10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(9):973-982. doi:10.7150/ijbs.9088 This issue Cite
Research Paper
Aberrantly Up-regulated miR-20a in Pre-eclampsic Placenta Compromised the Proliferative and Invasive Behaviors of Trophoblast Cells by Targeting Forkhead Box Protein A1
1. Department of Gerontology, The First Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang 150081, P. R. China;
2. Department of Pharmacology (Key Laboratory of Cardiovascular Medicine Research, Ministry of Education; State-Province Key Laboratories of Biomedicine-Pharmaceutics of China), Harbin Medical University, Harbin, Heilongjiang 150081, P. R. China.
* Equal contributions to the work.
Received 2014-3-12; Accepted 2014-8-1; Published 2014-8-30
Abstract
Preeclampsia is a serious complication in pregnancy. Dysregulation of trophoblast cell proliferation and invasion is a major pathological alteration observed in preeclampsia. Recently, microRNAs were shown to participate in the pathogenesis of preeclampsia. In this study we explored the effect of miR-20a on the proliferation and invasion of trophoblast cells and the underlying mechanism.
We verified the distribution of miR-20a in human placenta by in situ hybridization. Real time PCR data showed that the level of miR-20a increased by 2.6 folds in human preeclampsia than normal tissues. We then cultured trophoblast-like JEG-3 cells and evaluated the effect of miR-20a on JEG-3 cell proliferation, migration and invasion. Overexpression of miR-20a significantly inhibited the proliferation, migration and invasion of cultured JEG-3 cells, which were abolished by co-transfection of AMO-20a. Transfection of miR-20a also inhibited JEG-3 cell xenograft tumor growth in nude mice. Luciferase assay technique was used to identify the direct regulation of miR-20a on Forkhead Box Protein A1(FOXA1). Transfection of miR-20a markedly reduced the luciferase activity of the chimeric plasmid containing the 3'UTR of FOXA1, indicating FOXA1 is the target of miR-20a. Furthermore, transfection of miR-20a inhibited both protein and mRNA expression of FOXA1 in JEG-3 cells. In summary, the upregulated miR-20a in human preeclampsia tissue can inhibit the proliferative and invasive activities of trophoblast cells by repressing the expression of FOXA1.
Keywords: FOXA1, invasion, miR-20a, migration, proliferation.
Introduction
Preeclampsia is one of the leading causes for maternal and neonatal mortality and morbidity, which is characterized by high blood pressure and significant amounts of protein in the urine of a pregnant woman (1, 2). Defective embryo implantation plays a critical role in the development of preeclampsia. Trophoblast cells of the human placenta proliferate, differentiate and invade the endometrium leading to successful implantation of the embryo. Trophoblast invasion into the decidual stroma and spiral arteries is one of the crucial steps in human placental development, which provide an anchor to the developing embryo and regulate the temporal changes that occur in the fetal-maternal circulation during early pregnancy. Aberrant trophoblast invasive behaviors such as failure of the trophoblasts to appropriately invade the uterus, shallow trophoblast invasion, fewer invasive trophoblasts and failure to convert the spiral arteries are often observed in the placental pathology of preeclampsia (3-5). Immune-system alterations have been shown to be associated with the origin of preeclampsia (6, 7). Moreover, proinflammatory cytokines, neutrophil activation and endothelial dysfunction are major contributors of the pathogenesis of this syndrome (6, 7). Despite of these advancements, the molecular mechanism in the process of trophoblast cell proliferation and invasion remains elusive.
MicroRNAs are a group of single strand, noconding RNAs which functions in transcriptional and post-transcriptional regulation of gene expression (8). A host of studies demonstrated that microRNAs play critical roles in the process of development and the pathogenesis of various human diseases via regulating the protein expression of target genes (9, 10). Modulating on the aberrant expressed miRNAs has been proved to be a promising strategy in the treatment of human diseases (11-13). Recent studies confirmed the involvement of microRNAs in the pathogenesis of preeclampsia. Zhang et al. showed that miR-155 was up-regulated in preeclampsia tissue and overexpression of miR-155 contributes to preeclampsia development by down-regulating angiogenic regulating factor CYR61 (14). Zhang et al. found that ectopic expression of mir-210, an up-regulated microRNA in patients with pre-eclampsia, inhibited the migration and invasion capability of trophoblast cells by targeting Ephrin-A3 and Homeobox-A9(15). They further confirmed that the overexpression of mir-210 under hypoxia was regulated by NF-κB transcription factor. Bai et al found that miR-195 was downregulated in preeclamptic placenta tissues and overexpression of miR-195 promote the invasion of human placental trophoblast cells via repressing ActRIIA(16). In a previous study, we identified the dysregulated microRNAs in preeclamptic placenta tissues by quantitative PCR based array. Among them, miR-20a was greatly upregulated(17). However, the detailed role of miR-20a in the pathogenesis of preeclampsia remains unclear. In this study, we will employ JEG-3 cells to explore the effects of miR-20a on the proliferation and invasion of trophoblast cells and the underlying mechanism.
