Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(7):689-701. doi:10.7150/ijbs.8327 This issue Cite
Research Paper
Amoebozoa Possess Lineage-Specific Globin Gene Repertoires Gained by Individual Horizontal Gene Transfers
1. Institute of Bioinformatics, Faculty of Medicine, University of Muenster, Niels Stensen Str. 14, 48149 Muenster, Germany
2. Institute of Molecular Biology and Biotechnology, A. Mickiewicz University, Poznan, Poland
3. Department of Medical Genomic Sciences, University of Tokyo, Tokyo, Japan
Received 2013-12-10; Accepted 2014-3-24; Published 2014-6-19
Abstract
The Amoebozoa represent a clade of unicellular amoeboid organisms that display a wide variety of lifestyles, including free-living and parasitic species. For example, the social amoeba Dictyostelium discoideum has the ability to aggregate into a multicellular fruiting body upon starvation, while the pathogenic amoeba Entamoeba histolytica is a parasite of humans. Globins are small heme proteins that are present in almost all extant organisms. Although several genomes of amoebozoan species have been sequenced, little is known about the phyletic distribution of globin genes within this phylum. Only two flavohemoglobins (FHbs) of D. discoideum have been reported and characterized previously while the genomes of Entamoeba species are apparently devoid of globin genes. We investigated eleven amoebozoan species for the presence of globin genes by genomic and phylogenetic in silico analyses. Additional FHb genes were identified in the genomes of four social amoebas and the true slime mold Physarum polycephalum. Moreover, a single-domain globin (SDFgb) of Hartmannella vermiformis, as well as two truncated hemoglobins (trHbs) of Acanthamoeba castellanii were identified. Phylogenetic evidence suggests that these globin genes were independently acquired via horizontal gene transfer from some ancestral bacteria. Furthermore, the phylogenetic tree of amoebozoan FHbs indicates that they do not share a common ancestry and that a transfer of FHbs from bacteria to amoeba occurred multiple times.
Keywords: Amoebozoa, globin genes
Introduction
Globins (Gbs) are small heme proteins that have been found in all kingdoms of life in a wide range of different species [1-3]. Gbs are able to bind various gaseous ligands, such as oxygen and nitric oxide, and have diverse functions, e.g. in respiration and nitric oxide detoxification [4]. The globin superfamily can be divided into three lineages, namely the S, F, and T globins, which belong to two structural classes [5-7]. The members of the F and S lineages possess the typical globin fold, i.e. a 3-over-3 (3/3) α-helical fold consisting of seven or eight α-helices, designated A through H [8]. In contrast, the members of the T lineage exhibit a 2/2 structure characterized by a shortened or completely deleted A helix, a missing D helix, and the substitution of the proximal F helix by a polypeptide segment [9]. The F globin family consists of flavohemoglobins (FHbs), FHb-like globins with N- and C-terminal extensions, and related single-domain globins (SDFgbs). The chimeric FHb proteins possess an N-terminal globin domain and a C-terminal FAD- and NAD(P)H-binding reductase domain. Increasing evidence indicates that FHbs protect bacteria and simple eukaryotes, including yeast, against the toxic effects of nitric oxide likely via the nitroxylation of oxygen [10-13]. The S globin family comprises chimeric globin-coupled sensors (GCSs), protoglobins (Pgbs), and related single-domain globins (SDSgbs). The GCSs can be further categorized as either aerotactic or gene regulating [14]. The globins of the T lineage were named truncated hemoglobins (trHbs) because of their shortened primary structure. Based on phylogenetic analyses and structural differences the trHbs can be further divided into three groups, i.e. group I (trHbN), II (trHbO), and III (trHbP) [15, 16]. Several distinct functions for the trHbs have been proposed, including nitric oxide detoxification, oxygen/nitric oxide sensing, ligand/substrate storage, etc. [15, 17].
A model of globin evolution has been suggested in which the three lineages descent from a single common ancestor that likely resembled an extant SDFgb [3, 7]. It is assumed that globins emerged only in bacteria [7]. Later, the eukaryote and archaeal 3/3 and 2/2 globin genes originated from horizontal gene transfers (HGT) of bacterial SDFgb and trHb genes, respectively [5-7]. This is supported by several studies that demonstrate that HGT events shaped the phyletic distribution of globin genes in plants, fungi, and unicellular eukaryotes [3, 5, 6, 16, 18-24]. Finally, it is known that HGT events played a major role in the evolution of various species [25, 26].
Although the evolution of globins has been intensively studied in several species from all kingdoms of life, only little is known about their origins and distribution in unicellular eukaryotes, such as the Amoebozoa [27]. The Amoebozoa are a phylum of amoeboid protozoa that move by the means of pseudopodia. They are closely related to Opisthokonts (Metazoa/fungi group) and can be divided into six monophyletic clades within two main groups, the Lobosea and the Conosea [28, 29]. They adopted different habitats and life styles, such as free-living unicellular amoeba, obligate parasitic amoeba, social amoeba, and true slime molds. The free-living Amoebozoa are common inhabitants of soils and water, where they represent one of the main predators of bacteria [30]. Acanthamoeba castellanii is the most frequently found amoeba in soil and plays an important role in a variety of environments. One of the best-known amoebas is the social amoeba Dictyostelium discoideum that has been studied for decades. D. discoideum is a solitary amoeba that can achieve multicellularity under starvation by aggregation and morphogenesis into a fruiting body [31]. Some amoebas are known to cause infectious diseases in humans, for instance the intestinal parasite Entamoeba histolytica which can induce amebic colitis and amebic liver abscess [32]. Likewise, also some members of the genus Acanthamoeba have been identified as origin of amebic infections in humans [33].
The presented study aims to define the globin gene repertoire of amoebozoan species. Eleven different species of the subphyla Conosa (infraphyla Mycetozoa, Archamoebae) and Lobosa (infraphyla Tubulinea, Acanthopodina) were scanned for the presence of globin genes. We show that the examined Amoebozoa possess lineage-specific globin gene repertoires, composed of either FHbs, SDFgbs, or trHbs that have been likely gained through multiple horizontal gene transfer events from ancestral bacteria.
Methods
Identification of amoebozoan globin genes
Initially, the two previously described FHbs of D. discoideum [34] were used to search the non-redundant protein database of NCBI for homologous amoebozoan globin proteins employing the BLASTp algorithm with default parameters [35]. The retrieved sequences and the globin sequences of two fungi, Schizophyllum commune and Saccharomyces cerevisiae, were used to search the genomes of A. castellanii, D. discoideum, D. fasciculatum, D. purpureum, E. dispar, E. histolytica, E. invadens, E. moshkovskii, P. polycephalum and Polysphondylium pallidum for the presence of additional globin genes. This was done by a tBLASTn search applying varying parameters: e-value 0.1, 0.001, matrix: BLOSUM62, BLOSUM45, soft masking of query sequence. Likewise, a tBLASTn search was conducted against TBestDB [36]. Additionally, the transcriptome of A. castellanii was analyzed for the presence of globin sequences (Buczek et al., unpublished data). Subsequently, the similarity search was repeated with the newly found sequences. The intron/exon structures of the novel genes were manually annotated, guided by tBLASTn results, GENSCAN [37], and NetGene2 predictions [38, 39]. Intron positions with respect to the coding sequences were reported with reference to the secondary structure of the protein. This was done by aligning the amoebozoan globin proteins to myoglobin of the sperm whale (UniProtKB: P02185.2). FUGUE [40] and SMART [41] were used to validate the annotated sequences as globin genes. A table of sequences used in the subsequent analyses is provided in the supplementary materials (Table S1). All genes used in the consecutive analyses were labeled with first two letters of the binary species name and a suffix in cases multiple copies are present in a single genome.
