. 2004;239:142-149
12. Roayaie S, Frischer JS, Emre SH. et al. Long-term results with multimodal adjuvant therapy and liver transplantation for the treatment of hepatocellular carcinomas larger than 5 centimeters. Ann Surg. 2002;235:533-539
13. Yao FY, Ferrell L, Bass NM. et al. Liver transplantation for hepatocellular carcinoma: expansion of the tumor size limits does not adversely impact survival. Hepatology. 2001;33:1394-1403
14. Liuzzi JP, Cousins RJ. Mammalian zinc transporters. Annu Rev Nutr. 2004;24:151-172
15. Kim BE, Wang F, Dufner-Beattie J. et al. Zn2+-stimulated endocytosis of the mZIP4 zinc transporter regulates its location at the plasma membrane. J Biol Chem. 2004;279:4523-4530
16. Liuzzi JP, Bobo JA, Lichten LA. et al. Responsive transporter genes within the murine intestinal-pancreatic axis form a basis of zinc homeostasis. Proc Natl Acad Sci U S A. 2004;101:14355-14360
17. Eide DJ. The SLC39 family of metal ion transporters. Pflugers Arch. 2004;447:796-800
18. Guerinot ML. The ZIP family of metal transporters. Biochim Biophys Acta. 2000;1465:190-198
19. Murgia C, Vespignani I, Rami R. et al. The Znt4 mutation inlethal milk mice affects intestinal zinc homeostasis through the expression of other Zn transporters. Genes Nutr. 2006;1:61-70
20. Taylor KM, Morgan HE, Johnson A. et al. Structure-function analysis of LIV-1, the breast cancer-associated protein that belongs to a new subfamily of zinc transporters. Biochem J. 2003;375:51-59
21. Hogstrand C, Kille P, Nicholson RI. et al. Zinc transporters and cancer: a potential role for ZIP7 as a hub for tyrosine kinase activation. Trends Mol Med. 2009;15:101-111
22. Zhao L, Chen W, Taylor KM. et al. LIV-1 suppression inhibits HeLa cell invasion by targeting ERK1/2-Snail/Slug pathway. Biochem Biophys Res Commun. 2007;363:82-88
23. Taylor KM, Hiscox S, Nicholson RI. Zinc transporter LIV-1: a link between cellular development and cancer progression. Trends Endocrinol Metab. 2004;15:461-463
24. Kagara N, Tanaka N, Noguchi S. et al. Zinc and its transporter ZIP10 are involved in invasive behavior of breast cancer cells. Cancer Sci. 2007;98:692-697
25. Mao X, Kim BE, Wang F. et al. A histidine-rich cluster mediates the ubiquitination and degradation of the human zinc transporter, hZIP4, and protects against zinc cytotoxicity. J Biol Chem. 2007;282:6992-7000
26. Dufner-Beattie J, Weaver BP, Geiser J. et al. The mouse acrodermatitis enteropathica gene Slc39a4 (Zip4) is essential for early development and heterozygosity causes hypersensitivity to zinc deficiency. Hum Mol Genet. 2007;16:1391-1399
27. Wang FD, Kim BE, Dufner-Beattie J. et al. Acrodermatitis enteropathica mutations affect transport activity, localization and zinc-responsive trafficking of the mouse ZIP4 zinc transporter. Hum Mol Genet. 2004;13:563-571
28. Dufner-Beattie J, Wang F, Kuo YM. et al. The acrodermatitis enteropathica gene ZIP4 encodes a tissue-specific, zinc-regulated zinc transporter in mice. J Biol Chem. 2003;278:33474-33481
29. Dufner-Beattie J, Langmade SJ, Wang F. et al. Structure, function, and regulation of a subfamily of mouse zinc transporter genes. J Biol Chem. 2003;278:50142-50150
30. Kury S, Dreno B, Bezieau S. et al. Identification of SLC39A4, a gene involved in acrodermatitis enteropathica. Nat Genet. 2002;31:239-240
31. Kury S, Devilder MC, Avet-Loiseau H. et al. Expression pattern, genomic structure and evaluation of the human SLC30A4 gene as a candidate for acrodermatitis enteropathica. Hum Genet. 2001;109:178-185
32. Li M, Zhang Y, Bharadwaj U. et al. Down-regulation of ZIP4 by RNA interference inhibits pancreatic cancer growth and increases the survival of nude mice with pancreatic cancer xenografts. Clin Cancer Res. 2009;15:5993-6001