Materials and Methods
Cell Culture and Transfection
Human JEG-3 cell line was cultured in RPMI-1640 (Hyclone, USA) supplemented with 10% fetal bovine serum. When the cells grew to 60% confluent, they were transfected with hsa-miR-20a mimics (miR-20a), hsa-miR-20a inhibitor (AMO20a) or negative control (Scramble) (RIBOBIO, Guangzhou) using X-tremeGENE siRNA Transfection Reagent (Roche, Germany). Forty eight hours after transfection, the total protein was extracted for Western Blot and the total RNA was isolated for qRT-PCR.
Proliferation Assay
Cell counting was used to measure JEG-3 cell proliferation. 2×104 cells were seeded in 24-well culture plate 24 hours before transfection. miR-20a, miR20a+AMO, and scramble sequences were transfected into JEG-3 cells by using lipofectamine 2000 (Invitrogen, USA). After 48h's transfection, the cells were trypsinized and counted with hemocytometer. The experiments were repeated for 4 times.
Cell Migration and Invasion Assay
The ability of cell migration was measured by wound healing assay. JEG-3 cells were seeded in 6-well culture plates. When the cells grew to 70% confluent, they were transfected with miR-20a, miR20a+AMO, or scramble sequence by lipofectamine 2000. A sterile pipette tip of 10μl was used to draw straight lines on the confluent monolayers. Photographs of the wounds were taken at 0, 24, 48, and72 hours after transfection. The width of the wound was measured to test the migration ability.
Alternatively, transwell system (Corning, USA) was prepared for migration and invasion assay. After being transfected for 36h, the cells were trypsinized, centrifuged, and then resuspended by 1640 with 0.1% FBS (8×105 cells/ml). 3×105 cells were seeded into the upper chambers and allowed to migrate through an 8-µm pore size membrane or invade through a Matrigel-coated membrane. The chambers were then placed into 24-well culture plates contained 1640 supplemented with 20% FBS. After 24 hours (migration assays) or 48 hours (invasion assays), the medium was discarded, and the chambers with cells were fixed with 4% PFA for 30 min and stained by crystal violet for 20 min. The cells on the upper surface of the membrane were removed using cotton swab gently and cells that had migrated or invaded to the lower surface of the membrane were counted at three random fields.
Total RNA extraction and real-time RT-PCR
Total RNA was extracted from JEG-3 cells after treatment by using Trizol reagent (Invitrogen, USA). Total RNA(0.5μg) was then reverse transcribed using TransScript Reverse Transcriptase (TransGene Biotech, Beijing) to obtain cDNA. The level of miR-20a was determined using TransStart Top Green qPCR SuperMix method on ABI 7500 fast Real-time PCR Systerm, with U6 as an internal control.
Western blot analysis
The total protein was extracted from cultured JEG-3 cells 48h after treatment. The protein sample (75µg) was fractionated by SDS-PAGE (10% polyacrylamide gels) and transferred to nitrocellulose membrane. Then the membrane with samples were blocked with 5% nonfat milk for 1.5h at room temperature. After that, the membranes were incubated overnight at 4℃ with primary antibodies FOXA1 (1:1000 dilution, Abcam, USA) and β-actin (1:1000 dilution, Kangcheng Inc, China) on shaking bed. Next day, the membranes were washed with PBS-T for 3 times and incubated with secondary antibodies for 1h at room temperature. Finally, the membranes were rinsed with PBS-T before scanned by Imaging System (LI-COR Biosciences, Lincoln, NE, USA) and quantified with odyssey v1.2 software by measuring intensity (area×OD) in each group with β-actin as internal control.
Luciferase assay
Firstly, we employed RT-PCR technique to amplify the 3'UTRs of FOXA1 containing the binding sites for miR-20a. The sequence of the 3'UTR fragment is TTATAGGGAGCTGGATTTCAAAACGTGGTCCAAGATTCAAAAATCCTATTGATAGTGGCCATTTTAATCATTGCCATCGTGTGCTTGTTTCATCCAGTGTTATGCACTTTCCACAGTTGGACATGGTGTTAGTATAGCCAGACGGGTTTCATTATTATTTCTCTTTGCTTTCTCAATGTTAATTTATTGCATGGTTTATTCTTTTTCTTTACAGCTGAAATTGCTTTAAATGATGGTTAAAATTACAAATTAAAT.