Shared synteny is a reliable criterion to prove orthology between genomic segments and to trace back the evolution of those segments. The neighboring genes of the FHbs from the social amoebas D. discoideum, D. purpureum, D. fasciculatum, and P. pallidum were identified using the genome browser and the tBLASTn tool provided by dictyBase [42, 43]. Orthologous genes were defined as reciprocal best hits (RBHs) via tBLASTn and BLASTp searches. Additionally, the direct neighbors of the trHbs of A. castellanii were determined via GENSCAN and subsequent BLASTp searches. We did not conduct a synteny analysis of the FHb genes of P. polycephalum due to the highly fragmented nature of the current genome assembly.
Function inference analyses
Several bacterial and metazoan globins are known to associate with the cell membrane [44-50]. To determine if the amoebozoan globins are potentially linked to the membrane, posttranslational modifications and transmembrane domains were predicted. The Myristoylator [51] and CSS-Palm 3.0 [52] servers were used to predict myristoylation and palmitoylation sites, respectively. The identification of transmembrane regions was done with TMpred [53] and TMHMM [54].
The secondary and tertiary structures of the amoebozoan globin proteins were modeled to further support inference of their functionality. Modeling was done using Swiss-Model and SwissPDBViewer 4.04 [55, 56], as described in [57]. Percent identity values of query and template sequences and e-values of the PSI-BLAST searches are provided in the supplementary materials (Table S2).
Phylogenetic analysis
In a genomic survey from 2009 among the three kingdoms of life more than 550, 253, and 668 globins of the F, S, and T lineages, respectively, were identified [3]. With ongoing sequence projects these numbers have further increased. Thus, the selection of a representative set of globin sequences for phylogenetic analyses is a challenging task. To receive a set of globin sequences that likely provide an insight into the evolution of amoebozoan globin proteins, the following steps were conducted. First, homologous globin proteins closely related to the amoebozoan globins were identified conducting BLASTp searches against the non-redundant protein database of NCBI. Thereby, for each amoebozoan globin the closest relative in bacteria, plants, fungi, archaea, and in eukaryotes other than plants, fungi, and Amoebozoa, was identified. This search was iterated for each received globin protein, till no new sequences could be added to the data set. Next, the sequence identity and similarity values of the received sequences were analyzed via MatGat [58]. Subsequently, redundant sequences were removed from the data set, e.g. highly similar globins from different subspecies. Based on ubiquitous clustering with high statistical support and short branches, sequences with identity equal or higher than 85 percent were considered as redundant and consequently only one from a given cluster was used for the phylogenetic inference.
Next, the sequences were divided into two data sets, one comprising FHbs (66 sequences), the other one trHbs (76). To identify overrepresented groups, i.e. clades containing several closely related species that do not provide any information on the origins and relationships of amoebozoan globins, a neighbor-joining tree was created employing PHYLIP 3.69 with default settings and 1,000 bootstrap replications [59]. This reduced the FHb and trHb data sets to 38 and 27 sequences, respectively. Multiple sequence alignments were conducted using MUSCLE 3.8.31 [60], MUSCLE 4.0 (preliminary, experimental version), the L-INS-i, G-INS-i and FFT-NS-i strategies of MAFFT [61, 62], and COBALT [63]. The best scoring alignment was chosen based on MUMSA scores [64]. The program packages RAxML 7.0.4 [65, 66] and MrBayes 3.1.2 [67, 68] were used for phylogenetic tree reconstructions. The best-fitting model of amino acid substitution was selected by the analysis of the alignment with ProtTest3 [69]. Phylogenetic analyses of the FHb and trHb data sets were based on the WAG [70] model of amino acid evolution, assuming gamma-distributed rate variation among sites. Maximum likelihood analyses were performed using the rapid bootstrapping RAxML algorithm with 1,000 bootstrap replications. The Bayesian interference was conducted using MrBayes 3.1.2. Metropolis-coupled Markov chain Monte Carlo sampling was performed with one cold and three heated chains that were run for 5,000,000 generations in two independent runs. The trees were sampled every 1,000th and 500th generation in the FHb and trHb analysis, respectively, and the 'burn in' was set to 25 %. Convergence of the runs was verified by assessing the average standard deviation of split frequencies, which reached values of 0.006193 and 0.001289 for the trHb and FHb data sets, respectively. Additionally, the parameters of the Bayesian inference were analyzed using Tracer [71]. For the calculation of the Bayesian trees, the CIPRES Science Gateway V.3.1 was used [72], while the phylogenetic trees were visualized with iTOL [73, 74].
CONSEL was used to test phylogenetic hypotheses [75]. The site-likelihoods for each tested tree topology were calculated applying TREE-PUZZLE [76]. Subsequently, the approximately unbiased (AU) test [77] was performed using CONSEL with default parameters.
Genomic localization, gene length, number of exons, and protein length of annotated amoebozoan globin genes.
| Species | Globin type | Location | Coordinates | Strand | Gene length | Number of exons | Protein length |
|---|---|---|---|---|---|---|---|
| Dictyostelium discoideum | FHbA | chromosome 6 | 1,649,565 to 1,650,758 | - | 1194 bp | 1 | 397 |
| Dictyostelium discoideum | FHbB | chromosome 6 | 1,651,520 to 1,652,908 | - | 1389 bp | 2 | 423 |
| Dictyostelium purpureum | FHbA | scaffold_485 | 14,681 to 16,277 | + | 1597 bp | 3 | 392 |
| Dictyostelium purpureum | FHbB | scaffold_530 | 9,247 to 10,772 | + | 1526 bp | 1 | 423 |
| Dictyostelium fasciculatum | FHb | DFA1501812 | 1,760,310 to 1,763,147 | - | 2837 bp | 2 | 400 |
| Polysphondylium pallidum | FHb | PPA1277996 | 2,450,684 to 2,451,898 | + | 1215 bp | 1 | 404 |
| Physarum polycephalum | FHb-1 | contigs 10755, 8539 | N/D | N/D | N/D | 71 | 3752 |
| Physarum polycephalum | FHb-2 | contigs 8539, 3993 | N/D | N/D | N/D | 71 | 3742 |
| Physarum polycephalum | FHb-3 | contigs 8539, 3993 | N/D | N/D | N/D | 9 | 376 |
| Hartmannella vermiformis | SDFgb | N/D | N/D | N/D | N/D | N/D | 159 |
| Acanthamoeba castellanii | trHbN | GL877269 | 354,379 to 354,915 | - | 537 bp | 3 | 179 |
| Acanthamoeba castellanii | trHbO | GL877210 | 157,074 to 157,782 | + | 709 bp | 1 | 201 |
1likely exons missing
2estimated length
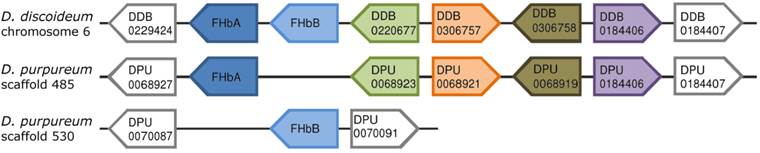
Comparison of the genomic neighborhood of the FHb genes from D. discoideum and D. purpureum. The direct neighboring genes of the FHbA and FHbB genes of D. discoideum and D. purpureum are shown. The directions of the boxes indicate the genomic orientations of the genes, i.e. a box directed to the right equates the plus strand, directed to the left equates the minus strand. Boxes with the same color represent orthologs, while grey boxes indicate that those genes do not possess an ortholog in this genomic location. For each gene either the gene symbol or the accession number provided by dictyBase is given. The FHbs of D. discoideum are located on chromosome 6 in a head-to-tail orientation. In contrast, the FHbs of D. purpureum are lying on two different scaffolds. The FHbA genes of the two amoebas lie in a short conserved syntenic block.
