Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(3):309-320. doi:10.7150/ijbs.7475 This issue Cite
Research Paper
CARP, a Myostatin-downregulated Gene in CFM Cells, Is a Novel Essential Positive Regulator of Myogenesis
1. Institute of Neurology, Guangdong Medical College, Zhanjiang 524001, China;
2. Guangdong Key Laboratory of Age-Related Cardiac and Cerebral Diseases, Affiliated Hospital of Guangdong Medical College, Zhanjiang 524001, China;
3. Department of General Surgery, First Affiliated Hospital of Harbin Medical University, Harbin 150001, China;
4. National Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100005, China.
* These authors contributed equally to this work.
Received 2013-8-22; Accepted 2014-2-18; Published 2014-3-6
Abstract
Myostatin, a member of the TGF-β superfamily, has been shown to act as a negative regulator of myogenesis. Although its role in myogenesis has been clearly documented through genetic analysis, few gene cascades that respond to myostatin signaling and regulate myogenesis have been characterized, especially in avian species. In a previous study, we screened for such genes in chicken fetal myoblasts (CFMs) using the differential display PCR method and found that cardiac ankyrin repeat protein (CARP) was downregulated by myostatin and specifically expressed in chicken skeletal muscle. However, little is known about the potential functions of CARP in chicken skeletal myogenesis. In this study, the expression patterns of chicken CARP and the possible function of this gene in skeletal muscle growth were characterized. Our data showed that CARP was predominantly expressed in postnatal skeletal muscle, and its expression increased during myogenic differentiation in CFM cells. When CARP was overexpressed, CFM cell growth was enhanced by accelerating the cell cycle at the G1 to S phase transition and increasing cyclin D1 expression. CARP knockdown had the opposite effect: while myoblasts underwent differentiation, knockdown of CARP expression induced extensive cell death, suppressed the formation of myotubes, and markedly decreased the expression of differentiation-related genes such as myosin heavy chain (MHC), myoD, and caveolin-3. Our findings indicate that CARP may play a key role in the myostatin signaling cascade that governs chicken skeletal myogenesis through promoting proliferation and avoiding apoptosis during CFM cell differentiation.
Keywords: CARP, myostatin, chicken fetal myoblast, skeletal muscle, development.
Introduction
During skeletal muscle development, various hormones, growth factors, and cytokines regulate the expression of myogenic regulatory factors and muscle-specific genes that determine the fate of myogenic precursors, myoblast proliferation, and muscle cell differentiation [1, 2]. Myostatin, a member of the TGF-β superfamily, is a well-known endogenous cytokine that modulates muscle growth [3]. Deletion of myostatin in mice induces a dramatic and widespread increase in skeletal muscle mass due to both muscle hypertrophy and hyperplasia [4]. The double-muscling phenotype observed in some cattle and sheep breeds is caused by mutations in the myostatin gene [5, 6]. Therefore, a central function of myostatin is inhibition of myogenesis. However, few genes that respond to myostatin signaling in order to control myogenesis have been characterized, especially in avian species.
In our previous work, we found that cardiac ankyrin repeat protein (CARP) is a target gene of myostatin that is downregulated in CFMs [7]. CARP was initially identified in a screen for cytokine-inducible factors in human microvascular endothelial cells [8]. CARP is specifically expressed in mammalian cardiac muscle and plays important roles in transcriptional regulation [9], the cytoskeletal architecture, and stretch sensing [10] during heart development and pathological processes [11-14]. Although several reports have identified CARP-induced expression in skeletal muscle in various human muscle diseases [15], the physiological role of CARP in skeletal muscles is poorly understood. In contrast to its mammalian homologs, we found that the expression of chicken CARP is restricted to skeletal muscle [7]. These results suggest that CARP might play a role in the growth of chicken skeletal muscle, but not in the heart. However, the potential role that CARP plays in chicken myogenesis remains unknown.
In this study, the expression pattern of chicken CARP was evaluated in detail to gain insight into the function of this protein. Our data show that CARP is expressed at high levels in postnatal skeletal muscle from chickens and in differentiated CFM cells. When CFM cells were in the proliferation stage, forced expression of CARP enhanced proliferation by promoting the G1-S phase transition of the cell cycle. On the other hand, depletion of CARP via RNAi had the opposite effect on cell growth and induced cell cycle arrest. During differentiation of myoblasts, knockdown of CARP expression resulted in extensive cell death and impaired myotube formation as well as downregulating the expression of MHC, MyoD, and cavolin-3.
Based on these observations, we concluded that CARP, as a downstream target of myostatin, governs chicken skeletal myogenesis through promoting proliferation, and acts as a survival protein to avoid apoptosis during CFM cell differentiation.
Materials and Methods
Tissue collection
All experimental procedures were conducted in conformity with institutional guidelines for the care and use of laboratory animals, and protocols were approved by the Institutional Animal Care and Use Committee in Guangdong Medical College. Male adult White Leghorn chickens were sacrificed via cervical dislocation. Various tissues including pectoralis muscle, heart, liver, lung, kidney, intestine, stomach, spleen, and brain, were collected separately and stored at -70°C for RNA and protein isolation. Additionally, pectoralis muscles were obtained from different developmental stages (on embryonic days 10, 12, 14, 16, and 18 as well as 1 day and in 1, 2, 3, 4, 5, 6, 7, and 10 weeks after birth) and stored at -70°C until use.
Cell culture and treatment
CFMs were prepared from the pectoralis muscles of 10-day-old White Leghorn chicken embryos and cultured as described previously [7]. They were maintained under 5% CO2 at 37°C in growth medium (GM) composed of Dulbecco's modified Eagle's medium (DMEM; Hyclone) supplemented with 20% fetal bovine serum (FBS; Hyclone). At 12-18 h post-plating, CFMs were treated with purified recombinant myostatin at a final concentration of 25, 50, 100, or 200 ng/ml for another 24 h or with the same volume of PBS as a control.
Differentiation of myoblasts into myotubes was induced after CFMs had achieved 70% confluence by switching the medium from GM to DM, which consisted of DMEM supplemented with 2% horse serum (Hyclone). To perform myogenic differentiation experiments, CFMs were cultured in DM for 2 days, and the cells were collected for RNA and protein extraction.
Sequence analysis of the chicken CARP gene
The CARP cDNA sequence (NM_204405) was compared with the chicken genomic database (http://genome.ucsc.edu) through BLAST analysis. Based on these data, we determined exon/intron sizes and precise boundaries. The 5'-flanking region was analyzed using online software (http://cbrc.jp/research/db/TFSEARCH.html) to predict the putative regulatory elements. miRNAs targeting the CARP 3'-UTR were predicted using miRBase (http://miRBase.org), with an E-value cut-off of 6 for the predictions. AREsite (http://rna.tbi.univie.ac.at/cgi-bin/AREsite.cgi) was employed to identify AU -rich elements (AREs) in the CARP mRNA 3'-UTR sequence.