33. Li M, Zhang Y, Liu Z. et al. Aberrant expression of zinc transporter ZIP4 (SLC39A4) significantly contributes to human pancreatic cancer pathogenesis and progression. Proc Natl Acad Sci U S A. 2007;104:18636-18641
34. Zhang Y, Bharadwaj U, Logsdon CD. et al. ZIP4 regulates pancreatic cancer cell growth by activating IL-6/STAT3 pathway through zinc finger transcription factor CREB. Clin Cancer Res. 2010;16:1423-1430
35. Chen QG, Zhang Z, Yang Q. et al. The role of zinc transporter ZIP4 in prostate carcinoma. Urol Oncol. 2011
36. Weaver BP, Zhang Y, Hiscox S. et al. Zip4 (Slc39a4) expression is activated in hepatocellular carcinomas and functions to repress apoptosis, enhance cell cycle and increase migration. PLoS One. 2010 5
37. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408
38. Cheng J, Xie HY, Xu X. et al. NDRG1 as a biomarker for metastasis, recurrence and of poor prognosis in hepatocellular carcinoma. Cancer Lett. 2011;310:35-45
39. Wittekind C. [The new WHO classification of liver tumors--what is really new?]. Verh Dtsch Ges Pathol. 2001;85:212-218
40. Greene FL. American Joint Committee on Cancer, American Cancer Society. AJCC cancer staging handbook: from the AJCC cancer staging manual. 6th ed. New York: Springer. 2002
41. Chen KJ, Lin SZ, Zhou L. et al. Selective recruitment of regulatory T cell through CCR6-CCL20 in hepatocellular carcinoma fosters tumor progression and predicts poor prognosis. PLoS One. 2011;6:e24671
42. Yau WL, Lam CS, Ng L. et al. Over-Expression of miR-106b Promotes Cell Migration and Metastasis in Hepatocellular Carcinoma by Activating Epithelial-Mesenchymal Transition Process. PLoS One. 2013;8:e57882
43. Xue YJ, Xiao RH, Long DZ. et al. Overexpression of FoxM1 is associated with tumor progression in patients with clear cell renal cell carcinoma. J Transl Med. 2012;10:200
44. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70
45. Marsh JW, Dvorchik I, Bonham CA. et al. Is the pathologic TNM staging system for patients with hepatoma predictive of outcome? Cancer. 2000;88:538-543
46. Yao FY, Ferrell L, Bass NM. et al. Liver transplantation for hepatocellular carcinoma: comparison of the proposed UCSF criteria with the Milan criteria and the Pittsburgh modified TNM criteria. Liver Transpl. 2002;8:765-774
47. Ma XJ, Patel R, Wang X. et al. Molecular classification of human cancers using a 92-gene real-time quantitative polymerase chain reaction assay. Arch Pathol Lab Med. 2006;130:465-473
48. Taylor KM, Morgan HE, Smart K. et al. The emerging role of the LIV-1 subfamily of zinc transporters in breast cancer. Mol Med. 2007;13:396-406
49. Huang L, Kirschke CP, Zhang Y. et al. The ZIP7 gene (Slc39a7) encodes a zinc transporter involved in zinc homeostasis of the Golgi apparatus. J Biol Chem. 2005;280:15456-15463
50. Wang F, Dufner-Beattie J, Kim BE. et al. Zinc-stimulated endocytosis controls activity of the mouse ZIP1 and ZIP3 zinc uptake transporters. J Biol Chem. 2004;279:24631-24639
Author contact
![]() Corresponding author: Prof. Shu-Sen Zheng, PhD, MD, FACS, Department of Surgery, Division of Hepatobiliary and Pancreatic Surgery, The First Affiliated Hospital, ZhejiangUniversity School of Medicine, 79 QingChun Road, Hangzhou, 310003, China, Tel: +86 57187236616, Fax: +86 57187072577, e-mail: zyzssedu.cn.
Corresponding author: Prof. Shu-Sen Zheng, PhD, MD, FACS, Department of Surgery, Division of Hepatobiliary and Pancreatic Surgery, The First Affiliated Hospital, ZhejiangUniversity School of Medicine, 79 QingChun Road, Hangzhou, 310003, China, Tel: +86 57187236616, Fax: +86 57187072577, e-mail: zyzssedu.cn.

 Global reach, higher impact
Global reach, higher impact