Results
Identification of amoebozoan globin genes
The previously characterized globins from D. discoideum were used to search the protein database of NCBI for additional amoebozoan globin proteins. Two FHbs of D. purpureum, an FHb of D. fasciculatum and an FHb of P. pallidum were identified. Additionally, an SDFgb of Hartmannella vermiformis was found in the EST database TBestDB. The obtained sequences were used to search against available amoebozoan genomes and the transcriptome of A. castellanii for further globin genes. As already reported, no globins are present in the genomes of Entamoeba parasites [5]. The slime mold P. polycephalum seems to possess three FHb genes. Although we could not detect any FHb genes, two putative trHb genes were found in the genome of A. castellanii. Table 1 summarizes detailed information of the analyzed globin genes.
The Flavohemoglobins of social amoebas and the slime mold P. polycephalum
The FHb genes of D. discoideum (DidiA and DidiB) are located next to each other on chromosome 6 in a head-to-tail orientation (Figure 1) [34]. DidiA is a single exon gene while DidiB contains two coding exons interrupted by an intron at position H2.1, i.e. between the first and second base of codon two in globin helix H, of 117 bp lengths (Figure 2). In contrast, the FHb genes of D. purpureum (DipuA, DipuB) are present on two different scaffolds. The DipuA gene lies on scaffold_485 and is disrupted by two introns at positions E1.1 and H9.1 of 74 and 78 bp lengths (Figure 2). The DipuB gene is a single exon gene, located on scaffold_530. Interestingly, the FHbA genes of both species lie at the 5' end of a short genomic block of conserved synteny (Figure 1). A total of five genes are conserved in order and orientation between the two Dictyostelium species. The shared synteny indicates that the FHbA genes are orthologs, despite their different intron/exon structure.
Alignment of amoebozoan FHbs to the Hmp protein of Escherichia coli. The alignment was created with MUSCLE. Conserved residues are shaded in different levels of grey. Residues that are conserved in all sequences are in dark grey. The secondary structure of the Hmp protein is given above the alignment (PDB: 1gvh). Predicted α-helices and β-strands are indicated as red and yellow lines, respectively, below the corresponding sequences. The positions of the introns are marked with green boxes and by arrows below the alignment. The topological positions of the introns as compared to sperm whale Mb are indicated below the alignment. The helix structure of the sperm whale myoglobin was superimposed on the alignment and indicated by violet bars above the alignment.
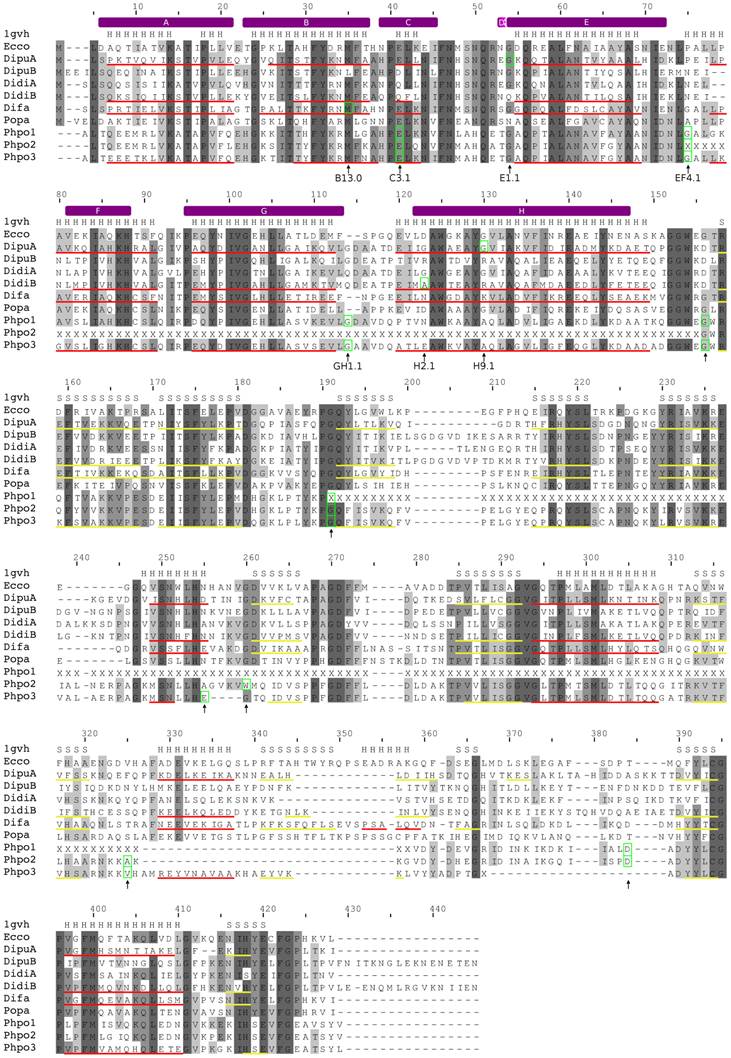
In contrast, D. fasciculatum and P. pallidum possess only single FHb genes, which were found on supercontigs DFA1501812 and PPA1277996, respectively. The FHb of P. pallidum (Popa) is a single exon gene while the FHb of D. fasciculatum (Difa) likely consists of two coding exons, which are separated by an intron of approximately 1.7 kb. The first exon codes for the first 32 amino acids of the globin domain. Although the sequence is highly conserved to other amoebozoan FHbs and comprises the A and B helix of the globin domain, no canonical splice sites were found. The current annotation places the intron at position B13.0 (Figure 2). No synteny conservation among the FHbs of D. fasciculatum and P. pallidum and to the other Dictyostelium species was observed.
The true slime mold P. polycephalum seems to possess three FHb genes (Phpo-1, Phpo-2, and Phpo-3), which span three potentially overlapping contigs (10755, 8539, and 3993). Thus, they are likely located next to each other in head-to-tail orientations in the genome. However, due to the highly fragmented nature of the current assembly, parts of the FHb genes are still missing. All globins are incomplete at their N-terminal ends. Additionally, in Phpo-1 a large part of the flavin-containing oxidoreductase domain of about 125 amino acids is absent while in Phpo-2 half of the globin domain (helices F to H) is missing. Nevertheless, it is highly likely that the missing exons lie in the still undetermined regions of the assembly. The Phpo-3 gene consists of nine exons and seems to be almost complete. However, the predicted splice sites of the last intron would result in two exons in different reading phase, which would consequently lead to a frame shift. Since the acceptor splice site is conserved among the Phpo genes, we assume that the second to last exon of Phpo-3 is incomplete at its 3' end. Three of the eight introns of Phpo-3 are lying in the globin domain at positions C3.1, EF4.1, and GH1.1 (Figure 2). The partial Phpo-1 and Phpo-2 genes each consist of seven exons interrupted by introns at the same positions as Phpo-3, except for introns four and six of Phpo-2 and Phpo-3, respectively. However, the position of the intron may change once undetermined regions are resolved.