Plasmid construction and small interfering RNA (siRNA)
To generate chicken CARP fusion protein construct with a Myc tag at the C-terminus of CARP, the pcDNA4.0-Myc vector (Invitrogen) was used. The coding sequence of CARP was obtained by PCR amplification using chicken skeletal muscle cDNA as a template. The primers used in this study list in Table 1. The forward primer included an EcoRI site, and the reverse primer included an XhoI site. The obtained PCR product was subsequently digested and cloned into the pcDNA4.0-Myc vector to express CARP. The construct was confirmed through automated sequencing.
The siRNA sequence targeting CARP was designed using Invitrogen's BLOCKiT RNAi designer (http://rnaidesigner.invitrogen.com/sirna). The primer sequence targeting CARP corresponding to the coding region from 834 to 854 bp was 5'-AACCAUUAAGAAUGUCGAUGG-3'. The siRNA and scramble RNA were purchased from Invitrogen in the desalted, pre-annealed duplex form.
RNA extraction and real-time PCR
Total RNA was isolated from the CFMs using the TRIzol Reagent (Invitrogen), digested with DNase I and then reverse-transcribed at 42°C for 1 h in a 10 μl reaction mixture with SuperScript II transcriptase (Invitrogen). For quantitative comparison of mRNA levels, real-time PCR was performed according to the instructions of the one-step SYBR primeScrip RT-PCR II kit (TAKARA), with β-actin as an endogenous control. The obtained first-strand cDNA (100 ng) was used as template for PCR amplification of the chicken CARP MyoD, p21, p27, cyclinD1, Caveolin-3, MHC and β-actin genes with the primers listed in Table 1. Amplification was performed in an ABI7300 real-time PCR thermocycler (Applied Biosystems).
Expression, purification and production of a monoclonal antibody against chicken CARP
An N-terminally truncated CARP (Met1-Pro110) construct was PCR cloned using the primers listed in Table 1. The obtained PCR product was digested and ligated into the pET28b plasmid (Novagen) and introduced into E. coli strain BL21 (DE3) after sequence verification. The recombinant 6His-tagged, truncated chicken CARP protein was expressed and purified through Ni2+ metal-chelating chromatography (Ni-NTA, Qiagen), according to the manufacturer's instructions. To produce a monoclonal antibody against CARP, 6-week-old female BALB⁄C mice were immunized with the purified recombinant protein as described previously [16]. The culture supernatants or ascites from hybridomas were used for Western blot and immunofluorescence analysis.
Western blot analysis
The CFM cells and chicken tissues were lysed in lysis buffer (50 mM Tris⁄HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P40, 50 mM NaF, 1mM Na3VO4, 5mM β-glycerophosphate, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). Equal amounts of total protein were separated via 12% SDS-PAGE, transferred to a PVDF membrane, and probed with anti-chicken CARP, anti-MHC (MH-20, Developmental Studies Hybridoma Bank), anti-CyclinD1 (Cloud-clone Corp), anti-p21 (GeneTex), anti-p27 (Novus Biologicals), anti-Cavolin-3 (Abcam), anti-MyoD (LSBio) anti-myc (CellBiolabs) or an anti-actin antibody (Santa Cruz). The detected proteins were visualized with the ECL detection system (Amersham Biosciences).
Oligonucleotides used in this study.
| Purpose | Primer sequence (5'→3') | Restriction site |
|---|---|---|
| pcDNA4.0-myc-CARP | Fwd (CGGAATTC ATGATGACGATGAAAGTAGAAG) | EcoR I |
| Rev (CCGCTCGAGTCAAAACTTGTTTAGATGGGCG) | Xho I | |
| pET-28b-CARP | Fwd (CGCGGATCCGATGATGACGATGAAAGTAGA) | BamH I |
| Rev (CCGCTCGAGGGTTCCTTTAAAAGGGGAGC) | Xho I | |
| CARP real time PCR | Fwd (ATGATGACGATGAAAGTAGAAG ) | |
| Rev (TCAAAACTTGTTTAGATGGGCG ) | ||
| myoD real time PCR | Rev (ATGGACTTACTGGGCCCCATG ) | |
| Rev (CTGGAACAAGTCTTGAAGTCC) | ||
| caveolin-3 real time PCR | Fwd (ATGGCTGAGGAGCAGAGAGAGCTGGAG) | |
| Rev (GTGGTTGTGGTGCTGCTGGGATTTAGG) | ||
| MHC real time PCR | Fwd (AGCCCTGGAACAGACAGAGC) | |
| Rev (TGATCAAGCTGGTGTTCTGGG) | ||
| cyclin D1 real time PCR | Fwd (CTCACTAAAATGCCTCTGGC) | |
| Rev (CCTTCTCAGATGCCCTTTCT) | ||
| p21 real time PCR | Fwd (AGCGTGCAGGAACCTCTTC) | |
| Rev (GGGCTTATCGTGGACAAC) | ||
| p27 real time PCR | Fwd (TGTCAAACGTCCGCATTTC) | |
| Rev (TTTCGGCCTACACAGTGAG) | ||
| β-actin real time PCR | Fwd (CACCAACTGGGACGACAT) | |
| Rev (ATACTCCTGCTTGCTGATC) |
The restriction enzymes recognition sites are underlined. Fwd=forward primer, Rev=reverse primer.
Indirect immunofluorescence assays
CFM cells were fixed with 4% (w/v) paraformaldehyde in PBS. The cells were then washed three times with permeabilization buffer (0.3% Triton X-100 in PBS) for 10 min and blocked with 3% (w/v) BSA (Calbiochem) in PBS. The samples were incubated for 2 h at room temperature with an anti-CARP or anti-MHC antibody and for 1 h with a FITC-conjugated secondary antibody against mouse IgG (Santa Cruz). Finally, the cells were washed with PBS and mounted on glass microscope slides. 4', 6-diamidino-2-phenylindole (DAPI) staining was performed simultaneously to localize nuclei. Images were captured by fluorescence microscopy performed on a Leica DM5000B microscope (Wetzlar).