After sequencing to ensure the correct amplification of the 3'UTR fragment, we inserted the sequences into the multiple cloning site of the pMIR-REPORT TM luciferase plasmid (Ambion, Inc.) to construct the miR-20a target-sites containing reporting vector. The constructed luciferase reporting vector and miR-20a were transfected to cultured HEK-293 cells and JEG-3 cells, and the TK-driven Renilla luciferase expression vector was used as an internal control. Forty-eight hour after transfection, we measured the luciferase activities by using a dual luciferase reporter assay kit (Promega) on a luminometer (GloMaxTM 20/20). The influence of miR-20a on the FOXA1 mRNA translation was calculated by normalizing the luciferase activity to control group.
In situ hybridization
The expression of miR-20a in human placenta tissues was examined by in situ hybridization with miRCURY locked nucleic acid (LNA) detection probes (Exiqon, Vedbeak, Denmark) as described in our previous work (18). Briefly, human placenta samples were fixed with 4% paraformaldehyde at 4℃overnight. The samples were sectioned and hybridized with 3'-digoxigenin (DIG)-labeled LNA probes for miR-20a at 55℃ overnight. The sections were then treated with sheep anti-DIG-alkaline phosphatase-Fab fragments (1:2000 dilution; Roche, Basel, Switzerland) at 4℃ overnight. Lastly, the sections were visualized with nitroblue tetrazolium chloride/5-bromo-4-chloro-3'-indolylphosphatase p-toluidine salt (NBT/BCIP; Roche) at room temperature for 2 h. 3'-DIG-labeled LNA scramble-miR control probe (Exiqon) was applied to negative control samples. Images of the sections were obtained using a Spot RT SE6 CCD camera (Diagnostic Instruments, Sterling Heights, MI) equipped on a BX60 microscope (Olympus).
Immunofluorescent staining
Human placenta samples were cut into small pieces and put into 4% paraformaldehyde at 4℃overnight for fixation. After that, the samples were treated with 30% sucrose for 24 hours to dehydrate. The samples were then embedded in OCT and sliced into 12 μm sections by using cryostat (LEICA CM 1850). The sections were blocked with 10% BSA for 1 hour at room temperature, and then treated with the primary antibody anti-FOXA1 antibody (1:200; Abcam) and incubated overnight at 4°C. After washing with PBS for three times, the samples were incubated with the secondary antibody conjugated to Alexa Fluor 594 (Invitrogen) for 1 h at room temperature. The fluorescent images of the samples were obtained using a laser scanning confocal microscope (FV-300, Olympus, Japan).
Construction of the plasmid expressing short hairpin RNA for human FOXA1
The short hairpin RNA(shRNA) for human FOXA1 (NM_004496) was constructed by using plasmid GV115 Expression Vector (Shanghai GeneChem Co. Ltd., China), which contained a human U6 promoter, a GFP reporter gene and an ampicillin resistance gene to enable antibiotic selection in mammalian cell. The target sequence is 5'- GTACTACCAAGGTGTGTAT -3'. A control-shRNA vector (pGCsi.U6/amp/GFP RNAi-NC Plasmid) was constructed by insertion of a sequence that expresses a shRNA with no significant homology to FOXA1 mRNA gene.
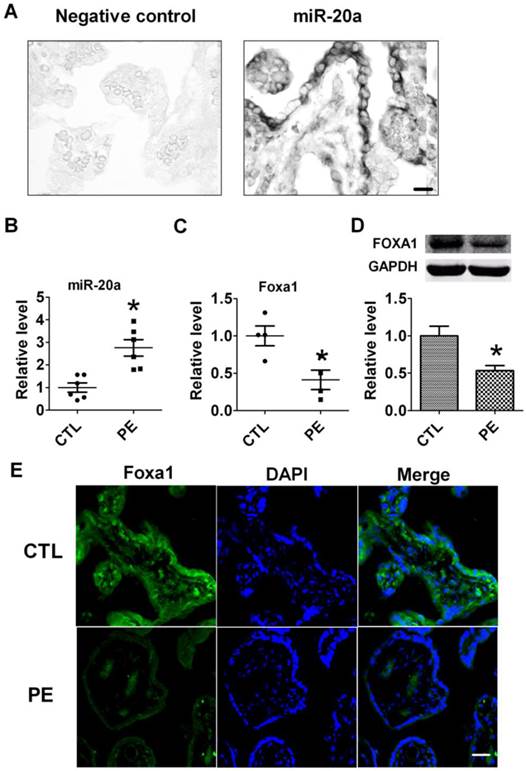
Expression of miR-20a in human placental tissues. A. Verification of miR-20a expression in human placenta tissue by In situ hybridization. B. miR-20a levels in human normal(CTL) and preeclampsia(PE) placental tissues. Data are expressed as mean±SEM. N=6. * P < 0.05 versus CTL. Scale bar=25µm. C, D. FOXA1 mRNA and protein levels in human normal(CTL) and preeclampsia(PE) placental tissues. Data are expressed as mean±SEM. N=4. * P < 0.05 versus CTL. E. Foxa1 expression in human normal(CTL) and preeclampsia(PE) placental tissues by immunofluorescent staining. Scale bar=25µm.