The modeling of the tertiary structure of DidiB, DipuA, Difa, and Phpo-3 revealed that these globins can most likely adopt the typical globin fold. Nevertheless, as in the case of the hmp protein of Escherichia coli, the D helix seems to be absent [78]. All globins contain the highly conserved residues of the heme pocket, such as TyrB10, PheCD1, and HisF8 (proximal histidine). Moreover, also residues known to be responsible for FAD binding, e.g. Tyr206, Ser207, Phe390, Gly391 of E. coli hmp, are conserved among amoebozoan FHbs (Figure 2). Thus, we conclude that the amoebozoan FHbs likely represent functional proteins. The FHb proteins show no characteristics of membrane-bound proteins, and thus a membrane-association can be ruled out.
Phylogeny of the amoebozoan Flavohemoglobins
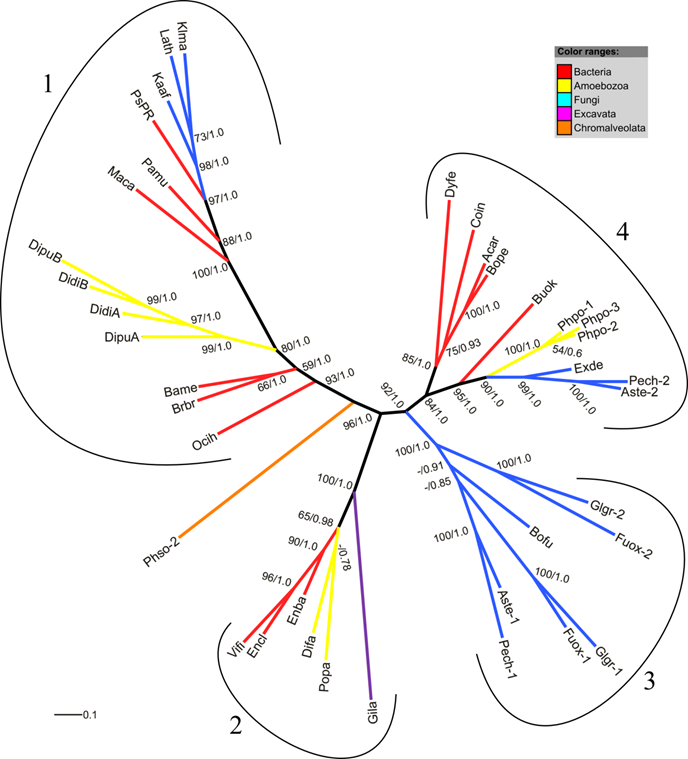
Phylogenetic trees of the amoebozoan FHbs and their closest relatives in the different kingdoms of life were reconstructed applying maximum likelihood and Bayesian interference algorithms. The MUSCLE 4.0 alignment (highest MUMSA score) was used for tree reconstruction and model selection. Both tree-building methods resulted in the same tree topology. Since no proper outgroup exists for our data set unrooted trees are presented. Figure 3 shows the maximum likelihood tree with superimposed bootstrap support and posterior probability values. Four highly supported clades can be identified of which three contain amoebozoan FHb proteins. The clade 1 comprises the FHbs of D. discoideum and D. purpureum, bacterial FHbs of some Firmicutes (Ocih, Brbr, Bame, Maca) and one γ-Proteobacterium (Pamu) as well as three fungal FHbs. Clade 2 encompasses the FHbs from P. pallidum and D. fasciculatum as well as the FHb of Gardia lamblia and FHbs of some γ-Proteobacteria (Enba, Encl, Vifi). The globins of P. polycephalum cluster together in clade 4 with FHbs from α- and β-Proteobacteria (Coin, Acar, Bope, Buok), one Cytophagium (Dyfe) and three fungal FHbs. This clade is sister to a group of fungal FHb proteins (clade 3). The FHb of P. sojae (Phso) seems to be unrelated to the other included FHbs.
The truncated hemoglobins of A. castellanii
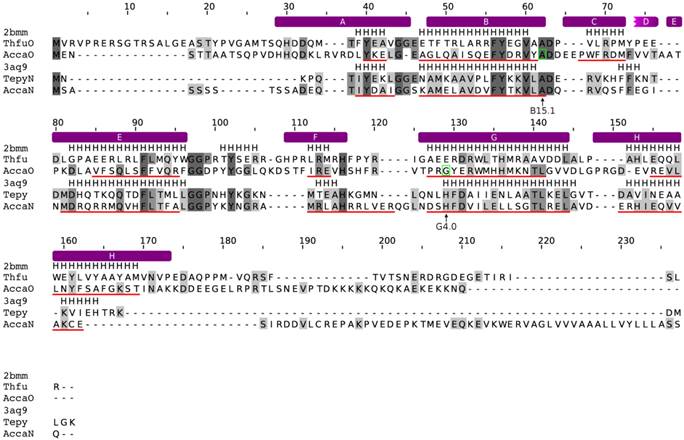
We identified and annotated two trHbs of A. castellanii that were found in the genome as well as in the transcriptomic sequence data. The trHbs, named AccaN and AccaO, are located on the whole genome shotgun (wgs) contigs GL877269 and GL877210, respectively. Strikingly, the last four nucleotides of the AccaO mRNA, derived from the transcriptomic data, do not align to the genome. Moreover, the coding sequence (CDS) of the predicted genomic gene is 57 nucleotides longer than the CDS of the transcriptomic mRNA. The C-termini of both translated peptides do not show any significant similarities to other known proteins. We were not able to determine which sequence represents the true AccaO gene. We decided to use the transcriptomic data in the subsequent phylogenetic analyses.
In contrast to AccaN, which is a single exon gene, AccaO contains two introns each 98 bp long at positions B15.1 and G4.0 (Figure 4). To check for similarities in the genomic localization of the trHbs, the direct neighboring genes of AccaN and AccaO were determined. tBLASTn searches revealed that the upstream and downstream neighboring genes of AccaN are similar to an oxidoreductase and to a serine/threonine kinase, respectively, while the direct neighbors of AccaO resemble proteins with an mscl domain and an N-acetylglucosamine-1-phosphodiester alpha-4-acetylglucosaminidase.
Similarly to the FHb case, the tertiary structure of the trHbs has been predicted as described in the method section. Both globins are able to adopt the typical fold of truncated globins, including a shortened A helix and absence of the D helix [9]. Furthermore, conserved residues of group I and II trHbs are present, such as the conserved glycine motifs, the Phe-Tyr pair at B9-10 and the proximal histidine (HisF8) (Figure 4). Thus, both globins likely represent functional proteins.
Interestingly, the AccaN protein may possess a transmembrane domain at its C-terminus (amino acids 159 - 176) as predicted by TMpred and TMHMM. Furthermore, CSS-Palm 3.0 predicted a potential palmitoylation site at Cys133. These findings indicate that AccaN may be a membrane-bound globin.
Radial maximum likelihood tree of FHb proteins. The colors of branches correspond to the taxonomic classification of the used sequences. Bootstrap support (bs) and posterior probability (pp) values equal or greater than 50 % are given (bs/pp). The FHb proteins cluster in four highly supported clades (1-4). For a description of used abbreviations please refer to Supplementary Material: table S1.