Cell proliferation assay
CFMs cultured in a six-well plate (2 x 105 per well) were transiently transfected with 10 μg of plasmid DNA or siRNA using LipofectamineTM 2000 (Invitrogen) and maintained in DMEM for 24, 36, or 48 h. The rate of 3H-thymidine incorporation into DNA was analyzed as previously described [17] and used as a measure of DNA synthesis. During the last 6 h, 5 μCi of 3H-thymidine was added to the culture medium. The cells were then washed two times with PBS and trypsinized to measure radioactivity in scintillation vials. In parallel, the number of viable cells was manually counted in a hemocytometer. The assay was performed in triplicate and repeated three times.
Flow cytometric analysis of the cell cycle distribution
Adherent cells were washed and harvested. Cellular pellets were resuspended in 100 ml of PBS, fixed via dropwise addition of 1 ml of 70% ethanol, and incubated overnight at 4°C. The fixed cells were then centrifuged, washed twice with cold PBS, resuspended in 200 ml of ribonuclease A (250 mg/ml) and incubated for 30 min at 37°C. The cells were next centrifuged again and resuspended in 200 ml of 50 mg/ml propidium iodide (PI). Finally, the cells were analyzed through flow cytometry to determine their DNA contents in a FACSCalibur flow cytometer (BD Biosciences). The obtained results are presented as histograms, showing the cell distribution in the different stages of the cell cycle due to fluorescent PI staining. The data were analyzed using the program WinMDI (version 2.8).
Myoblast fusion index
The myoblast fusion index was calculated as the ratio of the number of DAPI-positive nuclei located in MHC-positive myotubes (i.e., fused myoblasts) to the total number of nuclei in the same field. The fusion index was defined as the number of nuclei within MHC-stained myotubes containing more than two nuclei. This index was used as a read-out of myogenic differentiation and was determined from 10 non-overlapping areas on each coverslip.
Detection of cell death
CFM cells were collected following transfection with CARP, siRNA targeting CARP, or empty vector for 12, 24, 36, 48, or 60 hours in DM. Flow cytometry analysis was performed using the FACScan system (BD Biosciences) to count the number of cells, as described previously. A 630-nm long-band pass filter was used to measure the red fluorescence generated from PI-bound DNA. The data were analyzed as a single-parameter frequency histogram in an SFIT model.
Detection of apoptotic DNA fragmentation
Equal numbers of cells were cultured as described for the detection of cell death. Genomic DNA was extracted from both attached and detached cells after harvesting. The cells were washed twice with PBS, lysed in lysis buffer (50 mM Tris-HCl, pH 8.0, 20 mM EDTA and 1% NP-40), and incubated on ice for 10 min; and then centrifuged at 13,000 rpm for 5 min. The supernatant was digested with 0.3 mg/ml RNase A at 50°C for 2h then digested with proteinase K (0.5 mg/ml) and 1% SDS at 55°C for 2h. Each extract (20 μl) was subsequently subjected to electrophoresis in a 1.2% agarose gel in the presence of ethidium bromide.
Statistical analysis
The data were expressed as the mean ± SD and analyzed via one-way ANOVA (SPSS 13.0). Differences between groups were considered significant at P < 0.05.
Results
CARP is inhibited by myostatin in a dose-dependent manner in CFM cells
Previously, through screening for genes showing altered expression in CFMs in response to myostatin via the differential display PCR method, we determined that the chicken CARP gene was strongly inhibited in CFMs upon treatment with recombinant myostatin [7].
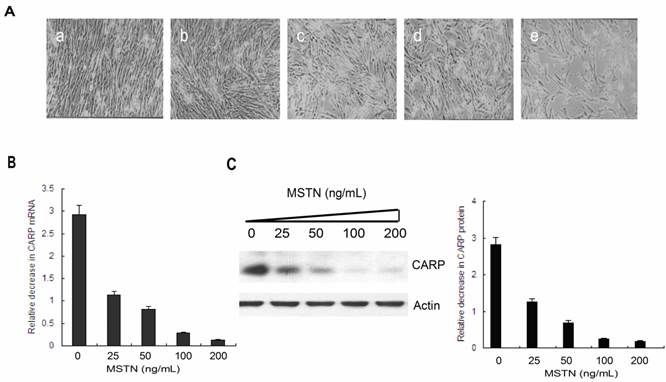
To confirm the inhibition of CARP by myostatin, we isolated mRNA and protein from CFMs treated with different concentrations of myostatin (Figure 1A) and performed real-time PCR (Figure 1B) and Western blotting analyses (Figure 1C). The results indicated that myostatin inhibited CARP expression in a dose-dependent manner at both the mRNA and protein levels.
Myostatin inhibits CARP expression in a dose-dependent manner in CFMs. A. Morphology of CFMs following treatment with 25 (b), 50 (c), 100 (d), or 200 ng/ml (e) of recombinant myostatin or PBS (a) for 24 h. B. Total RNA was prepared from myostatin- or PBS-stimulated CFMs. Real-time quantitative PCR analysis of the relative expression of CARP. C. A total of 20 μg of the cell lysate from each sample was separated via 12% SDS-PAGE and then transferred to a PVDF membrane for Western blot analysis with the monoclonal antibody against chicken CARP. The membrane was stripped and reprobed with an anti-actin antibody as an equal-loading control. The same experiment was performed three times for statistical analysis. Error bars represent standard deviation.
Genomic organization and sequence analysis of the chicken CARP gene
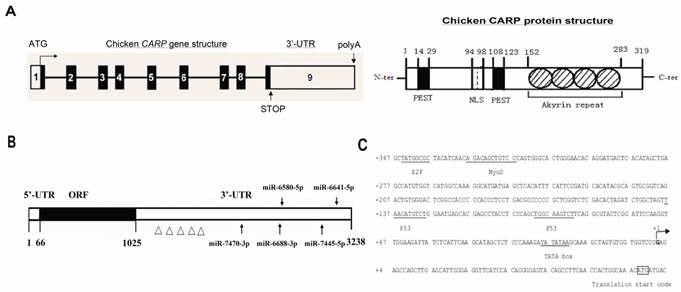
To determine the genomic structure of the chicken CARP gene, its cDNA sequence (NM_204405) was compared with sequences in the chicken genome database (http://genome.ucsc.edu). The results of BLAST analysis indicated that the CARP gene mapped to chromosome 6 and its cDNA sequence was collinear with the genomic sequence published in the NCBI database. Based on these data, we determined the precise nucleotide sequences of the CARP exon/intron boundaries. All exon/intron junctions were consistent with the AG/GT rule. The CARP gene contained nine exons ranging in size from 93 to 2,296 bp (Table 2 and Figure 2A). Exon 1 contained an ATG translation initiation codon and the 5' untranslated region (5'-UTR). Exons 5 to 8 encodes for the four ankyrin-like domains. Exon 9 contains a stop codon and the 3' untranslated region (3'-UTR). Like its human counterpart, chicken CARP protein have four conserved ankyrin repeats, one nuclear localization signal (NLS), and two PEST sequences (Figure 2A).