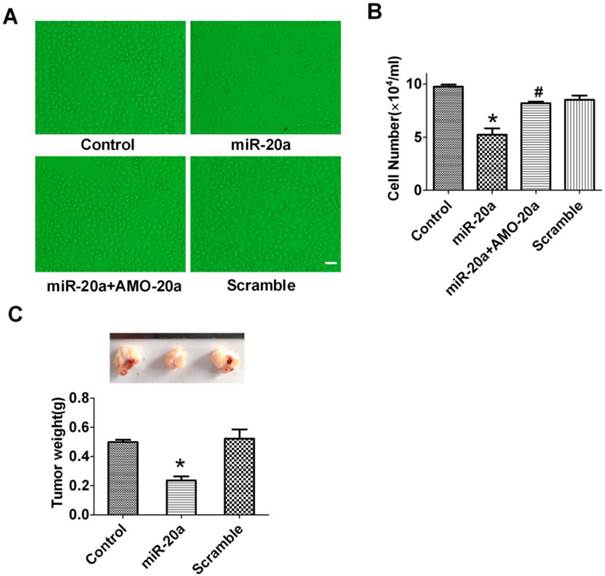
Tumorigenesis in Nude Mice
Male BALB/cA-nu nude mice (5-week old) were purchased from Vital River Laboratories (China), and maintained in pathogen-free conditions. H460 cells overexpressing miR-153 were injected subcutaneously into both flanks of nude mice (5×106 cells in 100 µl). H460 cells expressing scramble sequences were used as the negative control. Animals were killed, and xenograft excised and weighed after 1 month from the injection. All tumorigenicity assays were carried out according to the guidelines of the Ethical Committee of Harbin Medical University.
Tumorigenesis in Nude Mice
Male BALB/cA-nu nude mice (5-week old) were purchased from Vital River Laboratories (China), and maintained in pathogen-free conditions. JEG-3 cells overexpressing miR-20a were injected subcutaneously into both flanks of nude mice (5×106 cells in 100 µl). JEG-3 cells expressing scramble sequences were used as the negative control. Animals were killed, and xenograft excised and weighed after 1 month from the injection. All tumorigenicity assays were carried out according to the guidelines of the Ethical Committee of Harbin Medical University.
Statistical analysis
All data were expressed as mean ± SEM. Statistical analysis was performed using Kruskal Wallis H test. Differences were considered as statistically significant when P < 0.05.
Results
miR-20a and Foxa1 in human preeclamptic placental tissues
To verify the distribution of miR-20a in human placental tissues, we performed in situ hybridization to detect miR-20a. We found that miR-20a was abundantly expressed in normal human placental tissues (Fig 1A). We then tested the expression of miR-20a in human preeclampsia tissues by qRT-PCR. Consistent with our previous findings (17), the level of miR-20a significantly increased by 2.6 folds in human preeclampsia than normal tissues (P<0.05). In contrast to the upregulation of miR-20a, we found both mRNA and protein expression of FOXA1 were decreased in human preeclampsia than normal tissues (P<0.05) (Fig 1C, D). We also performed immunofluorescent staining of FOXA1 in human tissues to further confirm its dysregulation during preeclampsia. As shown in Figure 1E, the fluorescent intensity of FOXA1 was shown to be lower in human preeclampsia than normal tissues.
Effects of miR-20a on proliferation of JEG-3 cells and xenograft tumor growth of nude mice
JEG-3 trophoblast-like cells have been widely employed to investigate the invasion and proliferation of trophoblastic cells (19). In this study, we cultured JEG-3 cells and test the effect of miR-20a on its proliferation. We found that overexpression of miR-20a inhibited the proliferation of JEG-3 cells (Figure 2A, B) as counted by cell number. The anti-proliferative effects of miR-20a were abrogated by co-transfection of AMO-20a(Figure 2A, B). Transfection of the scramble sequence produced no effects on JEG-3 cell proliferation (Figure 2A, B). We then performed an in vivo study in nude mice to investigate the influence of miR-20a on JEG-3 cell growth. Consistent with in vitro finding, we found transfection of miR-20a inhibited the growth of xenograft JEG-3 cell tumor in nude mice (Figure 2C).