Alignment of the trHbs of A. castellanii to trHbN of Tetrahymena pyriformis and trHbO of Thermobifida fusca. The alignment was created with MUSCLE. Conserved residues are shaded in different levels of grey. Residues that are conserved in all sequences are in dark grey. The secondary structure of the trHb proteins of T. pyriformis (PDB: 3aq9) and T. fusca (PDB: 2bmm) are given. Predicted α-helices are indicated as red lines, below the corresponding sequences. The positions of the introns are marked with green boxes and by arrows below the alignment. The topological positions of the introns as compared to sperm whale Mb are indicated below the alignment. The helix structure of the sperm whale myoglobin was superimposed on the alignment and indicated by violet bars above the alignment.
Phylogeny of the trHbs of A. castellanii
Akin to the analysis of FHbs, phylogenetic trees of the trHbs of A. castellanii and its closest relatives were reconstructed. Here, the FFT-NS-i strategy of MAFFT produced the highest scoring MUMSA alignment. It is assumed that group I and group III globins likely represent the products of a duplication event of an ancestral group II gene [16]. The maximum likelihood and Bayesian interference analyses resulted in slightly different trees. However, clustering of the major clades was recovered in all analyses (Figure 5, Supplementary Material: Figure S1). Figure 5 shows the Maximum likelihood tree with superimposed bootstrap support values and posterior probability values of the Bayesian analysis. The trHbs cluster in accordance to their classifications in three monophyletic groups (I, II, III) (Figure 5). AccaO is positioned between the clades consisting of group II and group I/III globins while AccaN clusters together with a trHb of the castor oil plant (RicoN) and a putative trHb of a fungus (BadeN) in the clade comprising group I globins. Although, the clustering of AccaN with RicoN and BadeN is not well supported, it was recovered in all analyses.
The single-domain SDFgb of Hartmannella vermiformis
We identified an EST sequence of H. vermiformis in the TBestDB that shares high sequence similarity to F globin genes and most likely represents an SDFgb. Our analysis indicates that the EST sequence contains the complete CDS. The presence of a single globin domain in the translated peptide was verified via SMART. As for the FHbs, a membrane association of the SDFgb can be ruled out. BLASTp searches against the protein database of NCBI revealed that the closest relatives of the globin of H. vermiformis are bacterial SDFgbs and FHbs. Our phylogenetic trees further support this finding. In a tree comprising the different types of single-domain globins (SDFgbs, SDSgbs, Pgbs) the SDFgb of H. vermiformis (Have) clusters with two bacterial SDFgb proteins (Figure 6).
Discussion
Evolution of amoebozoan globin genes
Figure 7 summarizes our findings in the phylogenetic connects. The identification and characterization of amoebozoan globin genes revealed that two of the three major globin lineages are present in Amoebozoa, represented by FHbs, SDFgbs, and trHbs. The absence of members of the S globin family is not surprising given that GCSs seem to be completely missing in eukaryotes and that SDSgbs have so far only been described in some bacteria, archaea, and fungi [5, 7]. All analyzed species of the infraphylum Mycetozoa possess at least one FHb gene, while trHb genes and an SDFgb gene were only found in A. castellanii (infraphylum Acanthopodina) and H. vermiformis (infraphylum Tubulinea). These findings hint at lineage-specific adaptations of the globin gene repertoires although further research must be done to corroborate this belief.
Radial maximum likelihood tree of trHb proteins. The colors of branches correspond to the taxonomic classification of the used sequences. Bootstrap support (bs) and posterior probability (pp) values equal or greater than 50 % are given (bs/pp). The trHb proteins cluster in three highly supported clades (I-III), in accordance to their classification. For a description of used abbreviations please refer to Supplementary Material: table S1.
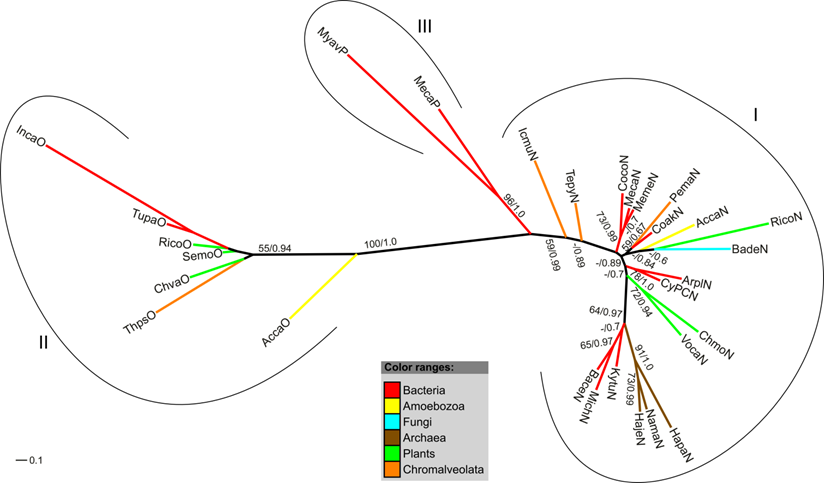
Radial maximum likelihood tree of single-domain globins and the SDFgb of H. vermiformis. The colors of branches correspond to the taxonomic classification of the used sequences. Bootstrap support (bs) and posterior probability (pp) values equal or greater than 50 % are indicated (bs/pp). The SDFgb of H. vermiformis (HaveSDFgb) clusters with two bacterial SDFgbs. For a description of used abbreviations please refer to Supplementary Material: table S1.
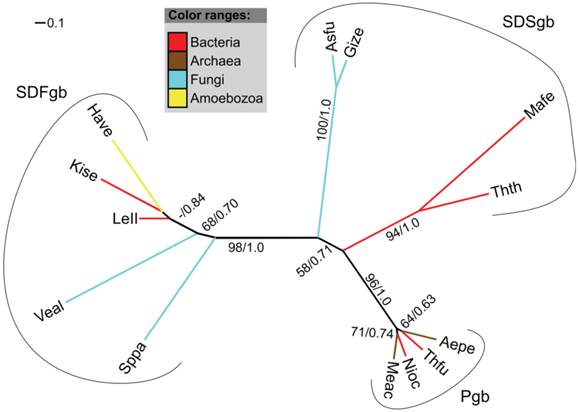
Phylegenetic relationships between studied Amoebozoa organisms. The tree is based on Adl et al. [91] Please note that branch length is not to scale. Type of globin found in a given group is indicated in dark blue inside the group circles.
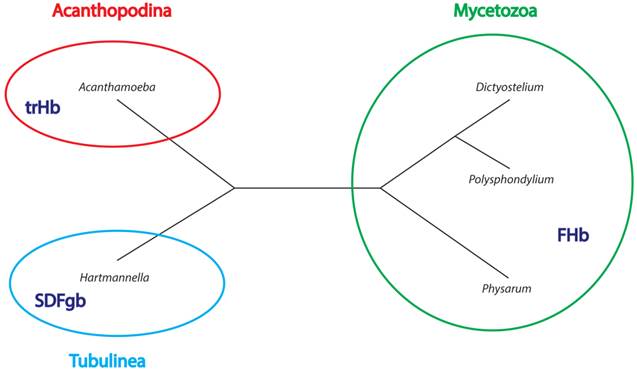
The phylogenetic tree of FHbs suggests that the three FHb genes of P. polycephalum (Phpo-1, Phpo-2, Phpo-3) arouse as a result of lineage-specific gene duplication events (Figure 3). Additionally, it can be inferred that the FHbs of D. fasciculatum and P. pallidum, as well as the FHbs of D. discoideum and D. purpureum share a common origin (Figure 7). The placement of AccaO in our phylogenetic trees is ambiguous and could be either basal to group II or to group I/III trHbs (Figure 5). Given its closer clustering to group II trHbs and the recognition of group II trHbs ahead of other trHbs in BLAST searches, we propose that AccaO represents a group II trHb.