One notable feature of chicken CARP mRNA is its long 3'-UTR (more than 2.2 kb), which contains many AREs (Figure 2B) related to mRNA stability [18] and may be a target of several regulatory miRNAs. We used miRBase (www.mirbase.org) to predict miRNAs that might target the CARP 3'-UTR. Five miRNAs were found to be excellent candidates for targeting CARP mRNA: gga-miR-7470, gga-miR-6688-3p, gga-miR-6580-5p, gga-miR-7445-5p, and gga-miR-6641-5p. The positions of their predicted target sites in the CARP 3'-UTR are shown in Figure 2B.
The 5'-flanking region of the CARP gene was analyzed with online software, which identified a TATA box and multiple consensus sites for transcription factors. The TATA box was located 29 bp upstream of the transcriptional start site (position -29). Several putative regulatory elements were identified, including binding sites for E2F, p53, and myoD (Figure 2C). Notably, we identified a myoD consensus sequence, which is widely recognized as a critical regulatory component in muscle-specific gene expression.
Spatio-temporal expression patterns of the CARP gene
To gain insight into the function of CARP, we examined its expression in adult chicken tissues via Western blotting analysis using a monoclonal antibody against the CARP protein. These experiments revealed that CARP was expressed specifically in chicken skeletal muscle (Figure 3A). No signal was observed in other tissues, including the brain, spleen, liver, lung, kidneys, intestine, stomach, and heart.
Organization of the chicken CARP gene, mRNA, promoter and protein. A. Genomic and protein organization of the chicken CARP gene. Numbered boxes denote each exon. White boxes indicate the 5'- and 3'-UTRs. Black boxes indicate coding regions. The conserved domains of CARP protein are: NLS, nuclear localization signal; PEST, PEST sequence; Ankyrin repeat, ankyrin repeat domain. B. Five miRNA target sites and several AREs are predicted within the CARP 3'-UTR. miRBase was used to predict five miRNAs that target the CARP 3'-UTR (gga-miR-7470-3p, gga-miR-6688-3p, gga-miR-6580-5p, gga-miR-7445-5p, and gga-miR-6641-5p). The Δ symbols stand for ARE motifs. C. Sequence analysis of the genomic region (from -347 to +73) of the CARP promoter. The putative binding sites for transcription factors are underlined. The transcriptional start site is indicated with an arrow.
Temporal and spatial expression patterns of chicken CARP. A. Spatial expression pattern of CARP. Male adult White Leghorn chickens were maintained on a standard diet. Total proteins were isolated from cardiac muscle (H), brain (B), spleen (Sp), liver (L), skeletal muscle (M), Lung (Lu), kidney (K), intestine (I), and stomach (St) and analyzed via Western blotting. B. Western blot analysis of CARP expression during skeletal muscle development. The pectoralis muscles of White Leghorn chickens were obtained from different developmental stages (10, 12, 14, 16, and 18 embryonic days as well as from chicks 1 day and 1, 2, 3, 5, 7 and 10 weeks posthatch). Actin was used as an equal loading control. C. The expression of chicken CARP increases during in vitro myogenesis. Left: typical morphology of chicken fetal myoblasts (MB) and myotubes (MT). Right: Western blot analysis of CARP expression of CARP during proliferation (0 to 48 h) and differentiation (24 to 72 h). CFMs were isolated and cultured in GM for 24 and 48h, or in DM 24, 48 and 72h to induce differentiation. Immunoblots for myosin heavy chain (MHC) and actin were used as a myogenic differentiation marker and an equal loading control, respectively. h = hour, P = proliferation and D = differentiation. D. Subcellular localization of CARP in chicken fetal myoblasts (24h in GD) and myotubes (24h in MD). CFM cells were fixed with paraformaldehyde and immunostained with the anti-CARP monoclonal antibody (green). Nuclei were visualized by DAPI (blue).

Genomic organization of the chicken CARP gene and donor/acceptor junction.
| Exon number | Exon length (bp) | 5'-Splice donor | Intron length (bp) | 3'-Splice acceptor |
|---|---|---|---|---|
| 1 | 93 | GAGCTG/gtaggt | 1,450 | ttacag/GTGACC |
| 2 | 180 | GGCAGA/gtgacc | 1,097 | ttggag/GACCTT |
| 3 | 138 | GTTATT/gtaagt | 88 | caatag/ACAGGG |
| 4 | 108 | GATGAG/gtatgc | 774 | tgacag/TACAAG |
| 5 | 99 | GATATG/gtatgt | 766 | ccatag/CTTGAA |
| 6 | 99 | GACAAG/gtatgt | 352 | actcag/CTACGC |
| 7 | 99 | ACAGAG/gtaagt | 133 | ccccag/GAAGGT |
| 8 | 99 | AATGCC/gtaagt | 920 | aatcag/GATGGG |
| 9 | 2,296 |
The pattern of CARP protein expression in skeletal muscle tissue during development was also analyzed. The pectoralis muscles were collected from chickens at different developmental stages. Western blot analysis indicated that CARP protein was highly expressed in postnatal skeletal muscle and was nearly undetectable in E10 to E18 embryos (Figure 3B). We cultured CFMs and confirmed their differentiation based on morphological examination (Figure 3C). Total proteins extracted from proliferating myoblasts and differentiated myotubes was analyzed to detect CARP expression by Western blotting. CARP protein levels were increased in myocytes during in vitro differentiation (Figure 3C). Hence, the expression of chicken CARP is high in differentiated myotubes and increases with myogenesis, as was observed during both in vivo and in vitro myogenesis.
To determine the subcellular distribution pattern of the CARP protein, immunofluorescent staining experiments with a monoclonal antibody against chicken CARP were performed in both proliferating and differentiating CFM cells. The results have revealed dynamic subcellular localization of CARP in CFMs: early proliferating myoblasts displayed some detectable CARP antigen in the nucleus, while differentiating myoblasts were stained predominantly in the cytoplasm (Figure 3D).