Effect of miR-20a on migration and invasion of cultured JEG-3 cells
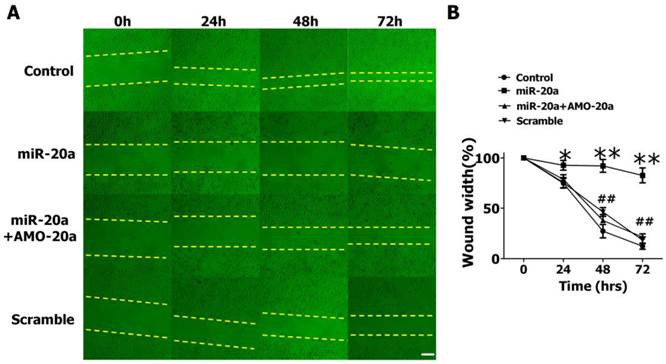
We then evaluated the effects of miR-20a on JEG-3 cell migration by wound healing assay. We found that the introduction of miR-20a into JEG-3 cells resulted in a significant reduction of cell migration during the closing of an artificial wound created over a confluent monolayer (Figure 3A, B). Cotransfection of AMO-20a abolished the inhibitory effects of miR-20a on wound healing (Figure 3A, B). We also performed transwell assay to further investigate the effect of miR-20a on JEG-3 cell migration. Consistent with wound healing assay, the cells passing the first layer well reduced dramatically after transfection with miR-20a, which were inhibited by co-transfection of AMO-20a. These data showed that miR-20a can inhibit JEG-3 cell migration(Figure 4A,B). miR-20a treatment also decreased the invasion of JEG-3 cells into a Matrigel-coated membrane (Figure 4C, D).
Effect of miR-20a overexpression on the growth of JEG-3 cells. A. Representative pictures of cultured JEG-3 cells after miR-20a treatment. B. Statistical analysis of JEG-3 cell proliferation after miR-20a treatment. Data are expressed as mean±SEM. N=4. * P < 0.05 versus Control. # P < 0.05 versus miR-20a. Scale bar=50µm. C. Effect of miR-20a on the growth of xenograft tumors of JEG-3 cells in nude mice. Data are expressed as mean±SEM. N=5. * P < 0.05 versus Control.
Effect of miR-20a on migration of cultured JEG-3 cells by wound healing assay. A. Representative pictures. B. Statistical analysis of wound width. Data are expressed as mean±SEM. N=5. * P < 0.05, ** P<0.01 Control versus miR-20a. # P < 0.05, ## P<0.01 miR-20a versus miR-20a+AMO-20a. Scale bar=100µm
Effect of miR-20a on migration and invasion of cultured JEG-3 cells by transwell assay. A. Representative pictures of cells migrating through an 8-µm pore size membrane. B. Statistical analysis of cells migrated. C. Representative pictures of cells invading through a Matrigel-coated membrane. D. Statistical analysis of cells invaded. Data are expressed as mean±SEM. N=3. * P < 0.05, versus Control. # P < 0.05 versus miR-20a. Scale bar=50µm

Effect of miR-20a on the expression of FOXA1. A. Sequence alignment between miR-20a and the 3'UTRs of human FOXA1 and the chimeric plasmid construct containing the 3'UTRs of human FOXA1. B, C. Verification of interactions between miR-20a and the 3'-UTR of FOXA1 in HEK293 and JEG-3 cells by luciferase assay. D. Effect of miR-20a on the protein expression of FOXA1. E. Effect of miR-20a on the mRNA expression of FOXA1. Data are expressed as mean±SEM. N=4. *P< 0.05 vs control, # P<0.05 vs miR-20a.

FOXA1 is the target of miR-20a
Bio-informatic analysis indicated that FOXA1 is a candidate target of miR-20a. The sequence complimentarity was shown in Figure 5A. We then constructed a reporter plasmid harboring the wild-type 3′-UTR region of FOXA1 downstream of the luciferase coding region (Figure 5A). Luciferase assay showed that overexpression of miR-20a suppressed luciferase activity, while co-transfection of AMO-20a alleviated the reduction luciferase activity caused by miR-20a in both HEK293 (Figure 5B) and JEG-3 cells (Figure 5C). Transfection of scramble sequence produced no effects (Figure 5B, C). These data indicated that miR-20a inhibited the translation of FOXA1.