Of the examined Amoebozoa species, H. vermiformis is the only one that possesses an SDFgb. Its closest relative is a globin of a Leptospirillum bacterium from the phylum Nitrospirae. The presence of an SDFgb in H. vermiformis and absence in all other analyzed amoebozoan genomes could be explained by several independent losses in the other lineages. However, in light of its close relationship to bacterial SDFgbs, a horizontal gene transfer (HGT) event from an ancient bacterium seems to be more plausible. Moreover, increasing evidence suggests that HGT played a major role in the evolution of many species [25, 26] and it is assumed that HGT is an important force that shaped the phyletic distribution of globin genes [5-7, 18-21].
Likewise, the free-living amoeba A. castellanii is the only examined species that contains trHbs. In our phylogenetic trees (Figure 5) the trHbs cluster in accordance to their classification in three distinct groups that are highly supported. However, support for other clades is rather low and clustering of some proteins varies among the different methods. Therefore, it is difficult to draw precise conclusion from the phylogenetic analyses. However, the strong deviation of the inferred tree from the species tree and the absence of trHbs in the closely related genomes indicate a horizontal inheritance [79] of trHbs to an ancestor of the extant A. castellanii. Supportingly, several previous studies have already emphasized the importance of HGT of trHbs from prokaryotes to eukaryotes [16, 19-21]. Amoeba not only feed on bacteria, but can also harbor bacteria either as transient or stable endosymbionts [30, 80, 81]. The various interactions between free-living amoebae and bacteria may be the source of the horizontal transfer of an SDFgb and of trHbs to H. vermiformis and A. castellanii, respectively.
The FHb proteins of the social amoebas and the slime mold P. polycephalum were expected to be tightly related in view of the close relationship of the species, all belonging to the infraphylum Mycetozoa. However, in the inferred phylogenetic trees the FHbs groups are paraphyletic present in three distinct clades of which each contains some bacterial FHbs (Figure 3). Thus, it appears that the amoebozoan FHbs are closer related to some bacterial sequences than to their mycetozoan counterparts. One explanation of this tree would be that the common ancestor of Mycetozoa possessed multiple paralogous FHbs and that each of the described clades only retained one of them. However, given the previous thoughts, we favor a scenario in which the FHb genes were individually gained by HGT. In addition, two independent studies also observed the nesting of the FHbs from D. discoideum within a clade of some Firmicutes, β- and ε-Proteobacteria and proposed the possibility of a prokaryote-to-eukaryote HGT event [6, 22]. It should be added that based on the tree presented in Figure 3, we cannot exclude possibility of the lateral transfer of the FHb gene in the other direction, i.e. from Eumycetozoa to bacteria (see clade 2). Similar observations were made for the FHb gene of the diplomonad Giardia lamblia [22] that in our tree (Gila) clusters in a common clade with the FHbs of D. fasciculatum (Difa) and P. pallidum (Popa). To further support such an evolutionary scenario, the likelihoods of the presented tree topology and of a topology in which the amoebozoan FHb cluster as a monophyletic group were compared, applying the AU test implemented in CONSEL [75, 77]. The monophyly of amoebozoan FHbs was rejected at a high confidence level (p-value 3e-74), substantiating our assumption of three independent HGT events.
Shared synteny among D. discoideum and D. purpureum
The shared synteny among the FHbA genes of D. discoideum (DidiA) and D. purpureum (DipuA) supports an orthologous relationship of these two genes (Figure 1). In contrast, their orthology is not clearly evident from the phylogenetic tree (Figure 3). However, inference of phylogenetic trees is known to be error-prone, while shared synteny represents a reliable criterion for defining orthologs [82]. Apart from that, the phylogenetic tree (Figure 3) confirms the orthologous relationship of the FHbB genes (DidiB, DipuB) of these two Dictyostelium species. We hypothesize that the FHbA and FHbB genes emerged through the duplication of a pre-FHb gene in the common ancestor of D. discoideum and D. purpureum. This is supported by the retained linkage of the FHb genes in D. discoideum. Later the DipuB gene of D. purpureum got translocated to a new genomic location. Coincidently, Eichinger and colleagues observed that the genome of D. discoideum is enriched in relatively recently duplicated genes [83].
Evolution of introns
The presence or absence of introns and their positions vary widely among the analyzed globin genes. Although, D. discoideum and D. purpureum are closely related and possess highly conserved orthologous FHb genes, introns are not shared among the orthologs. While DidiB and DipuA harbor one and two introns, respectively, their orthologs are single exon genes (Figure 2). Thus, the introns have been either gained or lost after the divergence of D. discoideum and D. purpureum. The globin genes of almost all vertebrates, many invertebrates and several plants contain two introns at positions B12.2 and G7.0, which are considered as phylogenetically ancient [1, 84]. None of the amoebozoan globin genes contains introns at these ancestral positions. Therefore, lineage-specific intron gains seem more likely than several intron loss events. The length of the introns is rather short and ranges from 74 bp to 117 bp, with the exception of the intron in the FHb gene of D. fasciculatum. The genome analysis of for example D. discoideum and D. purpureum revealed only few and short introns with a mean length of 146 and 177 bp, respectively [83, 85]. Thus, the intron length of the globin genes does not deviate significantly from the overall intron length distribution in these species.
Potential association of AccaN of A. castellanii with the membrane
The in silico analysis of the trHbs of A. castellanii indicates that AccaN may be a membrane-bound globin protein. The presence of a potential transmembrane domain at its C-terminus was predicted by two independent tools, namely TMpred [53] and TMHMM [54]. Additionally, a possible palmitoylation site was found. Palmitoylation is a reversible lipid modification of proteins, which enhances the surface hydrophobicity and membrane affinity of proteins [86-88]. Although, palmitoylation occurs mainly close to the N-terminus of proteins, it has also been observed in other parts of proteins [87, 88]. Globin proteins associated with the membrane have been identified in some bacteria [44-47], in the nematode Caenorhabditis elegans [89], in the shore crab Carcinus maenas [48], and recently also in vertebrates [49, 50]. However, there is no evidence that these globins share common ancestry suggesting that membrane-associated globins arouse independently several times in the course of evolution. A respiratory function of globins associated with the membrane is highly unlikely. It has been proposed that such bacterial globins facilitate oxygen transfer to the terminal oxidases of the respiratory chain [44, 46, 90]. Though eukaryotes lack a respiratory chain in their cell membranes, eukaryotic membrane associated globins may perform a comparable function, i.e. they may protect the membrane lipids from oxidative stress [48-50]. Alternatively, as suggested for vertebrate globin X, AccaN may function as an O2 sensor or as a binding partner in a signal transduction pathway [49]. Any of these roles would be conceivable for AccaN and elucidating its function will also shed light upon its evolutionary history.