CARP enhances CFM cell proliferation
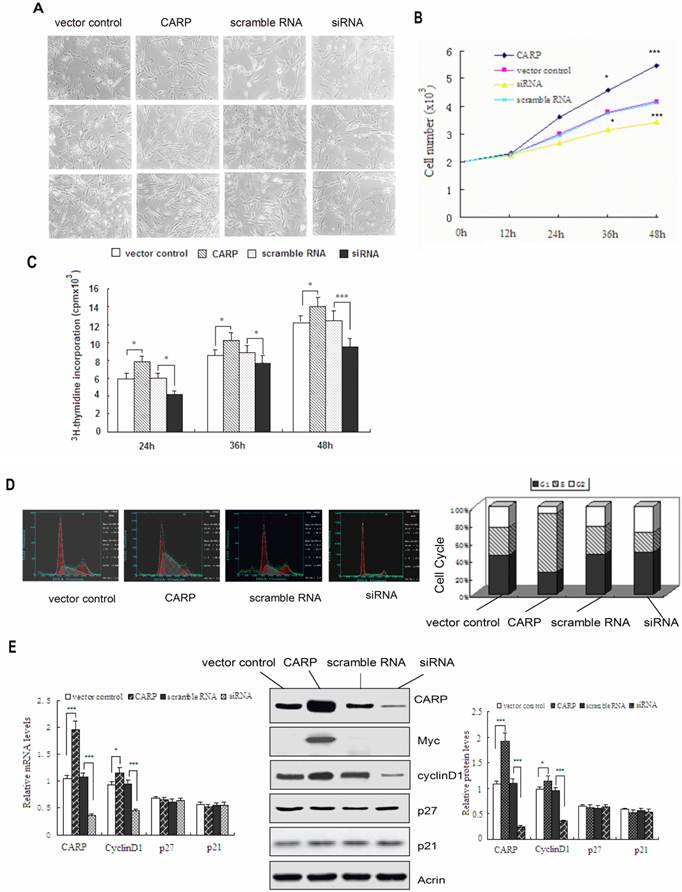
To address the question of whether CARP plays a role in myogenesis, we transfected CFM cells with an expression vector containing either the cDNA sequence or a siRNA for CARP gene. First, we confirmed the overexpression or knockdown of CARP in CFM cells. As shown in Figure 4E, CFM cells transfected with pcDNA4.0 myc-CARP expressed an approximately 1.8-fold amount of CARP protein than control cells (P < 0.001). Conversely, cells transfected with an siRNA targeting CARP showed decreases in endogenous CARP protein expression by approximately 75% compared to control cells at 48 h post-transfection (P < 0.001). We also confirmed CARP overexpression in cells transfected with CARP by Western blotting using anti-myc antibody (Figure 4E). We then examined the potential function of CARP during myoblast proliferation in CFMs. Cells were counted at 12, 24, 36, and 48 h after transfection and culturing in growth medium. Ectopic expression of CARP markedly increased the number of cells present in comparison to control cells. The maximal effect was obtained 48 h after transfection, reaching a 1.3-fold increase above the control. Knockdown of CARP expression inhibited CFM cell growth (Figure 4A and 4B). To further characterize the effects of CARP on the proliferation of CFM cells, DNA replication was measured via 3H-thymidine incorporation. Control cells showed increase in cell number and 3H-thymidine incorporation according to their basal growth rate. CFMs transfected with CARP cDNA exhibited a significant increase in the cell proliferation rate, as measured by 3H-thymidine incorporation. These results reinforce the hypothesis that CARP enhances cell proliferation (Figure 4C).
To address the molecular mechanism underlying the impact of CARP on cell proliferation, we next studied the distribution of cells in different phases of the cell cycle via FACS analysis. The proportion of cells in S-phase increased significantly, from 34.9% to 65.8% 24h after CFM cells were transfected with the plasmid expressing CARP, whereas this proportion decreased to 23.6% after CARP expression was knocked down in CFM cells via transfection with an siRNA targeting CARP (Figure 4D). These results indicate that CARP promotes CFM cell proliferation by modulating the transition of the cell cycle from G1 to S phase.
Since CARP plays a positive role in CFM cell proliferation by influencing the cell cycle, we assessed whether genes regulating the cell cycle, especially the G1-S transition, were concomitantly altered. As shown in Figure 4E, we found that the expression of cyclinD1 was downregulated when CARP expression was inhibited and it was upregulated upon CARP overexpression, consistent with the inhibition of G1 to S phase progression. There were no differences in the other cell cycle-related genes analyzed (Figure 4E).
CARP promotes proliferation of CFM cells. A. The morphology of CFMs was examined following transfection with CARP-myc construct, CARP siRNA or empty vector and scramble RNA (as controls) and growth for 24, 36, or 48 h. Each transfection was independently repeated three times. B. The number of CFMs cells, transfected with empty vector, CARP, scramble RNA or the CARP siRNA was counted at indicated time points. C. 3H-thymidine incorporation was measured as indicated in the Materials and Methods. D. CARP increases the percentage of CFM cells in S phase. DNA content frequency histograms CFM cells transfected with CARP, scramble RNA, empty vector and siRNA transfectants. Twenty-four hours after transfection, the myoblasts were stained with PI and analyzed by flow cytometry. A total of 1.0×106 cells were analyzed for each group and were distributed graphically into the G1, S, and G2/M phases of the cell cycle (left). The corresponding percentages for each phase are reported on the right. E. The effects of CARP on cell cycle-related genes. Twenty-four hours after transfection with the indicated constructs, total RNA and protein extracts were prepared, and the expression levels of cyclinD1, p21, p27, CARP were measured by Real-time PCR (left) and Western blotting (middle and right), respectively. Actin was used as an internal control. Each value represents the mean ±SD, *P<0.05 and *** P<0.001 compared to the control.