To further confirm the regulatory effect of miR-20a on FOXA1 expression, we measured the protein level of FOXA1 in JEG-3 cells after miR-20a overexpression. A significant decrease in FOXA1 protein level was detected in miR-20a-treated compared with untreated and scramble-treated cells, which was abolished by co-transfection of AMO-20a (Figure 5D). The same qualitative alteration of FOXA1 mRNA level after miR-20a treatment was detected in Figure 5E.
Knockdown of FOXA1 by RNA interfering inhibited the migration and invasion of JEG-3 cells
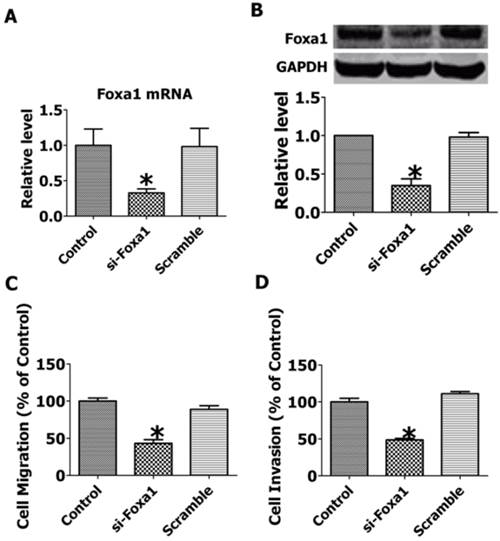
To explore the role of FOXA1 in the migration and invasion of JEG-3 cells, we employed RNA interfering technique to inhibit the expression of FOXA1. As shown in Figure 6A, B, transfection of short hairpin FOXA1 interfering plasmids successfully decreased the mRNA and protein expression of FOXA1, while the scrambled control sequence produced no effects. The migration and invasion of JEG-3 cells were significantly inhibited by knocking down of FOXA1(Figure 6 C, D), indicating that FOXA1 is a key regulator in the migration and invasion of JEG-3 cells.
Discussion
In this study, we found that the dysregulated miR-20a in human preeclampsia placenta tissue produced inhibitory effects on the proliferation, migration and invasion of trophoblast-like JEG-3 cells. The direct target that mediates the effect of miR-20a is FOXA1.
The pathogenesis role of microRNAs in various human diseases has been extensively explored, which highlights the great potential of microRNAs as novel therapeutic targets and diagnostic or prognostic markers(10, 11, 20, 21). microRNAs are also critical factors that affect the development of preeclampsia. A host of microRNAs have been shown to be dyregulated in the placenta with preeclampsia. Pineles et al. found that microRNAs (miR-210, miR-182 etc.) are differentially expressed in the normal human placentas and those with preeclampsia(22). The dysregulation of microRNAs in the human placentas of preeclampsis was further demonstrated by several other studies(23-26). Zhu et al. found that thirty-four microRNAs were expressed differentially in preeclamptic placentas in comparision with normal placentas, with 11 microRNAs up-regulated, and 23 down-regulated in preeclamptic pregnancies. Hu et al. found that in the placentae of Chinese patients with severe preeclampsia miR-16, miR-29b, miR-195, miR-26b, miR-181a, miR-335 and miR-222 were significantly increased(25). In a previous study of our group, we also screened the aberrantly expressed microRNAs in the placenta of preeclampsia. Among all the dysregulated microRNAs, miR-20a is greatly increased. Consistently, in this study we confirmed the upregulation of miR-20a in the placentae of Chinese patients with preeclampsia using quantitative RT-PCR technique. Similarly, Wang et al. also showed the increased expression of miR-20a in preeclampsia placentas(27).
Further studies showed that the dysregulated miRNAs are deeply involved in the pathogenesis of preeclampsia, especially in the dysregulation of trophoblast cell proliferation, migration and invasion(14, 15, 28). Overexpression of miR-155 contributed to preeclampsia development by down-regulating angiogenic regulating factor CYR61(14), which also decreased human-trophoblast-derived cell line (HTR-8/SVneo) proliferation and invasion and increased cell number at the G1 stage via targeting on cyclin D1(15, 28). Ectopic expression of up-regulated miRNA in preeclampsia, mir-210 inhibited the migration and invasion capability of trophoblast cells by suppressing the expression of Ephrin-A3 and Homeobox-A9, which related with cell migration and vascular remodeling(15). In cultured trophoblast JEG-3 cell line, we confirmed that the up-regulated miR-20a was able to inhibit the proliferation, migration and invasion of trophoblast cells, implying the pathological role of miR-20a in the development of preeclampsia. To our knowledge this is the first study investigating the influence of miR-20a on trophoblast cell activities.