Conclusion
Globin genes are present in almost all eukaryotes, except for some unicellular parasites, such as Entamoeba histolytica and Plasmodium falciparum [5]. Although the globin genes of the social amoeba D. discoideum have been described and analyzed several years ago [34], nothing was known about the globin genes of the closely related species. This survey aimed to characterize the globin gene repertoire of amoebozoan species. Our results suggest lineage-specific adaptations of the globin gene repertoires; however presently there is no strong evidence of the adaptive processes in amoebozoan globin evolution and further studies are required to elaborate the issue. FHb genes were identified in several social amoebas and the true slime mold P. polycephalum of the infraphylum Mycetozoa, while trHb genes and an SDFgb gene were only found in A. castellanii (infraphylum Acanthopodina) and H. vermiformis (infraphylum Tubulinea), respectively. Intriguingly, the trHbN of A. castellanii might be associated with the membrane and thus may protect membrane lipids against oxidative stress.
Based on the phylogenetic analyses we propose that these globin genes are products of ancient HGTs from bacteria, though we cannot entirely rule out a scenario of ancient duplications and subsequent losses. These horizontal transfers could have been easily achieved given the tight interconnection between Amoebozoa and bacteria [80]. Nevertheless, our knowledge on the globin distribution in many other taxonomic groups of unicellular eukaryotes is still very limited. It would be captivating to examine the impact of horizontal gene transfer events on the globin gene diversity in these lineages.
Supplementary Material
Table S1: Table of sequences used in this study. Table S2: Templates used for tertiary structure modeling employing Swiss-Model and SwissPDBViewer.Figure S1: Bayesian tree of trHb proteins. The colors of branches correspond to the taxonomic classification of the used sequences. Posterior probability values equal or greater than 50 % are indicated. The trHb proteins cluster in accordance to their classification in three distinct clades. For a description of used abbreviations please refer to table S1.
Acknowledgements
This work was supported by the Institute of Bioinformatics funds and partially by the
FP7-People-2009-IRSES Project ''EVOLGEN'' No. 247633, the Asia-Africa S & T Strategic Cooperation Promotion Program, and a Grant-in-Aid for Scientific Research on the Priority Area “Genome Science” from the Ministry of Education, Culture, Sports, Science and Technology of Japan. We are grateful to Marcin Jąkalski for his help in figure preparation.
Author's contribution
JD designed the study, carried out the bioinformatic analyses, and drafted the manuscript. DB conducted the transcriptome sequencing and assembly. YS conceived the transcriptome sequencing. WM conceived the study, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hardison RC. A brief history of hemoglobins: plant, animal, protist, and bacteria. Proc Natl Acad Sci U S A. 1996;93:5675-5679
2. Freitas TA, Hou S, Dioum EM, Saito JA, Newhouse J, Gonzalez G, Gilles-Gonzalez MA, Alam M. Ancestral hemoglobins in Archaea. Proc Natl Acad Sci U S A. 2004;101:6675-6680
3. Vinogradov S, Hoogewijs D, Vanfleteren J, Dewilde S, Moens L, Hankeln T. Evolution of the globin superfamily and its function. In Homoglobin: Recent Developments and Topics. Edited by Nagai M. Kerala, IND: Research Signpost. 2011:231-254
4. Vinogradov SN, Moens L. Diversity of globin function: enzymatic, transport, storage, and sensing. The Journal of biological chemistry. 2008;283:8773-8777
5. Vinogradov SN, Hoogewijs D, Bailly X, Arredondo-Peter R, Gough J, Dewilde S, Moens L, Vanfleteren JR. A phylogenomic profile of globins. BMC Evol Biol. 2006;6:31
6. Vinogradov SN, Hoogewijs D, Bailly X, Arredondo-Peter R, Guertin M, Gough J, Dewilde S, Moens L, Vanfleteren JR. Three globin lineages belonging to two structural classes in genomes from the three kingdoms of life. Proc Natl Acad Sci U S A. 2005;102:11385-11389
7. Vinogradov SN, Hoogewijs D, Bailly X, Mizuguchi K, Dewilde S, Moens L, Vanfleteren JR. A model of globin evolution. Gene. 2007;398:132-142
8. Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, Phillips DC. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958;181:662-666
9. Pesce A, Couture M, Dewilde S, Guertin M, Yamauchi K, Ascenzi P, Moens L, Bolognesi M. A novel two-over-two alpha-helical sandwich fold is characteristic of the truncated hemoglobin family. The EMBO journal. 2000;19:2424-2434
10. Hausladen A, Stamler JS. Is the flavohemoglobin a nitric oxide dioxygenase? Free radical biology & medicine. 2012;53:1209-1210 author reply 1211-1202
11. Forrester MT, Foster MW. Protection from nitrosative stress: a central role for microbial flavohemoglobin. Free radical biology & medicine. 2012;52:1620-1633
12. Bonamore A, Boffi A. Flavohemoglobin: structure and reactivity. IUBMB life. 2008;60:19-28
13. Frey AD, Kallio PT. Bacterial hemoglobins and flavohemoglobins: versatile proteins and their impact on microbiology and biotechnology. FEMS microbiology reviews. 2003;27:525-545
14. Freitas TA, Hou S, Alam M. The diversity of globin-coupled sensors. FEBS letters. 2003;552:99-104
15. Wittenberg JB, Bolognesi M, Wittenberg BA, Guertin M. Truncated hemoglobins: a new family of hemoglobins widely distributed in bacteria, unicellular eukaryotes, and plants. The Journal of biological chemistry. 2002;277:871-874
16. Vuletich DA, Lecomte JT. A phylogenetic and structural analysis of truncated hemoglobins. Journal of molecular evolution. 2006;62:196-210
17. Ascenzi P, Bolognesi M, Milani M, Guertin M, Visca P. Mycobacterial truncated hemoglobins: from genes to functions. Gene. 2007;398:42-51
18. Vinogradov SN, Hoogewijs D, Arredondo-Peter R. What are the origins and phylogeny of plant hemoglobins? Communicative & integrative biology. 2011;4:443-445
19. Vinogradov SN, Fernandez I, Hoogewijs D, Arredondo-Peter R. Phylogenetic relationships of 3/3 and 2/2 hemoglobins in Archaeplastida genomes to bacterial and other eukaryote hemoglobins. Molecular plant. 2011;4:42-58
20. Vazquez-Limon C, Hoogewijs D, Vinogradov SN, Arredondo-Peter R. The evolution of land plant hemoglobins. Plant science: an international journal of experimental plant biology. 2012;191-192:71-81
21. Hoogewijs D, Dewilde S, Vierstraete A, Moens L, Vinogradov SN. A phylogenetic analysis of the globins in fungi. PLoS One. 2012;7:e31856
22. Andersson JO, Sjogren AM, Davis LA, Embley TM, Roger AJ. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Current biology: CB. 2003;13:94-104
23. Watts RA, Hunt PW, Hvitved AN, Hargrove MS, Peacock WJ, Dennis ES. A hemoglobin from plants homologous to truncated hemoglobins of microorganisms. Proc Natl Acad Sci U S A. 2001;98:10119-10124
24. Moens L, Vanfleteren J, Van de Peer Y, Peeters K, Kapp O, Czeluzniak J, Goodman M, Blaxter M, Vinogradov S. Globins in nonvertebrate species: dispersal by horizontal gene transfer and evolution of the structure-function relationships. Molecular biology and evolution. 1996;13:324-333
25. Brown JR. Ancient horizontal gene transfer. Nature reviews Genetics. 2003;4:121-132
26. Keeling PJ. Functional and ecological impacts of horizontal gene transfer in eukaryotes. Current opinion in genetics & development. 2009;19:613-619
27. Vinogradov SN, Bailly X, Smith DR, Tinajero-Trejo M, Poole RK, Hoogewijs D. Microbial eukaryote globins. Advances in microbial physiology. 2013;63:391-446
28. Smirnov AV, Chao E, Nassonova ES, Cavalier-Smith T. A revised classification of naked lobose amoebae (Amoebozoa: lobosa). Protist. 2011;162:545-570
29. Cavalier-Smith T. A revised six-kingdom system of life. Biological reviews of the Cambridge Philosophical Society. 1998;73:203-266