Knockdown of CARP expression interferes with myogenic differentiation and causes death of CFM cells
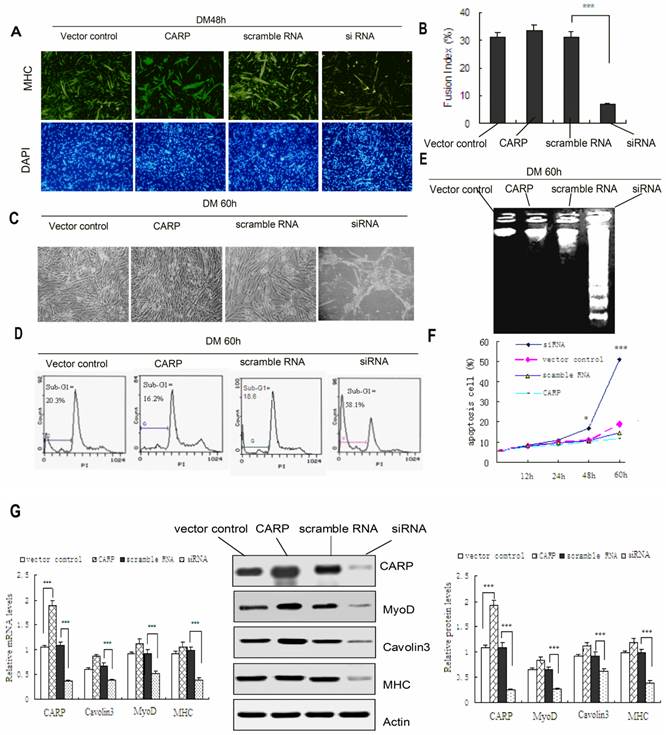
Since CARP expression increased with the differentiation of CFM cells and skeletal muscle tissue, we speculated that CARP might play a role in myogenic differentiation. When confluent cell cultures were incubated in DM, CFM control cells fused to become extremely elongated multinucleated myotubes. Since the morphological conversion of myoblasts into myotubes is accompanied by the accumulation of muscle-specific proteins we used an antibody for muscle-specific MHC to estimate the relative myogenic differentiation potential in the CARP-overexpressing and silencing cells. Fig. 5A shows the transfected cells immunostained with the MHC antibody at 48h after myogenic induction. The bars in Figure 5B represent the ratio of the number of DAPI-positive nuclei located in MHC-positive myotubes to the total number of nuclei in the same field and counted in 10 microscopic fields randomly chosen for each CFM cell culture. As can be seen, CFM cells in which CARP expression was knocked down exhibited impaired myotube formation, expressed skeletal MHC at a very low frequency and level, and generally exhibited little detectable CARP protein (Figures 5A and 5G). In addition to MHC, the levels of some myogenic differentiation-related proteins, including myoD and cavolin-3, were downregulated, suggesting that CARP may affect the expression level or the assembly of myofibril components in CFM cells. These data were confirmed by real-time PCR experiments (Figure 5G).
CFM cells underwent morphological changes typical for apoptosis following CARP inhibition (Figure 5C). The apoptotic population was quantified via flow cytometry. PI staining of the sub-G1 cell fraction showed that 58.1% of transfected cells were apoptotic (P<0.001) at 60 h after transfection, compared to 20.3% of the control cells (Figure 5D). Flow cytometry analysis revealed that the apoptotic sub-G1 fraction of cells also increased with the time following transfection (Figure 5F). We also isolated low-molecular weight DNA and performed an apoptosis DNA laddering assay. As shown in Figure 5E DNA fragmentation was observed only in CFMs transfected with siRNA for 60h. The accumulation of CARP protein in CFM cells transfected with CARP siRNA declined to less than 80% of that in the controls (Figure 5G). These results suggest that CARP could be required for cell survival in CFM Cells. On the other hand, the cell morphology, biochemical and survival data obtained from cells overexpressing CARP in differentiating CFM cells are similar to those from control CFM cells (transfected with vector or scramble RNA) (Figure 5).
In conclusion, we have found that overexpression of chicken CARP in CFM cells enhances skeletal muscle cells proliferation, whereas knockdown of CARP expression blocks myoblast differentiation and induces cellular apoptosis. These results strongly suggest that CARP is an essential positive regulator of chicken skeletal muscle development.
Discussion
Myostatin is a specific negative regulator of skeletal muscle growth and is regarded as one of the most important cytokines affecting myogenesis [3]. Screening for and identification of genes that are regulated by myostatin is important for understanding the molecular mechanisms of the myostatin signaling pathway during muscle development. In this study, we demonstrated that myostatin inhibited CARP expression in a dose-dependent manner. In addition, CARP mRNA and protein levels were synchronously downregulated by myostatin, indicating an effect on mRNA turnover, rather than on protein translation or degradation.
The molecular mechanisms associated with specific CARP expression in chicken skeletal muscle and the inhibition of CARP by myostatin in CFM cells still remain unclear. Our results showed that the CARP promoter region contains potential binding sequences for the muscle-specific transcription factor myoD, which suggests that CARP expression in skeletal muscle cells may be governed by this tissue-specific regulator, consistent with the increased CARP expression observed in differentiating CFM cells. Moreover, it was already demonstrated that MyoD was able to activate human CARP promoter [19].
The 3'-UTR of the chicken CARP gene is longer than the homologous sequences found in humans (GenBank NM_014391.2) and mice (GenBank NM_013468), which are 1,427 bp and 1,417 bp in length, respectively. We also predicted that several AREs related to mRNA stability and at least five candidate miRNA target sites exist within the long 3'-UTR (2.2 kb) of CARP. In addition, it has been reported that TGF-β1 [20] and the β-adrenoceptor agonist isoprenaline [21] upregulate human CARP, partly by increasing its mRNA stability. Considering these findings, we propose that the effects of myostatin inhibition on CARP mRNA levels may be mediated by more than one mechanism, and the potential for posttranscriptional control requires additional attention.
In addition to chickens, we detected CARP expression in the skeletal muscle of turkeys, geese, ducks, and other poultry species (data not shown). However, there are no available reports documenting any function for CARP during avian skeletal muscle growth. We speculate that there may be obvious differences in CARP function in skeletal myogenesis between mammalian and avian species. This hypothesis is based on the following observations. First, in contrast to mammals, CARP is specifically expressed in skeletal muscle in avian species, which means that CARP functions in chicken skeletal muscle rather than in the heart. Second, in mammals, CARP, together with Ankrd2 [22] and diabetes-related ankyrin repeat protein (DARP) [23], is part of a family of transcription regulators known as muscle ankyrin repeat proteins (MARPs). In addition, no obvious phenotype was observed in the skeletal muscle of single or triple MARPs knockout mice [24], suggesting the possible existence of yet unknown mechanisms able to compensate for MARPs physiological functions. In contrast, CARP is the only known member of the MARP family in avian species. Finally, alignment of the CARP amino acid sequence revealed that the chicken CARP protein shows only moderate similarity (approximately 70%) to its mammalian homologs [25], suggesting that the function of this protein may also have diverged over evolutionary time.