Knockdown of FOXA1 by RNA interfering inhibited the migration and invasion of JEG-3 cells. A. FOXA1 mRNA level after RNA interference. B. FOXA1 protein level after RNA interference. C. Migration of JEG-3 cells after RNA interference. D. Invasion of JEG-3 cells after RNA interference. Data are expressed as mean±SEM. N=3. *P< 0.05 vs control.
As microRNAs take effects by fine-tuning the expression of target proteins, we bioinformatically analyzed the potential targets of miR-20a to explore the mechanism underlying its effects on trophoblast cell proliferation, migration and invasion. We found FOXA1 is a good candidate and verified the regulatory role of miR-20a with biological techniques. FOXA1 is the founding member of the FOX family of transcription factors, which has been demonstrated to be a critical factor in various organ developments. Compound conditional ablation of both Foxa1 and Foxa2 in the pancreatic primordium results in complete loss of Pdx1 expression and severe pancreatic hypoplasia(29). The Foxa1-deficient mouse prostate shows a severely altered ductal pattern that resembles primitive epithelial cords surrounded by thick stromal layers, with no differentiated or mature luminal epithelial cells in epithelium. (30). Li et al. found that knockout of Foxa1 terminates bile duct expansion in the adult liver through inhibition of IL-6 expression(31). FOXA1 is also an important factor in several human cancers. Although both oncogenic and tumor suppressive roles have been proposed, it was demonstrated to be oncogenic in all the cancer tissues with FOXA1 overexpression(32). FOXA1 silencing by siRNAs reduced invasion and migration of esophageal squamous cell carcinomas cells(33). These studies imply that FOXA1 plays a key role in cell proliferation and migration, either during normal organ development or cancer development. Our RNA interfering study confirmed the involvement of FOXA1 in the migration and invasion of JEG-3 trophoblast cells. Therefore, we propose that the effect of miR-20a on trophoblast cell is due to its suppression on FOXA1 expression. Although various organ developmental abnormalities have been observed FOXA1 deficient mice, it is unclear whether FOXA1 ablation has any influence on placenta development. It will be interesting to investigate the developmental condition of the placenta by employing FOXA1 null mice in future studies.
In summary, we demonstrated that miR-20a may be a critical regulator in the pathogenesis of preeclampsia by altering the properties of trophoblast cells, which represents a potential therapeutic target in the development of novel drugs for pre-eclampsia.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81070216 to Luo Shanshun), New Century Excellent Talents Program of Ministry of Education(to Zhenwei Pan), New Century Excellent Talents Program of Heilongjiang Province (1251-ncet-012 to Pan Zhenwei) and Yu Weihan Excellent Youth Foundation of Harbin Medical University(001000004 to Pan Zhenwei).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Huppertz B. Placental origins of preeclampsia: challenging the current hypothesis. Hypertension. 2008;51(4):970-975
2. Goldman-Wohl D, Yagel S. Regulation of trophoblast invasion: from normal implantation to pre-eclampsia. Mol Cell Endocrinol. 2002;187(1-2):233-238
3. Khong TY, De Wolf F, Robertson WB, Brosens I. Inadequate maternal vascular response to placentation in pregnancies complicated by pre-eclampsia and by small-for-gestational age infants. Br J Obstet Gynaecol. 1986;93(10):1049-1059
4. Meekins JW, Pijnenborg R, Hanssens M, McFadyen IR, van Asshe A. A study of placental bed spiral arteries and trophoblast invasion in normal and severe pre-eclamptic pregnancies. Br J Obstet Gynaecol. 1994;101(8):669-674
5. Zhou Y, Damsky CH, Fisher SJ. Preeclampsia is associated with failure of human cytotrophoblasts to mimic a vascular adhesion phenotype. One cause of defective endovascular invasion in this syndrome? J Clin Invest. 1997;99(9):2152-2164
6. Perez-Sepulveda A, Torres MJ, Khoury M, Illanes SE. Innate immune system and preeclampsia. Front Immunol. 2014;5:244
7. Laresgoiti-Servitje E. A leading role for the immune system in the pathophysiology of preeclampsia. J Leukoc Biol. 2014;94(2):247-257
8. Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8(2):93-103