30. Rodriguez-Zaragoza S. Ecology of free-living amoebae. Critical reviews in microbiology. 1994;20:225-241
31. Kessin RH. Dictyostelium: Evolution, Cell Biology, and the Development of Multicellularity. Reissue edn. Cambrige, UK: Cambridge University Press. 2010
32. Stanley SL Jr. Amoebiasis. Lancet. 2003;361:1025-1034
33. Marciano-Cabral F, Cabral G. Acanthamoeba spp. as agents of disease in humans. Clinical microbiology reviews. 2003;16:273-307
34. Iijima M, Shimizu H, Tanaka Y, Urushihara H. Identification and characterization of two flavohemoglobin genes in Dictyostelium discoideum. Cell structure and function. 2000;25:47-55
35. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403-410
36. O'Brien EA, Koski LB, Zhang Y, Yang L, Wang E, Gray MW, Burger G, Lang BF. TBestDB: a taxonomically broad database of expressed sequence tags (ESTs). Nucleic acids research. 2007;35:D445-451
37. Burge CB, Karlin S. Finding the genes in genomic DNA. Curr Opin Struct Biol. 1998;8:346-354
38. Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic acids research. 1996;24:3439-3452
39. Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991;220:49-65
40. Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243-257
41. Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857-5864
42. Gaudet P, Fey P, Basu S, Bushmanova YA, Dodson R, Sheppard KA, Just EM, Kibbe WA, Chisholm RL. dictyBase update 2011: web 2.0 functionality and the initial steps towards a genome portal for the Amoebozoa. Nucleic acids research. 2011;39:D620-624
43. Kreppel L, Fey P, Gaudet P, Just E, Kibbe WA, Chisholm RL, Kimmel AR. dictyBase: a new Dictyostelium discoideum genome database. Nucleic acids research. 2004;32:D332-333
44. Liu C, He Y, Chang Z. Truncated hemoglobin o of Mycobacterium tuberculosis: the oligomeric state change and the interaction with membrane components. Biochemical and biophysical research communications. 2004;316:1163-1172
45. Rinaldi AC, Bonamore A, Macone A, Boffi A, Bozzi A, Di Giulio A. Interaction of Vitreoscilla hemoglobin with membrane lipids. Biochemistry. 2006;45:4069-4076
46. Ramandeep Hwang KW, Raje M Kim KJ, Stark BC Dikshit KL, Webster DA. Vitreoscilla hemoglobin. Intracellular localization and binding to membranes. The Journal of biological chemistry. 2001;276:24781-24789
47. Bonamore A, Farina A, Gattoni M, Schinina ME, Bellelli A, Boffi A. Interaction with membrane lipids and heme ligand binding properties of Escherichia coli flavohemoglobin. Biochemistry. 2003;42:5792-5801
48. Ertas B, Kiger L, Blank M, Marden MC, Burmester T. A membrane-bound hemoglobin from gills of the green shore crab Carcinus maenas. The Journal of biological chemistry. 2011;286:3185-3193
49. Blank M, Wollberg J, Gerlach F, Reimann K, Roesner A, Hankeln T, Fago A, Weber RE, Burmester T. A membrane-bound vertebrate globin. PLoS One. 2011;6:e25292
50. Blank M, Burmester T. Widespread occurrence of N-terminal acylation in animal globins and possible origin of respiratory globins from a membrane-bound ancestor. Molecular biology and evolution. 2012;29:3553-3561
51. Bologna G, Yvon C, Duvaud S, Veuthey AL. N-Terminal myristoylation predictions by ensembles of neural networks. Proteomics. 2004;4:1626-1632
52. Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein engineering, design & selection: PEDS. 2008;21:639-644
53. Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166
54. Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings International Conference on Intelligent Systems for Molecular Biology; ISMB International Conference on Intelligent Systems for Molecular Biology. 1998 6:175-182
55. Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195-201
56. Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714-2723
57. Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nature protocols. 2009;4:1-13
58. Campanella JJ, Bitincka L, Smalley J. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC bioinformatics. 2003;4:29
59. Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2). Cladistics. 1989;5:164-166
60. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792-1797
61. Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic acids research. 2005;33:511-518
62. Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic acids research. 2002;30:3059-3066
63. Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073-1079
64. Lassmann T, Sonnhammer EL. Automatic assessment of alignment quality. Nucleic acids research. 2005;33:7120-7128
65. Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758-771
66. Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688-2690
67. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572-1574
68. Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754-755
69. Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164-1165
70. Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Molecular biology and evolution. 2001;18:691-699
71. Rambaut A, Drummond AJ. Tracer v1.4. http://beast.bio.ed.ac.uk/Tracer
72. Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). 2010:1-8
73. Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127-128
74. Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic acids research. 2011;39:W475-478
75. Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246-1247
76. Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502-504
77. Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492-508
78. Ilari A, Bonamore A, Farina A, Johnson KA, Boffi A. The X-ray structure of ferric Escherichia coli flavohemoglobin reveals an unexpected geometry of the distal heme pocket. The Journal of biological chemistry. 2002;277:23725-23732
79. Philippe H, Douady CJ. Horizontal gene transfer and phylogenetics. Current opinion in microbiology. 2003;6:498-505
80. Horn M, Wagner M. Bacterial endosymbionts of free-living amoebae. The Journal of eukaryotic microbiology. 2004;51:509-514
81. Schmitz-Esser S, Toenshoff ER, Haider S, Heinz E, Hoenninger VM, Wagner M, Horn M. Diversity of bacterial endosymbionts of environmental acanthamoeba isolates. Applied and environmental microbiology. 2008;74:5822-5831
82. Droge J, Pande A, Englander EW, Makalowski W. Comparative genomics of neuroglobin reveals its early origins. PLoS One. 2012;7:e47972
83. Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q. et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43-57
84. Dixon B, Pohajdak B. Did the ancestral globin gene of plants and animals contain only two introns? Trends Biochem Sci. 1992;17:486-488
85. Sucgang R, Kuo A, Tian X, Salerno W, Parikh A, Feasley CL, Dalin E, Tu H, Huang E, Barry K. et al. Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum. Genome biology. 2011;12:R20
86. Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochimica et biophysica acta. 1999;1451:1-16
87. Nadolski MJ, Linder ME. Protein lipidation. The FEBS journal. 2007;274:5202-5210
88. Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nature reviews Molecular cell biology. 2007;8:74-84
89. Tilleman L, De Henau S, Pauwels M, Nagy N, Pintelon I, Braeckman BP, De Wael K, Van Doorslaer S, Adriaensen D, Timmermans JP. et al. An N-myristoylated globin with a redox-sensing function that regulates the defecation cycle in Caenorhabditis elegans. PLoS One. 2012;7:e48768
90. Pathania R, Navani NK, Rajamohan G, Dikshit KL. Mycobacterium tuberculosis hemoglobin HbO associates with membranes and stimulates cellular respiration of recombinant Escherichia coli. The Journal of biological chemistry. 2002;277:15293-15302
91. Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S. et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. The Journal of eukaryotic microbiology. 2005;52:399-451
Author contact
![]() Corresponding author: wojmakde
Corresponding author: wojmakde

 Global reach, higher impact
Global reach, higher impact