Downregulation of CARP through siRNA treatment inhibits the myogenic differentiation and induces apoptosis of CFM cells. A. Differentiated myocytes were detected by immunostaining with an MHC antibody (green). Nuclei were visualized using DAPI (blue). Incubation of control and CARP CFM cultures in differentiation medium (DM) resulted in a rapid increase in MHC synthesis and the formation of elongated, multinucleated myotubes. In contrast, CFM cells in which CARP expression was inhibited impaired to form multinucleated elongated myotubes. B. Calculation of the fusion index (the percentage of cells containing two or more nuclei among the MHC-positive myocytes) confirmed that CARP silencing in myoblasts resulted in a deficient fusion capacity. Each value represents the mean ±SD, and all experiments were performed at least three times. *** P<0.001 compared to the control. C. Inhibition of CARP induces apoptosis of CFM. Representative images of CFM cells 60 h after transfection with the CARP-myc, CARP siRNA, scramble RNA or with an empty vector and culturing in DM medium are shown. D. Quantification of the apoptotic fractions was performed using the FACScan system, as described in the Materials and Methods. E. DNA laddering assays. DNA was extracted from cells differentiated for 60 h and analyzed by 1.2% agarose gel electrophoresis in the presence of ethidium bromide. Low-molecular weight DNA was extracted from CFMs transfected with siRNA for CARP. No regular laddering pattern was observed in the Scramble RNA, vector control or CARP transfected myoblasts. F. Knockdown of CARP expression induced cell death in CFMs cells. The percentage of sub-G1 apoptotic cells was determined by FACS analysis using PI staining. CFM cells were transfected with the indicated expression plasmids, cultured in differentiation medium and collected at the indicated time points. G. Immunoblot and Real-time PCR analysis of CFM cells transfected with the indicated plasmids and cultured in differentiation medium for 48 hours. Total RNA and proteins were extracted and the expression levels of MHC, MyoD, Caveolin-3 and CARP were detected by real-time PCR (left) and Western blotting (middle and right). Actin was used as an internal control. Each value represents the mean ±SD, and all experiments were performed at least three times. *P<0.05, ** P<0.01 and *** P<0.001 compared to the control.
Taking into account the fact that myostatin acts as a negative regulator of muscle growth and inhibits CARP expression; we postulated that CARP might exert positive effects on skeletal muscle development. To explore the function of CARP in the growth of CFM cells, we performed both overexpression and knockdown experiments. Consistent with our hypothesis, our data showed that induced expression of CARP enhances CFM cell proliferation in GM by promoting the G1-S transition, while blocking CARP function delays CFM cell growth. In our investigation of the underlying mechanisms through which CARP may influence the cell cycle, we found that the expression of the cyclin D1 gene was consistent with the CARP levels observed in CFM cells. Precisely controlled synthesis and proteolysis of cyclins are essential for cell cycle progression [26]. Inhibition of cyclin D1 expression lengthens the G1 phase and suppresses cell proliferation [27, 28].
We also found that CARP expression is induced in differentiating CFM cells, suggesting that its function may be associated with differentiation. This hypothesis is supported by our observation that CFM cells transfected with CARP and control cells both undergo myogenic differentiation, forming multinucleate myotubes in DM. Under the same culture conditions, myoblasts transfected with CARP siRNA show extensive cell death and impaired myotube formation. Consistent with this observation, skeletal muscle differentiation markers, such as MHC, myoD, and caveolin-3, were inhibited in CFM cells transfected with CARP siRNA.
Myoblast differentiation involves a hierarchical series of events in which the myoblasts irreversibly exit the cell cycle, acquire an apoptosis-resistant phenotype, show upregulated expression of muscle-specific proteins, and finally form functional contractile myotubes. Differentiating muscle cells exhibit higher rates of apoptosis, and survival factors enhance muscle differentiation [29]. In this study, knockdown of CARP expression sensitized CFM cells to apoptosis. Thus, we propose that CARP could be required for CFM cell survival during differentiation. When CARP levels are low, homeostatic functions such as proliferation and differentiation are inhibited, and CFM cells undergo apoptosis. An anti-apoptotic function of CARP has been also reported in mammals, where the ectopic expression of CARP in H9c2 cells increased their resistance to GADD153-mediated, hypoxia-induced apoptosis [30].
As a multi-functional protein, the roles played by CARP may be affected by its intracellular location. In mammals, CARP acts as a structural protein localized to the I-region of sarcomeres where it binds titin/connectin, myopalladin, and calpain-3 and likely sends information to the nucleus concerning changes in the structure or function of the contractile machinery [10, 31]. When shuttled into the nucleus, CARP acts as a transcriptional cofactor and interacts with YB-1, P53, HAND2, or HEY1 to regulate cardiac muscle gene expression in mammals [9, 19]. Mammalian CARP negatively regulates many cardiac genes, including MLC-2v, ANF, BNP, and cTnC [9, 11, 32, 33]. Our results also demonstrate that the expression of CARP is restricted to the nucleus in proliferating CFM cells and that it shuttles from the nucleus to the sarcomere during myogenic differentiation. We would like to suggest that CARP localization in the nucleus of proliferating CFM cells promotes cell growth, while its expression at sarcomeres in differentiating cells maintains cell structure to avoid apoptosis. Taken together, these findings implicate that CARP coordinates the expression of genes involved in cell structure and proliferation and could play a role in the variation of avian muscle mass.
Skeletal muscle mass increases during the postnatal development of chickens through a process of hypertrophy, i.e., the enlargement of individual muscle fibers caused by the fusion of satellite cells [34]. Our data also demonstrated that the chicken CARP gene was specifically expressed in postnatal skeletal muscle tissue. We found that CARP expression was also induced in differentiated chicken satellite cells (data not shown), suggesting a role of CARP in chicken satellite cell development. Understanding the cellular and molecular mechanisms of CARP-regulated chicken satellite cell differentiation and the maintenance of muscle homeostasis may lead to promising approaches for optimizing the growth and composition of muscle, thus improving the production and quality of chicken meat.
To our knowledge, this is the first demonstration that CARP plays an important role in the coordination of proliferation and apoptosis during CFM cell differentiation. Further examination of the molecular mechanisms underlying CARP activity in skeletal myogenesis will be critical not only for understanding the control of skeletal myogenesis but also for improving yields in poultry production.
Abbreviations
CARP: cardiac ankyrin repeat ptotein; CFM: chicken fetal myoblast; MHC: myosin heavy chain; ARE: AU-rich element; PI: propidium iodide; GM: growth medium; DM: differentiation medium; siRNA: small interfering RNA; DARP: diabetes-related ankyrin repeat protein; MARP: muscle ankyrin repeat proteins; UTR: untranslated region; DAPI: 4', 6-diamidino-2-phenylindole.