9. Ullah S, John P, Bhatti A. MicroRNAs with a role in gene regulation and in human diseases. Mol Biol Rep. 2013
10. Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91(3):827-887
11. Soriano A, Jubierre L, Almazan-Moga A. et al. microRNAs as pharmacological targets in cancer. Pharmacol Res. 2013;75:3-14
12. Ishida M, Selaru FM. miRNA-Based Therapeutic Strategies. Curr Anesthesiol Rep. 2013;1(1):63-70
13. Deng JH, Deng Q, Kuo CH, Delaney SW, Ying SY. MiRNA targets of prostate cancer. Methods Mol Biol. 2013;936:357-369
14. Zhang Y, Diao Z, Su L. et al. MicroRNA-155 contributes to preeclampsia by down-regulating CYR61. Am J Obstet Gynecol. 2010;202(5):e461-467
15. Zhang Y, Fei M, Xue G. et al. Elevated levels of hypoxia-inducible microRNA-210 in pre-eclampsia: new insights into molecular mechanisms for the disease. J Cell Mol Med. 2012;16(2):249-259
16. Bai Y, Yang W, Yang HX. et al. Downregulated miR-195 detected in preeclamptic placenta affects trophoblast cell invasion via modulating ActRIIA expression. PLoS One. 2012;7(6):e38875
17. Ishibashi O, Ohkuchi A, Ali MM. et al. Hydroxysteroid (17-beta) dehydrogenase 1 is dysregulated by miR-210 and miR-518c that are aberrantly expressed in preeclamptic placentas: a novel marker for predicting preeclampsia. Hypertension. 2012;59(2):265-273
18. Luo SS, Ishibashi O, Ishikawa G. et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81(4):717-729
19. Bilban M, Tauber S, Haslinger P. et al. Trophoblast invasion: assessment of cellular models using gene expression signatures. Placenta. 2010;31(11):989-996
20. Xi JJ. MicroRNAs in Cancer. Cancer Treat Res. 2013;158:119-137
21. Khan S, Ansarullah, Kumar D, Jaggi M, Chauhan SC. Targeting microRNAs in Pancreatic Cancer: Microplayers in the Big Game. Cancer Res. 2013
22. Pineles BL, Romero R, Montenegro D. et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 2007;196(3):e261-266
23. Mayor-Lynn K, Toloubeydokhti T, Cruz AC, Chegini N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod Sci. 2011;18(1):46-56
24. Zhu XM, Han T, Sargent IL, Yin GW, Yao YQ. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol. 2009;200(6):661 e661-667
25. Hu Y, Li P, Hao S. et al. Differential expression of microRNAs in the placentae of Chinese patients with severe pre-eclampsia. Clin Chem Lab Med. 2009;47(8):923-929
26. Enquobahrie DA, Abetew DF, Sorensen TK. et al. Placental microRNA expression in pregnancies complicated by preeclampsia. Am J Obstet Gynecol. 2011;204(2):178 e112-121
27. Wang W, Feng L, Zhang H. et al. Preeclampsia up-regulates angiogenesis-associated microRNA (i.e, miR-17, -20a, and -20b) that target ephrin-B2 and EPHB4 in human placenta. J Clin Endocrinol Metab. 2012;97(6):E1051-1059
28. Dai Y, Qiu Z, Diao Z. et al. MicroRNA-155 inhibits proliferation and migration of human extravillous trophoblast derived HTR-8/SVneo cells via down-regulating cyclin D1. Placenta. 2012;33(10):824-829
29. Gao N, LeLay J, Vatamaniuk MZ. et al. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22(24):3435-3448
30. Gao N, Ishii K, Mirosevich J. et al. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development. 2005;132(15):3431-3443
31. Li Z, White P, Tuteja G. et al. Foxa1 and Foxa2 regulate bile duct development in mice. J Clin Invest. 2009;119(6):1537-1545
32. Bernardo GM, Keri RA. FOXA1: a transcription factor with parallel functions in development and cancer. Biosci Rep. 2012;32(2):113-130
33. Sano M, Aoyagi K, Takahashi H. et al. Forkhead box A1 transcriptional pathway in KRT7-expressing esophageal squamous cell carcinomas with extensive lymph node metastasis. Int J Oncol. 2010;36(2):321-330
Author contact
![]() Corresponding author: Dr. Luo Shanshun luoss63com Department of Gerontology, The First Affiliated Hospital of Harbin Medical University, or Dr. Pan Zhenwei panzwhrbmu.edu.cn. Department of Pharmacology, Harbin Medical University, Baojian Road 157, Harbin, Heilongjiang 150081, P. R. China. Fax: 86 451 86675769 or 86667511 Tel: 86 451 86671354.
Corresponding author: Dr. Luo Shanshun luoss63com Department of Gerontology, The First Affiliated Hospital of Harbin Medical University, or Dr. Pan Zhenwei panzwhrbmu.edu.cn. Department of Pharmacology, Harbin Medical University, Baojian Road 157, Harbin, Heilongjiang 150081, P. R. China. Fax: 86 451 86675769 or 86667511 Tel: 86 451 86671354.

 Global reach, higher impact
Global reach, higher impact