Acknowledgements
Support for this work includes funding from the National Nature Science Foundation of China (grant number 31171219, 81271213, 81350026 and 81271214), the Natural Science Foundation of Guangdong Province (No. S2012010008222), Natural Science Foundation of Heilongjiang Province (H201344) and US-China Biomedical Collaborative Research Program (NSFC grant number 81261120404). This work is also supported by the Science and Technology Innovation Fund of Guangdong Medical College (No. STIF 201101) and Doctor Startup Fund of Guangdong Medical College (No. XB1326).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12:349-361
2. Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012 4
3. Lee SJ. Extracellular Regulation of Myostatin: A Molecular Rheostat for Muscle Mass. Immunol Endocr Metab Agents Med Chem. 2010;10:183-194
4. McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83-90
5. Kambadur R, Sharma M, Smith TP. et al. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997;7:910-915
6. Clop A, Marcq F, Takeda H. et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813-818
7. Yang W, Zhang Y, Ma G. et al. Identification of gene expression modifications in myostatin-stimulated myoblasts. Bioch Biophys Res Commun. 2005;326:660-666
8. Chu W, Burns DK, Swerlick RA. et al. Identification and characterization of a novel cytokine-inducible nuclear protein from human endothelial cells. J Biol Chem. 1995;270:10236-10245
9. Zou Y, Evans S, Chen J. et al. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development. 1997;124:793-804
10. Miller MK, Bang ML, Witt CC. et al. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J Mol Biol. 2003;333:951-964
11. Kuo H, Chen J, Ruiz-Lozano P. et al. Control of segmental expression of the cardiac-restricted ankyrin repeat protein gene by distinct regulatory pathways in murine cardiogenesis. Development. 1999;126:4223-4234
12. Zolk O, Frohme M, Maurer A. et al. Cardiac ankyrin repeat protein, a negative regulator of cardiac gene expression, is augmented in human heart failure. Biochem Biophys Res Commun. 2002;293:1377-1382
13. Torrado M, Iglesias R, Nespereira B. et al. Intron retention generates ANKRD1 splice variants that are co-regulated with the main transcript in normal and failing myocardium. Gene. 2009;440:28-41
14. Torrado M, Nespereira B, Bouzamayor Y, Centeno A, Lopez E, Mikhailov AT. Differential atrial versus ventricular ANKRD1 gene expression is oppositely regulated at diastolic heart failure. FEBS Lett. 2006;580:4182-4187
15. Kojic S, Radojkovic D, Faulkner G. Muscle ankyrin repeat proteins: their role in striated muscle function in health and disease. Crit Rev Clin Lab Sci. 2011;48:269-294
16. Yu Y, Wang H, Zhao L. et al. Fine mapping of a foot-and-mouth disease virus epitope recognized by serotype-independent monoclonal antibody 4B2. J Microbiol. 2011;49(1):94-101
17. Zhang Q, Wang K, Zhang Y. et al. The myostatin-induced E3 ubiquitin ligase RNF13 negatively regulates the proliferation of chicken myoblasts. FEBS J. 2010;277:466-476
18. Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465-470
19. Kojic S, Nestorovic A, Rakicevic L. et al. A novel role for cardiac ankyrin repeat protein Ankrd1/CARP as a co-activator of the p53 tumor suppressor protein. Arch Biochem Biophys. 2010;502:60-67
20. Kanai H, Tanaka T, Aihara Y. et al. Transforming growth factor-beta/Smads signaling induces transcription of the cell type-restricted ankyrin repeat protein CARP gene through CAGA motif in vascular smooth muscle cells. Circ Res. 2001;88:30-36
21. Zolk O, Marx M, Jackel E. et al. Beta-adrenergic stimulation induces cardiac ankyrin repeat protein expression: involvement of protein kinase A and calmodulin-dependent kinase. Cardiovasc Res. 2003;59:563-572
22. Tsukamoto Y, Senda T, Nakano T. et al. Arpp, a new homolog of carp, is preferentially expressed in type 1 skeletal muscle fibers and is markedly induced by denervation. Lab Invest. 2002;82:645-655
23. Ikeda K, Emoto N, Matsuo M. et al. Molecular identification and characterization of a novel nuclear protein whose expression is up-regulated in insulin-resistant animals. J Biol Chem. 2003;278:3514-3520
24. Barash IA, Bang ML, Mathew L. et al. Structural and regulatory roles of muscle ankyrin repeat protein family in skeletal muscle. Am J Physiol Cell Physiol. 2007;293:C218-227
25. Ma G, Wang H, Li Y. et al. Cloning, expression, and bioinformatics analysis of the sheep CARP gene. Mol Cell Biochem. 2013;378:29-37
26. Ciemerych MA, Archacka K, Grabowska I. et al. Cell cycle regulation during proliferation and differentiation of mammalian muscle precursor cells. Results Probl Cell Differ. 2011;53:473-527
27. Ji M, Zhang Q, Ye J. et al. Myostatin induces p300 degradation to silence cyclin D1 expression through the PI3K/PTEN/Akt pathway. Cell Signal. 2008;20:1452-1458
28. Yang W, Zhang Y, Li Y. et al. Myostatin induces cyclin D1 degradation to cause cell cycle arrest through a phosphatidylinositol 3-kinase/AKT/GSK-3 beta pathway and is antagonized by insulin-like growth factor 1. J Biol Chem. 2007;282:3799-37808
29. Walsh K. Coordinate regulation of cell cycle and apoptosis during myogenesis. Prog Cell Cycle Res. 1997;3:53-58
30. Han XJ, Chae JK, Lee MJ. et al. Involvement of GADD153 and cardiac ankyrin repeat protein in hypoxia-induced apoptosis of H9c2 cells. J Biol Chem. 2005;280:23122-23129
31. Laure L, Daniele N, Suel L. et al. A new pathway encompassing calpain 3 and its newly identified substrate cardiac ankyrin repeat protein is involved in the regulation of the nuclear factor-kappaB pathway in skeletal muscle. FEBS J. 2010;277:4322-4337
32. Jeyaseelan R, Poizat C, Baker RK. et al. A novel cardiac-restricted target for doxorubicin. CARP, a nuclear modulator of gene expression in cardiac progenitor cells and cardiomyocytes. J Biol Chem. 1997;272:22800-22808
33. Aihara Y, Kurabayashi M, Saito Y. et al. Cardiac ankyrin repeat protein is a novel marker of cardiac hypertrophy: role of M-CAT element within the promoter. Hypertension. 2000;36:48-53
34. Dellavalle A, Maroli G, Covarello D. et al. Pericytes resident in postnatal skeletal muscle differentiate into muscle fibres and generate satellite cells. Nat Commun. 2011;2:499
Author contact
![]() Corresponding author: E-mail: zhaobinetom.com (Bin Zhao); keshenli1971com (Keshen Li). Tel & Fax: +86 759 2386 949.
Corresponding author: E-mail: zhaobinetom.com (Bin Zhao); keshenli1971com (Keshen Li). Tel & Fax: +86 759 2386 949.

 Global reach, higher impact
Global reach, higher impact